Introduction
Prostate cancer (PCa) is one of the most common
malignant tumors in the male worldwide, and the second leading
cause of cancer-related death among men in America (1–4).
Despite the development in PCa early diagnosis and treatment
methods (5), metastasis and tumor
relapse still remains a serious problem for the patients long-term
survival, and the mechanisms involved in PCa progression and
metastasis are not fully understood. Several studies have shown
that inhibition of apoptosis is the most critical factor for
tumorigenesis in PCa (6,7). It has been reported that
overexpression of Bcl-xL in PCa may suppress the activity of the
pro-apoptotic molecules Bax and Bak, and may contribute to androgen
deprivation resistance and progression of PCa (8–11).
However, apoptosis regulation is a very complicated process, with
many different signal pathways and molecules involved, therefore,
to better understand the anti-apoptosis mechanisms of PCa cells,
more apoptosis-related molecules that possibly are involved in the
PCa progression remains to be explored.
Bax-interacting factor-1 (Bif-1), also known as
SH3GLB1 or endophilin B1, is a member of Endophilin family,
contains an amino-terminal N-BAR (Bin-Amphiphysin-Rvs) domain and a
carboxy-terminal SH3 (Src-homology 3) domain and displays membrane
binding and extension activities (12–14).
Bif-1 was originally identified as a Bax-binding protein, and
represents a new type of Bax activator that controls the
mitochondrial pathway of apoptosis. However, the role of Bif-1 in
tumor development and progression remains to be elucidated. Various
results seem even contradictory. It is reported in PIN that,
knockout of Bif-1 could suppress Bax/Bak conformational change,
cytochrome c release, caspase activation and cell death
8,15–17.
However, it is also reported that in hepatocellular carcinoma,
Bif-1 expression is upregulated and correlates with shortened
patient survival (18). There are
studies suggested that Bif-1 gene may act as a tumor suppressor
16,19–21. It
is reported that Bif-1 mRNA levels are downregulated in lung
carcinoma (22), pancreatic
(16,23), breast (20,24)
and colon cancer (24–26). However, a study in Merkel cell
carcinoma (MCC) shows that the Bif-1 expression level is associated
with low levels of Bax, which indicated that upregulation of Bif-1
could in part be responsible for the tumorigenesis of MCC (13). There is scarce information
concerning the relationships between Bif-1 and PCa. The role of
Bif-1 in PCa tumorigenesis and progression remains unclear. A study
by Coppola et al (8) found
that Bif-1 expression was decreased in a subset of PCa as compared
to normal prostate, suggesting that Bif-1 may play an important
role in the early stage of PCa development, but they also found a
proportion of PCa cases that expressed high level of Bif-1
(8). Iacopino et al
(19) reported that both BCL-2 and
BAX expression levels were higher in PCa than in benign prostatic
hyperplasia (BPH) whereas the BCL-2/BAX ratio was lower in PCa than
in BPH, but the Bif-1 level was not analyzed at the same time.
Evidence suggests that Bif-1 may be involved in tumorigenesis of
PCa, but its effect in the development and progression of PCa
remains to be elucidated.
Therefore, to further understand the role of Bif-1
in PCa tumorigenesis, in the present study, we overexpressed Bif-1
by introducing the wild-type Bif-1 gene into an invasive human PCa
cell line LNCaP cells, which constitutively expressed very low
levels of Bif-1, to assess the possible role of overexpressed Bif-1
on the growth and apoptosis of PCa cells.
Materials and methods
Cell lines and tissue samples
The PCa cell lines 22RV1, PC3, LNCaP and DU145 were
all obtained from the American Type Culture Collection (ATCC;
Manassas, VA, USA) and cultured in RPMI-1640 medium (Gibco-BRL,
Invitrogen Corporation, Grand Island, NY, USA), 1 mM sodium
pyruvate, 100 U/ml penicillin G, 100 µg/ml streptomycin (Sigma
Corporation of America, Ronkonkoma, NY, USA), 10% fetal bovine
serum (FBS; Tianjin HaoYang Biological Manufacture Co., Ltd.,
Chinese Academy of Sciences, Beijing, China) at 37°C in a
humidified atmosphere of 5% CO2.
Prostate tissue samples were collected from
2010–2011 at the Department of Urology, The First Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China). In total,
200 samples were collected, including primary PCa (n=100) and
benign prostatic hypertrophy samples (BPH, n=100). The average age
of PCa patients was 70.66±8.18, and the average age of BPH patients
was 67.81±9.31. The 100 primary PCa specimens were divided into two
categories according to the Gleason score value, Gleason score 4, 5
and 6 (n=43), and Gleason score 7, 8 and 9 (n=57). All the samples
were confirmed by pathologic diagnosis. Prior written consent of
each patient for the use of clinical materials for research
purposes was obtained, and the approval was granted by the
Institutional Ethics Board (IRB) at The First Affiliated Hospital
of Sun Yat-sen University.
Construction and transfection of
pcDNA3.1(+)-Bif-1 vector
We amplified the full-length gene of wild-type human
Bif-1 from human genomic cDNA using forward primer,
5′-CCGGAATTCGCCTAGGATGAATATCATGGAC-3′ and reverse primer,
5′-CGCCTCGAGAGTCCACCTACTTAATTGAGCAG-3′. The PCR procedure was: 95°C
for 3 min, 1 cycle, 95°C for 30 sec, 58°C for 30 sec, 72°C for 90
sec, 35 cycles, 72°C, for 5 min. The length of the amplified
product was 1,087 bp. The cDNA fragments were then cloned into an
EcoRI-XhoI double restriction enzyme digested
pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA, USA). The selected
recombinant plasmids were partially sequenced to confirm a correct
Bif-1 open reading frame (ORF). The LNCaP cells were initially
seeded at a density of 5×106 cells/100 mm dish 24 h
before transfection. pcDNA3.1(+)-Bif-1 plasmid (10 µg) or the empty
vector pcDNA3.1(+) control was used to transfect LNCaP cells with
Lipofectamine 2000 reagent (Life Technologies, Gaithersburg, MD,
USA) according to the manufacturer's instructions. Forty-eight
hours after transfection, 0.5 mg/ml G418 was added for stable
positive clone selection on the basis of preliminary G418 serial
test results.
RT-PCR and real-time PCR
RT-PCR was used to confirm the expression of Bif-1
cDNA in pcDNA3.1(+)-Bif-1 recombinant plasmid transfected LNCaP
cells, and real-time PCR was used to detect the mRNA levels in PCa
cell lines. Briefly, total RNA was extracted from LNCaP cells using
TRIzol reagent (Invitrogen) according to the manufacturer's
instructions. The isolated total RNA was then mixed with oligo(dT)
primer and incubated at 65°C for 5 min. Then, cDNA was synthesized
using SuperScript III (Invitrogen) at 50°C for 50 min followed by
heating at 85°C for 5 min. The primers and PCR procedure were the
same as those used in Bif-1 cDNA cloning, as shown above. The
expected Bif-1 cDNA fragment length was 1,087 bp.
To determine relative Bif-1 mRNA expression levels
in PCa LNCaP and pcDNA3.1(+)-Bif-1 transfected LNCaP cells,
quantitative real-time PCR assays were performed on a Mastercycler
EP realplex real-time PCR detector (Eppendorf, Germany). Specific
PCR primers were designed based on Bif-1 cDNA gene (GenBank ID,
NM_016009). The forward primer sequence was,
5′-GCTTGGCCAGGCTGAGAAGACAG-3′ (nucleotides 417–439), and the
reverse primer was, 5′-TCCTGGCATTTGGATTTGGCTGCA-3′ (nucleotides
553–530). The amplified product length is 137 bp, which corresponds
to the ORF of Bif-1. The β-actin gene (GenBank ID, NM_001101.3) was
used as an internal control. The forward primer sequence of β-actin
was, 5′-CAGAGCCTCGCCTTTGCCGATCC-3′ (nucleotides 31–53), and its
reverse primer was, 5′-CCTTGCACATGCCGGAGCCGT-3′ (nucleotides
139–119), with the product length 109 bp. Real-time PCR was carried
out using the SYBR Premix Ex Taq™ kit (Perfect Real-Time; code,
DRR081A) (Takara, Dalian, China) in a 50 µl final volume,
containing 25 µl of reaction mixture, 0.5 µM of the forward and
reverse primers, and 3 µl of RNA. The real-time PCR procedure was:
cycle 1, 95°C for 3 min; cycle 2 through 35, 95°C for 5 sec, 60°C
for 34 sec, and fluorescence signal was detected at the end of each
cycle. Melting curve analysis was drawn to confirm the specificity.
Detection of each sample was repeated 3 times.
Immunohistochemistry
The slides were dewaxed by heating at 55°C for 30
min followed by 3 washes with xylene, each for 10 min. Tissues were
rehydrated by a series of 5-min washes in 100, 95 and 80% ethanol,
and distilled water. Standard cell conditioning [following the
Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing, China)
recommendations] was used for antigen retrieval. Then, the samples
were incubated with a Bif-1 goat anti-human antibody (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) with the dilution ratio 1:200
at 4°C overnight. The sections were then incubated with
biotin-labeled secondary antibody and streptavidin-horseradish
peroxidase for 30 min each (Zhongshan Golden Bridge Biotechnology
Co., Ltd.). The samples were developed with 3,3′-diaminobenzidine
tetrahydrochloride substrate and counterstained with hematoxylin
(both from Zhongshan Golden Bridge Biotechnology Co., Ltd.). The
slides were dehydrated and mounted. Negative controls were included
by omitting Bif-1 antibody during the primary antibody
incubation.
Western blotting
Cultured cells were harvested when 80% confluent.
Cell protein was extracted in a homogenization buffer:
phosphate-buffered saline (PBS) containing 1% Nonidet P-40, 0.5%
sodium deoxycholate, 0.1% SDS, 10 mg/ml phenylmethylsulfonyl
fluoride (PMSF), 1 mM sodium orthovanadate, 1 µg/ml leupeptin and 1
µg/ml aprotinin. The protein samples were separated by
SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a
nitrocellulose membrane. The membrane was blocked with 5% skim milk
in PBST (pH 7.4 PBS with 0.1% Tween-20) for 1 h and incubated with
the goat anti-human endophilin B1 polyclonal antibody S-20 (Santa
Cruz Biotechnology, Santa Cruz, CA, USA) and the antibody dilution
ratio was 1:1,000. The Bif-1 protein was then detected by rabbit
anti-goat horseradish peroxidase (HRP)-conjugated secondary
antibody (dilution ratio 1:2,000; Santa Cruz Biotechnology) coupled
with enhanced chemiluminescence (ECL) western blot detection
reagents (Amersham, Arlington, IL, USA). BPH sample BPH1 was used
as a control.
Immunofluorescence analysis
Briefly, 1×105 cells/well were seeded on
a sterile glass coverslip pre-coated with poly-L-lysine (Sigma, St.
Louis, MO, USA) in a 6-well culture plate. At 80% confluence, the
cells were washed twice with PBS, fixed by 4% paraformaldehyde in
PBS for 10 min, and permeablized with 0.1% Triton X-100 in PBS for
10 min at room temperature. Then, the coverslips were blocked by
incubation for 30 min with 5% normal goat serum (Jackson
Laboratory, Bar Harbor, ME, USA) in PBS. After washing with PBS 3
times, the cells were incubated with goat anti-human endophilin B1
antibody (dilution ratio 1:1,000; Santa Cruz Biotechnology) in PBS
in a humidified chamber overnight at 4°C. After 3 washes by PBS,
cells were incubated with FITC-conjugated secondary antibody
(1:2,000; Jackson Laboratory) for 1 h at room temperature. Then,
the cells were washed 3 times in PBS, and stained with 1 µg/ml
4′,6-diamidino-2-phenylindole (DAPI) for 5 min. After further 3
washes in PBS, the coverslips were sealed with nail polish and
observed, or stored in the dark at 4°C before observation.
Cell growth assay
For MTT assays, different groups of cells were
seeded into 96-well plates at the density of 2×104
cells/well. After 12, 24 and 48 h of culture,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma) was added to a final concentration of 0.5 mg/ml for 4 h at
37°C. MTT assays were carried out in 3 duplicate wells for each
group, and the absorbance at the wavelength of 570 nm was measured
on a Tecan Sunrise multiwell spectrophotometer (Tecan, Männedorf,
Switzerland). The experiments were repeated at least 3 times.
Cell apoptosis analysis
Flow cytometry was used to analyze cell apoptosis.
Briefly, cells were cultured in phenol red-free RPMI-1640 medium
containing 10% FBS. Flow cytometric analysis was performed
according to the methods of Mendonca et al (21). Briefly, 1×105 cells were
harvested and washed in PBS twice, then re-suspended in 195 µl
Annexin V-FITC buffer with 5 µl Annexin V-FITC. After 10 min of
staining in the dark, cells were washed 3 times with PBS, and
re-suspended in 190 µl Annexin V-FITC buffer with 10 µl of 25 µg/ml
propidium iodide. Cells (5×104) were used for cell
apoptosis analysis by flow cytometric analysis. Each analysis was
repeated at least 3 times with similar results.
Wound healing assay
Cell migration was evaluated by a scratched
wound-healing assay on plastic plate wells. In brief, cells were
seeded into a 6-well plate (5×105 cells/well) and grew
to 100% confluency. The monolayer culture was scratched with a
sterile micropipette tip to create a denuded zone (gap) of constant
width and the cellular debris was removed. The initial gap width
and the residual gap width at 12, 24 or 36 h after wounding were
observed under an inverted microscope and photographed. The wound
area was measured by the program ImageJ (http://rsb.info.nih.gov/ij/). The percentage of wound
closure was estimated by the formula: 1 - (wound area at Tt/wound
area at T0) × 100%, where Tt is the time after wounding and T0 is
the immediately wounding time.
Statistical analysis
Measurement data are presented as the mean ± SD.
Statistical analysis was performed using the unpaired Student's
t-test or analysis of variance (ANOVA). A two-sided value of
P<0.05 was considered statistically significant. All statistical
analyses were performed using SPSS 11.5 software (SPSS, Inc.,
Chicago, IL, USA).
Results
Bif-1 expression is downregulated in
both PCa cell lines and clinical PCa samples
Bif-1 levels in PCa cells and tissues were detected
by real-time PCR and western blot assays. BPH sample (BPH1) served
as a control. Real-time PCR results showed that Bif-1 mRNA level
was downregulated in PCa cells 22RV1, PC3, LNCaP and DU145
(P<0.01 compared with BPH1), and western blot analysis of Bif-1
protein expression revealed that Bif-1 protein expression was also
decreased in those PCa cell lines as compared to benign prostate
hypertrophy BPH1 (Fig. 1A and
B).
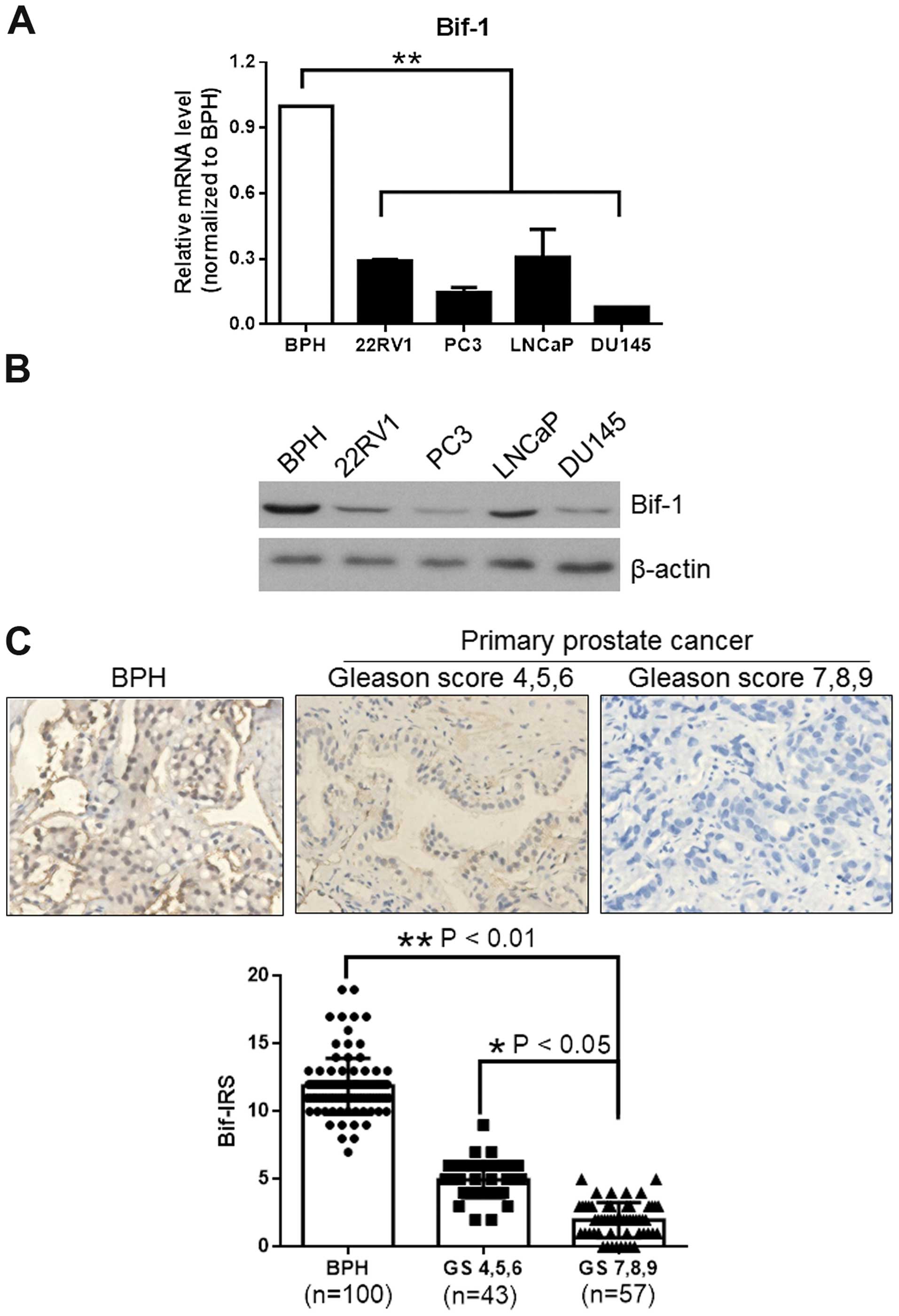 | Figure 1.Bif-1 level is downregulated in
prostate cancer cells and clinical PCa samples. Bif-1 mRNA levels
of prostate cancer cell lines 22RV1, PC3, LNCaP and DU145 were
analyzed by real-time PCR and the Bif-1 protein levels were
detected by western blotting, and β-actin was used as an internal
control. Bif-1 expression in BPH (n=100) and PCa (n=100) samples
was detected by immunohistochemistry and the results were analyzed
by immunoreactive score analysis. (A) Bif-1 mRNA expression level
was significantly decreased in all the prostate cancer cell lines
compared with benign prostatic hypertrophy sample BPH1. (B) Bif-1
protein expression level was markedly suppressed in all the
prostate cancer cells lines compared with BPH1. (C)
Immunohistochemistry staining of Bif-1 in benign prostatic
hyperplasia (BPH) and prostate cancer (PCa) samples, showed that
Bif-1 expression is higher in BPH than PCa, and high Gleason-scored
PCa samples (Gleason scores 7, 8 and 9, n=57) exhibited
significantly lower Bif-1 level than low Gleason-scored PCa samples
(Gleason scores 4, 5 and 6, n=43). Bif-IRS, Bif-1 immunoreactivity
scores; *P<0.05, **P<0.01. |
To verify the Bif-1 level in PCa samples, we carried
out immunohistochemistry in 100 PCa samples and 100 benign
prostatic hyperplasia (BPH) samples. The results showed that
significant intense nuclear immunoreactivity was seen in BPH
tissues and low-grade prostatic carcinoma tissues (Fig. 1C). However, few cells showed
positively-stained nuclei in high grade prostatic carcinoma
lesions. Immunoreactive score analysis using Student's t-test
further confirmed that Bif-1 expression is higher in BPH than PCa
(P<0.01), and higher Gleason-scored samples exhibited
significantly lower Bif-1 immunoreactivity scores (P<0.05).
Therefore, Bif-1 expression is suppressed with the cancer
progression, as shown in Fig. 1C
(BPH vs. high Gleason-scored PCa P<0.01, and low Gleason-scored
PCa vs. high Gleason-scored PCa P<0.05). The results confirmed
that compared with BPH samples, Bif-1 was significantly suppressed
in PCa cells and tissues, and the suppression was more obvious in
progressive PCa.
Overexpressed Bif-1 suppresses cell
proliferation in PCa cells
To further understand the role of Bif-1 in PCa
development, we constructed an ectopic Bif-1 expression vector, to
explore the effect of Bif-1 overexpression on PCa cell behavior.
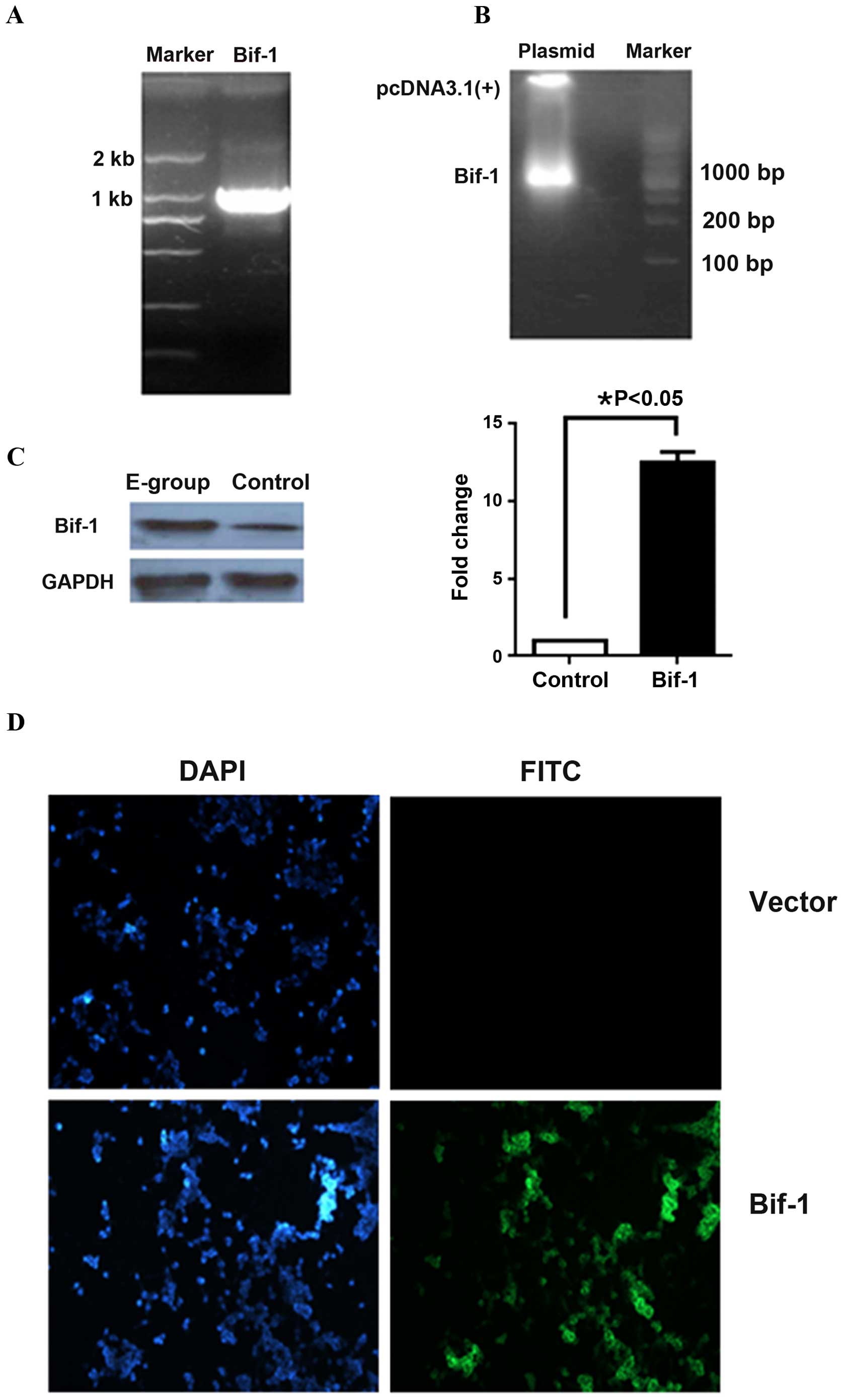
The full-length wild-type human Bif-1 was cloned from human genomic
cDNA (Fig. 2A) and the positive
recombinant vector was identified by double enzyme digestion and
sequencing (Fig. 2B). The
pcDNA3.1(+)-Bif-1 expression vector was then transfected into LNCaP
cells and the Bif-1 overexpression cell clones were identified by
G418 screening, real-time RT-PCR and western blotting (Fig. 2C). The indirect immunofluorescence
staining of Bif-1 also proved that Bif-1 was successfully expressed
in pCDNA3.1(+)-Bif-1 plasmid transfected LNCaP cells (Fig. 2D). Stable Bif-1 positive LNCaP cells
were used in the following functional analysis. LNCaP cells
transfected with the empty vector pcDNA3.1(+) served as a negative
control.
We found that after 24 h of Bif-1 overexpression,
the growth of Bif-1 overexpressing LNCaP cells (E-group) was
markedly inhibited, as shown in Fig.
3A, and the growth inhibition effect could be observed even
more obviously at 48 h of Bif-1 overexpression (P<0.05). As a
contrast, the proliferation of negative control (vector control)
and blank control (non-transfected control) groups was not markedly
changed (P>0.05). Although cell proliferation was significantly
inhibited, the morphology of Bif-1 overexpressing LNCaP cells
showed no change (Fig. 3B). The
results confirmed that Bif-1 could inhibit proliferation of PCa
cells, but did not affect the cell morphology.
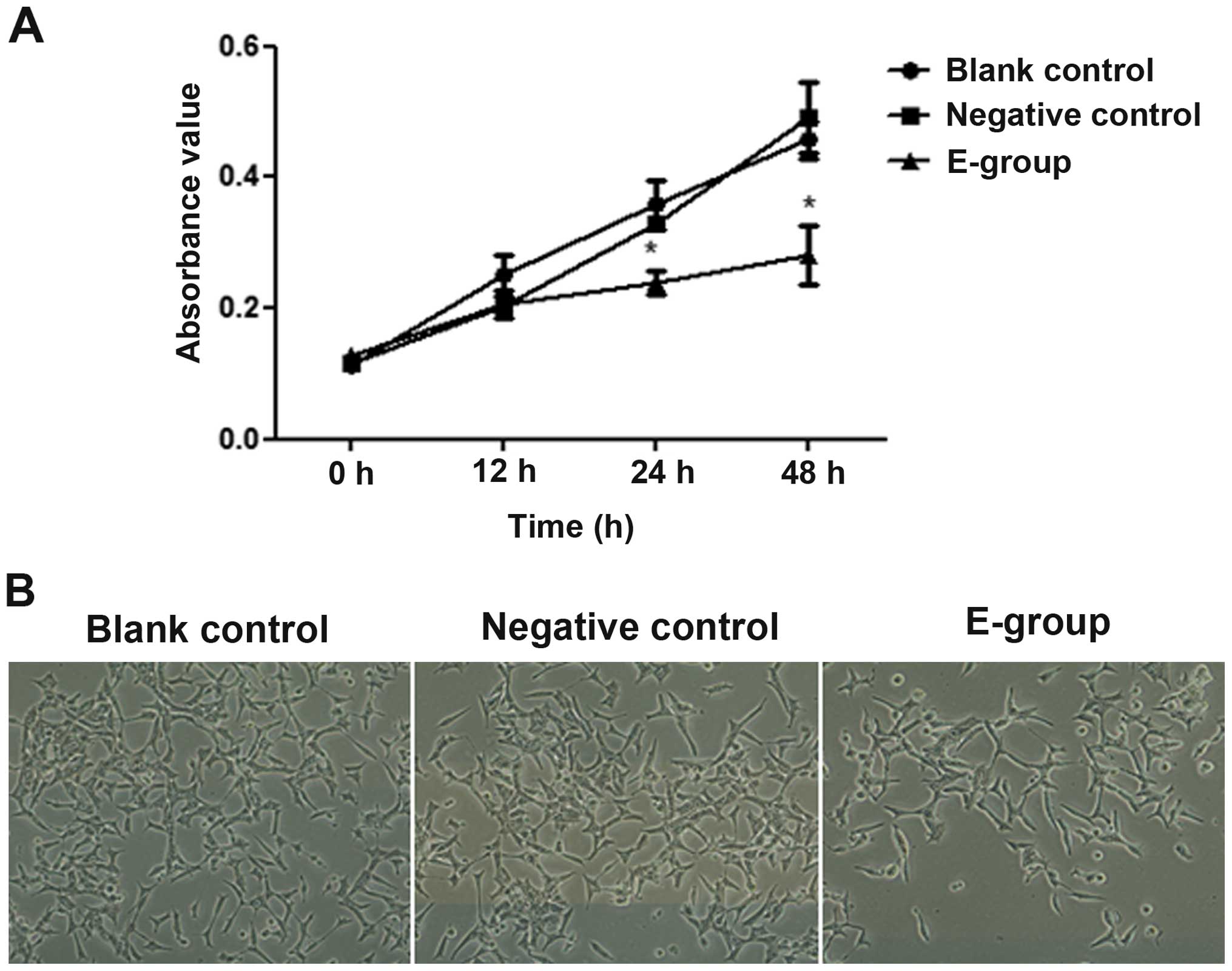 | Figure 3.Effect of Bif-1 overexpression on
LNCaP cell proliferation. Cells/well (2×104) were seeded
in triplicate into 96-well culture plates. At 12, 24 and 48 h of
pCDNA3.1(+)-Bif-1 transfection,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
was added to a final concentration of 5 mg/ml and incubated for 4 h
at 37°C, and cell proliferation was analyzed by MTT assay.
Absorbance of 570 nm was measured by a microplate reader at 0, 12,
24 and 48 h after Bif-1 overexpression. Cell morphology was
observed at 24 h of Bif-1 overexpression under a phase contrast
microscope. (A) Proliferation of E-group [pCDNA3.1(+)-Bif-1
transfected LNCaP cells], negative group (empty vector transfected
LNCaP cells) and blank group (non-transfected LNCaP cells) were
analyzed by MTT assay. Cell proliferation of E-group was
significantly inhibited after 24 h of Bif-1 overexpression compared
with negative and blank controls; *P<0.05. (B) The appearance of
negative and blank controls, and E-group cells at 24 h after Bif-1
overexpression. Although the number of E-group cells was fewer than
negative and blank control cells, cell morphology showed no obvious
change. (Inverted phase contrast microscope, magnification,
×100). |
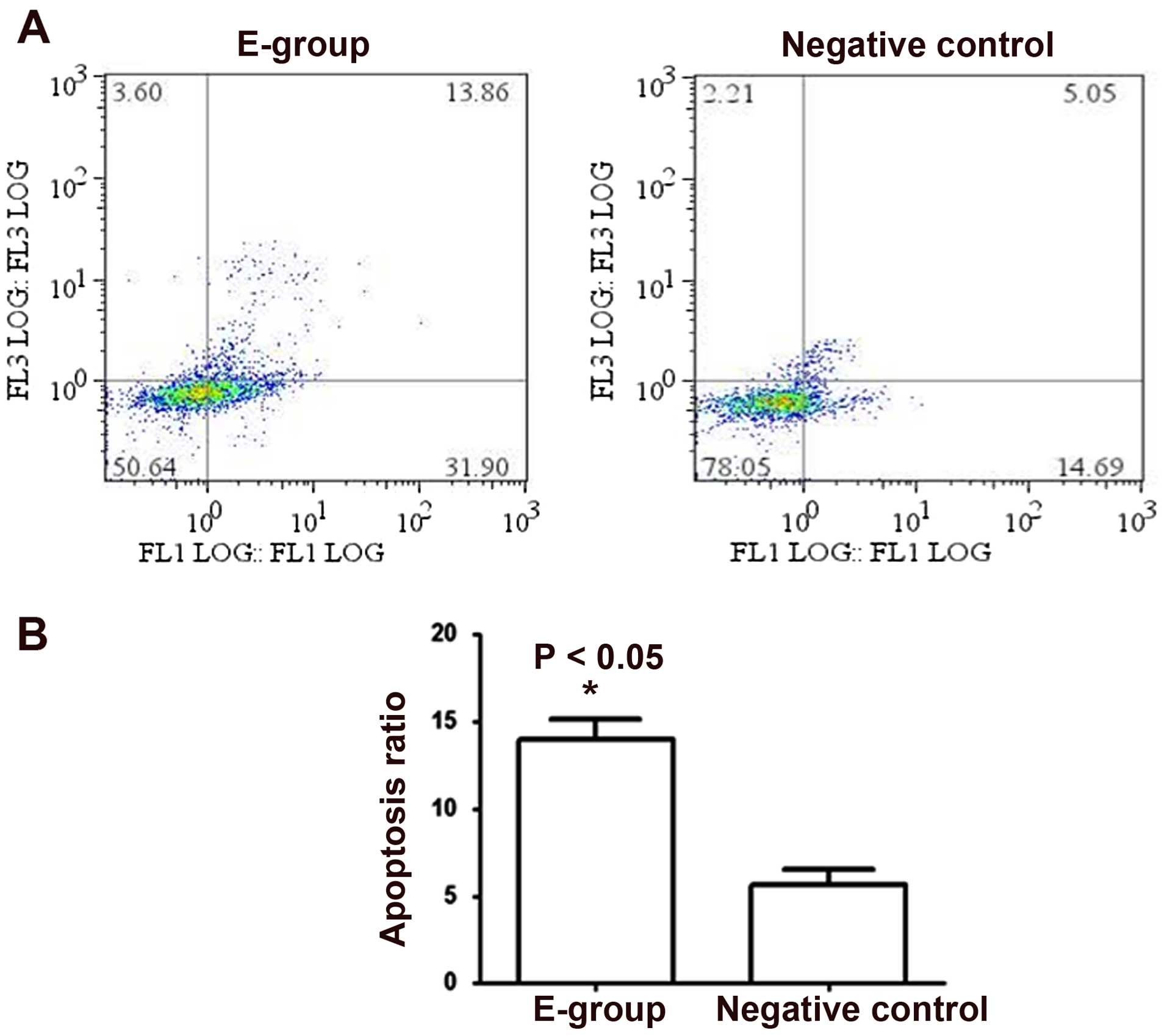
Bif-1 overexpression promotes cell
apoptosis in PCa cells
To further elucidate the molecular mechanism of the
proliferation inhibition effect of Bif-1 on PCa cells, we examined
the cell apoptosis change in Bif-1 overexpression cells. Flow
cytometric analysis showed that, in contrast to the negative
control cells, the apoptosis proportion in Bif-1 overexpression
LNCaP cells increased from 5.05±0.87 to 13.86±0.29 (P<0.05), as
shown in Fig. 4. These results
proved that Bif-1 overexpression in LNCaP cells induced a
significant enhancement of cell apoptosis.
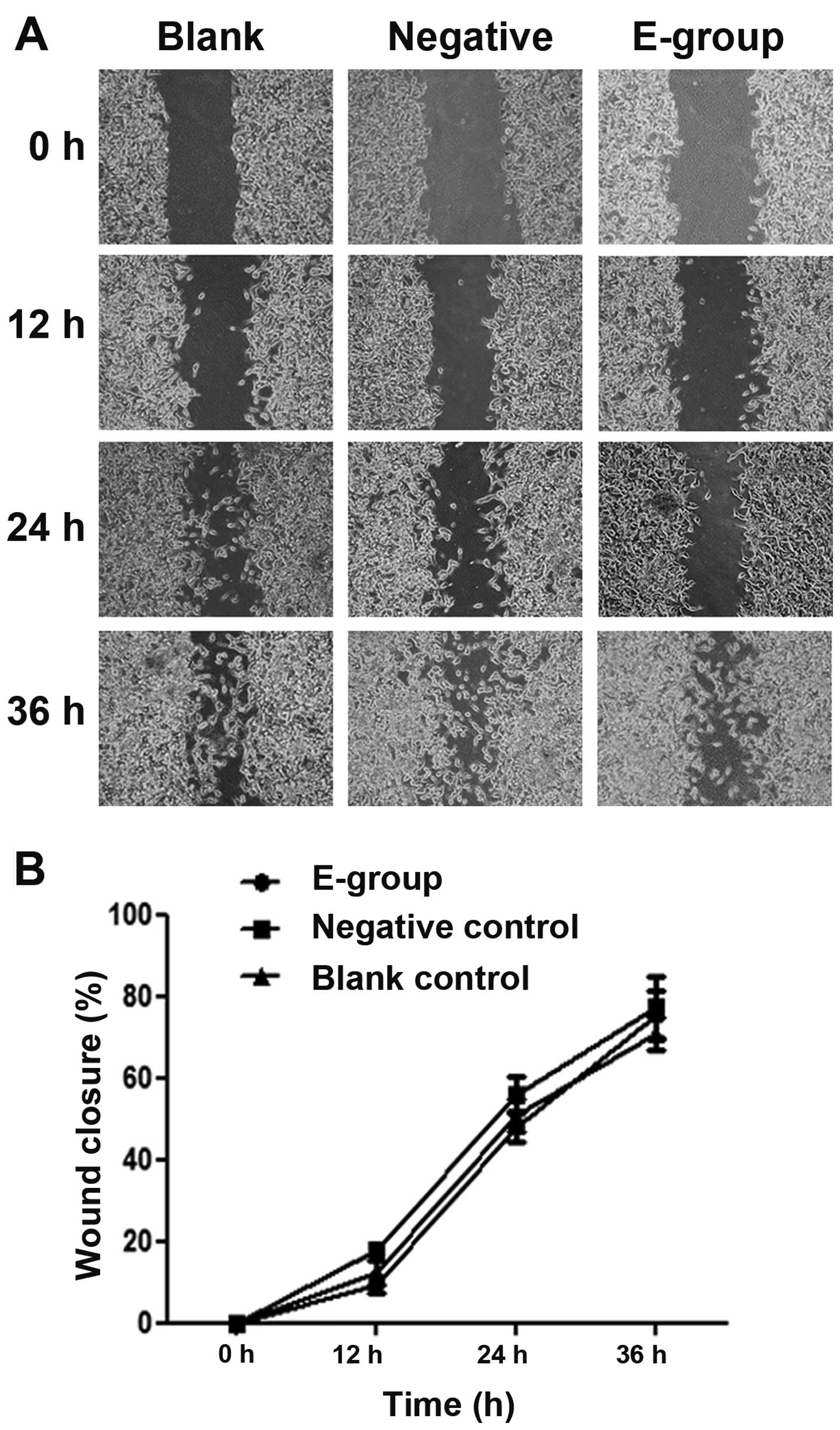
Bif-1 overexpression does not affect
the ability of PCa cell migration
To understand the role of Bif-1 overexpression in
cell migration, we used wound healing assay to compare the
migration ability change between Bif-1 overexpression LNCaP cells
(E-group) and the control cells, including the parental LNCaP cells
(blank control) and the pCNDA3.1(+) vector transfected cells
(negative control). By comparing the difference of the percentage
of wound closure between E-group and control groups, we found that
there is no significant change between the Bif-1 overexpression
LNCaP cells and the control cells (P>0.05; Fig. 5). Therefore, the wound-healing assay
results demonstrated that Bif-1 had no marked effect on prostate
cell migration capacity.
Discussion
Prostate cancer (PCa) is the most frequent
malignancy in males and one of the leading causes of cancer-related
death among males, particularly in more developed countries
(1,2,27–29).
Although high-risk locally advanced or metastatic patients who
receive androgen deprivation (AD) usually have durable remissions,
the cancer cells eventually develop a variety of pathways to
survive in a castrate environment and thus making tumor relapse
inevitable (27). It has been
reported that inhibition of apoptosis is critical in PCa and may
contribute to androgen resistance and progression of PCa (6,9).
Therefore, to elucidate the mechanisms behind apoptosis escape of
PCa cells is very important for the understanding of PCa
progression mechanisms.
BAX interacting factor 1 (Bif-1) is a member of the
endophilin B family, which binds to and activates the BAX protein
in response to the apoptosis signaling pathway. Recently,
mis-regulation of apoptosis has been reported as a new mechanism
that may contribute to the progression of PCa that involves BCL-2
and BAX regulation (9,19,30).
Although there is evidence that indicated that Bif-1 may play an
important role in the early stage of PCa (8), little is known about its effect on
prostate cell biological behavior and its role in PCa progression
remains unclear (8). It has been
reported that Bif-1 level is upregulated in a large proportion of
PCa cases (8), which seemed
inconsistent with the known pro-apoptotic function of Bif-1
(8,12,13,15,31).
The relationships between Bif-1 expression and PCa development are
not fully studied and the role of Bif-1 in PCa development remains
unclear.
To understand the role of Bif-1 in PCa development,
in the present study, we first detected the expression level of
Bif-1 in PCa cell lines including 22RV1, PC3, LNCaP and DU145, and
found that both Bif-1 gene and protein expression level was
decreased in these cell lines compared with benign prostate
hypertrophy (P<0.01). Further immunohistochemistry examinations
on clinical PCa and benign prostate hyperplasia (BPH) samples
confirmed that Bif-1 level was higher in BPH, suppressed in
low-grade prostatic carcinoma samples (P<0.05), and
significantly decreased in high grade prostatic carcinoma samples
(P<0.05). We presume that the mechanism of downregulated Bif-1
level in progressive PCa may be related to apoptosis escape of PCa
cells in cancer development. Our findings are consistent with those
of Coppola et al (8).
However, Coppola et al (8)
also found a proportion of PCa cases that express high level of
Bif-1, which we did not find in the present study. Since there is a
lack of studies reporting the Bif-1 level in PCa cases, the Bif-1
expression in PCa samples still needs more related studies for
complete elucidation.
To explore the relationships between Bif-1
expression level and PCa cell progression, we built a Bif-1
expression vector and Bif-1 overexpression LNCaP cells (E-group).
The effect of Bif-1 overexpression on cell apoptosis, proliferation
and migration was analyzed by flow cytometry, MTT and would healing
assays, respectively. The LNCaP cells were chosen for Bif-1 effect
analysis as it was an invasive human PCa cell line, and expressed
constitutively very low levels of Bif-1. It was found that
overexpression of Bif-1 significantly inhibited the proliferation
of LNCaP cells (P<0.05). Although the cell number was markedly
reduced, cell morphology was not affected. Further apoptosis
analysis by flow cytometry confirmed that Bif-1 could significantly
promote cell apoptosis (P<0.05), which proved that Bif-1 could
inhibit PCa cell proliferation by inducing cell apoptosis. These
results are consistent with the known function of proapoptosis of
Bif-1 (12,14) and the findings of Iacopino et
al (19), and support our
presumption that downregulation of Bif-1 may be involved in PCa
cell escape of apoptosis, and PCa cells may acquire apoptosis
resistance by downregulating Bif-1 expression. However, the PCa
apoptosis escape mechanism remains to be interpreted, and further
studies are needed to elucidate the signaling pathway that Bif-1 is
involved in. In the present study, we reported for the first time
that overexpression Bif-1 could significantly inhibit the
proliferation by inducing the apoptosis of PCa cells, and our
findings also give strong supports of the tumor suppressor
functions of Bif-1 in PCa progression. Since Bif-1 promoted
apoptosis, inhibited PCa cell proliferation and acted as a
suppressor in PCa progression, to restore the Bif-1 gene expression
may be a new potential strategy for PCa therapy.
We also explored the effect of Bif-1 overexpression
on PCa migration, since it was reported that Bif-1 could suppress
the migration of breast cancer cells by promoting the EGFR
endocytic degradation (20), and
whether or not Bif-1 can affect PCa cell migration is still
unknown. The wound healing assay showed that Bif-1 overexpression
had no significant effect on the migration capacity of PCa cells
(P>0.05). The biological significance of these finding remains
to be further determined.
In the present study, we detected the mRNA and
protein expression in PCa cell lines, BPH and PCa samples, proved
that Bif-1 is downregulated in both PCa cell lines and clinical
samples. By introducing the ectopically expressed Bif-1 into LNCaP
cells, we evaluated the role of Bif-1 on the growth and apoptosis
of PCa cells. We found that overexpression of Bif-1 could
significantly enhance PCa cell apoptosis and inhibit cell
proliferation, although it did not affect the migration ability.
Our findings give strong evidence that Bif-1 is involved in PCa
tumorigenesis and act as a suppressor in PCa progression. To
restore the Bif-1 gene expression may be a new potential strategy
for PCa therapy. Our findings may have significance in
understanding the process of PCa development.
Acknowledgements
The present study was supported by grants from the
National Natural Science Funds of China (nos. 81672556, 81071760
and 30772503).
References
|
1
|
McDavid K, Lee J, Fulton JP, Tonita J,
Thompson TD, Ferlay J, Brawley O and Bray F: Prostate cancer
incidence and mortality rates and trends in the United States and
Canada. Public Health Rep. 119:174–186. 2004.PubMed/NCBI
|
|
2
|
Bono AV: The global state of prostate
cancer: Epidemiology and screening in the second millennium. BJU
Int. 94:(Suppl 3). S1–S2. 2004. View Article : Google Scholar
|
|
3
|
Torre LA, Sauer AM, Chen MS Jr,
Kagawa-Singer M, Jemal A and Siegel RL: Cancer statistics for Asian
Americans, Native Hawaiians, and Pacific Islanders, 2016:
Converging incidence in males and females. CA Cancer J Clin.
66:182–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kontos CK, Adamopoulos PG and Scorilas A:
Prognostic and predictive biomarkers in prostate cancer. Expert Rev
Mol Diagn. 15:1567–1576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chao OS and Clément MV: Epidermal growth
factor and serum activate distinct pathways to inhibit the BH3 only
protein BAD in prostate carcinoma LNCaP cells. Oncogene.
25:4458–4469. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen S, Niu Y, Lee SO and Chang C: Androgen
receptor (AR) positive vs negative roles in prostate cancer cell
deaths including apoptosis, anoikis, entosis, necrosis and
autophagic cell death. Cancer Treat Rev. 40:31–40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Coppola D, Oliveri C, Sayegh Z, Boulware
D, Takahashi Y, Pow-Sang J, Djeu JY and Wang HG: Bax-interacting
factor-1 expression in prostate cancer. Clin Genitourin Cancer.
6:117–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Castilla C, Congregado B, Chinchón D,
Torrubia FJ, Japón MA and Sáez C: Bcl-xL is overexpressed in
hormone-resistant prostate cancer and promotes survival of LNCaP
cells via interaction with proapoptotic Bak. Endocrinology.
147:4960–4967. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shukla S, Fu P and Gupta S: Apigenin
induces apoptosis by targeting inhibitor of apoptosis proteins and
Ku70-Bax interaction in prostate cancer. Apoptosis. 19:883–894.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang X, Bi L, Ye Y and Chen J:
Formononetin induces apoptosis in PC-3 prostate cancer cells
through enhancing the Bax/Bcl-2 ratios and regulating the p38/Akt
pathway. Nutr Cancer. 66:656–661. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takahashi Y, Meyerkord CL and Wang HG:
Bif-1/endophilin B1: A candidate for crescent driving force in
autophagy. Cell Death Differ. 16:947–955. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schlauder SM, Calder KB, Khalil FK,
Passmore L, Mathew RA and Morgan MB: Bif-1 and Bax expression in
cutaneous Merkel cell carcinoma. J Cutan Pathol. 36:21–25. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cuddeback SM, Yamaguchi H, Komatsu K,
Miyashita T, Yamada M, Wu C, Singh S and Wang HG: Molecular cloning
and characterization of Bif-1. A novel Src homology 3
domain-containing protein that associates with Bax. J Biol Chem.
276:20559–20565. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi Y, Karbowski M, Yamaguchi H,
Kazi A, Wu J, Sebti SM, Youle RJ and Wang HG: Loss of Bif-1
suppresses Bax/Bak conformational change and mitochondrial
apoptosis. Mol Cell Biol. 25:9369–9382. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coppola D, Khalil F, Eschrich SA, Boulware
D, Yeatman T and Wang HG: Down-regulation of Bax-interacting
factor-1 in colorectal adenocarcinoma. Cancer. 113:2665–2670. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JW, Jeong EG, Soung YH, Nam SW, Lee
JY, Yoo NJ and Lee SH: Decreased expression of tumour suppressor
Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric
carcinomas. Pathology. 38:312–315. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan R, Miao Y, Shan X, Qian H, Song C, Wu
G, Chen Y and Zha W: Bif-1 is overexpressed in hepatocellular
carcinoma and correlates with shortened patient survival. Oncol
Lett. 3:851–854. 2012.PubMed/NCBI
|
|
19
|
Iacopino F, Angelucci C, Lama G, Zelano G,
La Torre G, D'Addessi A, Giovannini C, Bertaccini A, Macaluso MP,
Martorana G, et al: Apoptosis-related gene expression in benign
prostatic hyperplasia and prostate carcinoma. Anticancer Res.
26:1849–1854. 2006.PubMed/NCBI
|
|
20
|
Runkle KB, Meyerkord CL, Desai NV,
Takahashi Y and Wang HG: Bif-1 suppresses breast cancer cell
migration by promoting EGFR endocytic degradation. Cancer Biol
Ther. 13:956–966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mendonca MS, Howard KL, Farrington DL,
Desmond LA, Temples TM, Mayhugh BM, Pink JJ and Boothman DA:
Delayed apoptotic responses associated with radiation-induced
neoplastic transformation of human hybrid cells. Cancer Res.
59:3972–3979. 1999.PubMed/NCBI
|
|
22
|
Bonner AE, Lemon WJ, Devereux TR, Lubet RA
and You M: Molecular profiling of mouse lung tumors: Association
with tumor progression, lung development, and human lung
adenocarcinomas. Oncogene. 23:1166–1176. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Coppola D, Helm J, Ghayouri M, Malafa MP
and Wang HG: Down-regulation of Bax-interacting factor 1 in human
pancreatic ductal adenocarcinoma. Pancreas. 40:433–437. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ho J, Kong JW, Choong LY, Loh MC, Toy W,
Chong PK, Wong CH, Wong CY, Shah N and Lim YP: Novel breast cancer
metastasis-associated proteins. J Proteome Res. 8:583–594. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ko YH, Cho YS, Won HS, An HJ, Sun DS, Hong
SU, Park JH and Lee MA: Stage-stratified analysis of prognostic
significance of Bax-interacting factor-1 expression in resected
colorectal cancer. Biomed Res Int. 2013:3298392013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim SY, Oh YL, Kim KM, Jeong EG, Kim MS,
Yoo NJ and Lee SH: Decreased expression of Bax-interacting factor-1
(Bif-1) in invasive urinary bladder and gallbladder cancers.
Pathology. 40:553–557. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hoimes CJ and Kelly WK: Redefining hormone
resistance in prostate cancer. Ther Adv Med Oncol. 2:107–123. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Center MM, Jemal A, Lortet-Tieulent J,
Ward E, Ferlay J, Brawley O and Bray F: International variation in
prostate cancer incidence and mortality rates. Eur Urol.
61:1079–1092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang DB, Uo T, Kinoshita C, Sopher BL, Lee
RJ, Murphy SP, Kinoshita Y, Garden GA, Wang HG and Morrison RS: Bax
interacting factor-1 promotes survival and mitochondrial elongation
in neurons. J Neurosci. 34:2674–2683. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang J, Takahashi Y, Cheng E, Liu J,
Terranova PF, Zhao B, Thrasher JB, Wang HG and Li B: GSK-3beta
promotes cell survival by modulating Bif-1-dependent autophagy and
cell death. J Cell Sci. 123:861–870. 2010. View Article : Google Scholar : PubMed/NCBI
|