Introduction
Giant cell tumor of bone (GCT) is the most commonly
reported non-malignant bone tumor in Hong Kong (1). This kind of tumor usually affects
people aged 20–40 years (2). The
tumor leads to bone destruction near the major skeletal joints and
surgery is usually needed in order to remove the tumor and save the
joint. Moreover, GCT is well known for recurrence locally,
especially when the tumor cannot be removed completely. GCT is
comprised of 3 histological different cell types; the
multinucleated osteoclast-like giant cells, spindle-shaped
stromal-like cells and the monocytic round-shaped macrophage-like
cells (3–5). The stromal cells of GCT are the unique
primary neoplastic cells as well as the only proliferating cell
component in the cell culture of tumor cells (6). It is well known that macrophage-like
GCT cells are osteoclast precursors. GCT stromal cells (GCTSC) can
express osteoblastic lineage markers such as bone sialoprotein,
collagen type I and osteonectin proteins.
Filamins, which are actin-binding proteins, contain
three family members, filamin A, B and C. They are the products of
three different genes, FLNA, FLNB, and FLNC, which can generate
various transcript variants in different cell types (7). FLNA is predominantly expressed in the
brain and blood vessels while FLNB and FLNC can be found in bones
and muscles, respectively (8).
Filamins are vital to formation and maintenance of cell morphology,
motility for responding to the external stimuli and
differentiation. They are also able to interact with >90 binding
partners which include ion channels, receptors, intracellular
signaling molecules, transcription factors and other cytoskeleton
proteins. Therefore, they are mediators of many cellular processes
(9–11). Regarding the structure of filamin B,
it contains the N-terminal actin-binding domain (ABD), which
includes two calponin-homology domains (CH1 and CH2), followed by
24 immunoglobulin-like repeats. Repeats 1–15 represent the first
rod domain and are interrupted by a hinge region (hinge 1), then
repeats 16–23 form the second rod domain and interrupted by a
second hinge region (hinge 2). Finally, the C-terminal repeat 24 is
the dimerization domain (7,12,13).
The hinge 1 region is related to filamin flexibility and some
isoforms do not contain this region (14,15).
In this study, we focus on two FLNB isoforms: FLNB
variant2 (FLNBv2) and FLNB variant4 (FLNBv4). FLNBv2 is known as
ABP-278 and FLNBv4 is known as ABP-276 when they were being
discovered. FLNBv2 is the dominant isoform in prostate, uterus,
small intestine, liver, lymph node, stomach, lung, thyroid and
spleen, whereas, FLNBv4 is dominant in Daudi cells and spinal cord.
The placenta, bone marrow and brain express both isoforms with
comparable level (16). The only
difference of FLNBv2 and FLNBv4 is that FLNBv4 does not contain
hinge 1 region. In terms of their function, FLNBv4 accelerates
mouse myoblasts differentiation into myotubes (17). This may be due to the different
localization of FLNBv2 and FLNBv4 in the differentiating cells.
Also, FLNBv2 and FLNBv4 have very different binding affinity
towards integrins, which transduce signals through interactions of
their cytoplasmic tails with cytoskeletal and signaling proteins
(18). This difference may lead to
the alteration of signal transduction in several signaling events
critical for tumorigenesis (19).
We report on the differential expression of FLNB
splicing variants in GCTSC. Moreover, the clinical and functional
significance of the alteration was explored. We found that the
relative abundance of FLNBv4 varies among different GCT cell lines
while the expression level of FLNBv4 in normal osteoblast (OB) was
only marginally detectable. This phenomenon could also be observed
in liver cancer tissues and their adjacent normal counterparts. In
the functional aspect, overexpression of FLNBv4 led to upregulation
of RANKL, VEGFA and RUNX2, which are closely correlating with GCT
formation and cancer cell survival. In terms of proliferation,
overexpression of FLNBv2 increases cell viability and FLNBv4
decreased cell viability in some cell lines. In conclusion, FLNBv2
increase GCT cell proliferation and FLNBv4 is highly correlated
with differentiation genes.
Materials and methods
Patients information
GCT specimens were collected from the patients who
underwent surgical excision of the tumor at Prince of Wales
Hospital. All protocols were approved by the local Institutional
Ethics Committee. The information of patients involved in this
study is listed in Table I.
 | Table I.Characteristics of the 16 patients
with GCT. |
Table I.
Characteristics of the 16 patients
with GCT.
| Sample | Gender | Age | Tumor
localization | Stage |
|---|
| G25 | M | - | Left distal femur,
GCT | Stage III |
| G27 | M | 24 | Right distal radius,
GCT | Stage III |
| G28 | F | 26 | Right distal femur,
GCT | Stage II |
| G33 | M | 31 | Left distal radius,
GCT | Stage II |
| G34 | M | 47 | Right knee, GCT | Stage II |
| G35 | F | 34 | Left tibial, GCT | Stage II |
| G36 | F | 30 | Left calcaneum,
GCT | Stage II |
| G41 | F | 33 | Right femoral neck,
GCT | Stage II |
| G42 | F | 14 | Left tibia, GCT | Stage II |
| G44 | M | 35 | Left os calcis,
GCT | Stage II |
| G47 | M | 30 | Left distal femur,
GCT | Stage III |
| G51 | F | 36 | Left distal radius,
GCT | Stage II |
| G52 | F | 40 | Tibia, GCT | Stage II |
| G53 | F | 39 | Tibia, GCT | Stage II |
| G59 | F | 39 | Proximal tibia,
GCT | Stage III |
| G62 | F | 48 | Ischial bone
(pelvis), GCT | Stage II |
Primary culture of GCT SC
Primary culture of GCT stromal cells was established
as previously described (20).
Briefly, freshly extracted tissues were minced with scissors in
DMEM medium which contains 10% FBS (both from Thermo Fisher
Scientific, Waltham, MA, USA) and 100 U/ml penicillin (PSN). The
cell suspension was transferred to culture flasks and cultured at
37°C in a humidified atmosphere of 5% CO2 and 95% air.
Culture medium was changed every 2–3 days upon reaching confluence
and the cells were sub-cultured. GCTSC in cultures obtained after
the 5th passage, representing the proliferating homogeneous tumor
cell population, were used for the following assays.
Construction of FLNBv4 plasmid
The recombinant plasmid clone of FLNBv2 in pCR3.1(−)
vector driven by CMV promoter was a gift from Philip B. Daniel
(18). The DNA sequences of the
clones were in concordant with the reference sequence (NCBI
accession no. NM_001457.3). The cloning method and plasmid
construction were previously described (18). The FLNBv4 specific DNA fragment was
obtained from G33 cDNA by PCR amplification using FLNBv4 cloning
primers described in Table II. The
FLNBv4 specific DNA fragment was cloned into the FLNB variant 2
plasmid between MfeI and SacII (NEB, Hitchin, UK)
sites. The DNA sequence of the recombinant plasmid was confirmed by
Sanger sequencing and found to the same as the reported sequence of
FLNBv4 (NCBI accession no. NM_001164319.1).
 | Table II.Primer sequences designed for cloning
and real-time PCR amplification. |
Table II.
Primer sequences designed for cloning
and real-time PCR amplification.
| Primer
sequences |
| LNBv4 | F:
5′-AGATTCCTCGCAGTCCC-3′ |
|---|
| cloning |
|
| FLNBv4 | R:
5′-GCTGTTGATTTCTGGGATTTT-3′ |
| cloning |
|
| GAPDH | F:
5′-CGCCCCACTTGATTTTGGA-3′ |
| GAPDH | R:
5′-TTGCCATCAATGACCCCTTCA-3′ |
| FLNBv2 | F:
5′-GCCGAGGCCGATGTCATTGAGAA-3′ |
| FLNBv2 | R:
5′-CGGCTGTGACTTCCCCATCTGTG-3′ |
| FLNBv4 | F:
5′-GCCGAGGCCGATGTCATTGAGAA-3′ |
| FLNBv4 | R:
5′-ACATAGGCCTCTTCGGTCACCATGA-3′ |
| RANKL | F:
5′-AACAGGCCTTTCAAGGAGCTGTGC-3′ |
| RANKL | R:
5′-AAGAGGACAGACTCACTTTATGGG-3′ |
| OPG | F:
5′-CAAAGTAAACGCAGAGAGTGTAGA-3′ |
| OPG | R:
5′-GAAGGTGAGGTTAGCATGTCC-3′ |
| RUNX2 | F:
5′-GAGTCCTTCTGTGGCA-3′ |
| RUNX2 | R:
5′-GGCCGTTAGGGTTTGA-3′ |
| OCN | F:
5′-GCCTTTGTGTCCAAGC-3′ |
| OCN | R:
5′-GGACCCCACATCCATAG-3′ |
RNA extraction and cDNA synthesis
Total RNA was extracted from GCT stromal cell lines
using TRIzol reagent (Thermo Fisher Scientific) and the PureLink
RNA Mini kit (Invitrogen, Carlsbad, CA, USA), following the
manufacturer's instructions. Afterwards, 2.0 µg of total RNA from
each sample was subjected to cDNA synthesis using QuantiTect
Reverse Transcription kit (Qiagen, Hilden, Germany).
Semi-quantitative PCR
Real-time PCR was performed on a ViiA 7 Real-Time
PCR system using SYBR-Green master mix (both from Applied
Biosystems, USA), according to the manufacturer's instructions.
Fold-change was calculated using ΔΔCq method (21). Primer sequences are listed in
Table II.
Cell counting kit-8 (CCK-8) assay. Cultured GCTSC
were seeded on 96-well plates at a density of 7,000 cells/well
overnight. Subsequently, the cells were transfected by FLNBv2
plasmid, FLNBv4 plasmid and empty vector using Lipofectamine 3000
(Thermo Fisher Scientific) respectively. After 2 and 4 days of
transfection, CCK-8 assays were performed using the commercially
available assay kit (Dojindo, Shanghai, China). The absorbance was
measured at 450 nm using a microplate reader (Bio-Rad, Hercules,
CA, USA).
Results
Expression level of FLNB transcript
variants in GCTSC
To identify functionally important genes involved in
the tumorigenesis of GCT, the transcriptome analysis of three GCT
cell lines (G33, G35 and G53) and a control MSC cell line was
performed. The results revealed that expression levels of FLNB
variants in the three cell lines and MSC were significantly
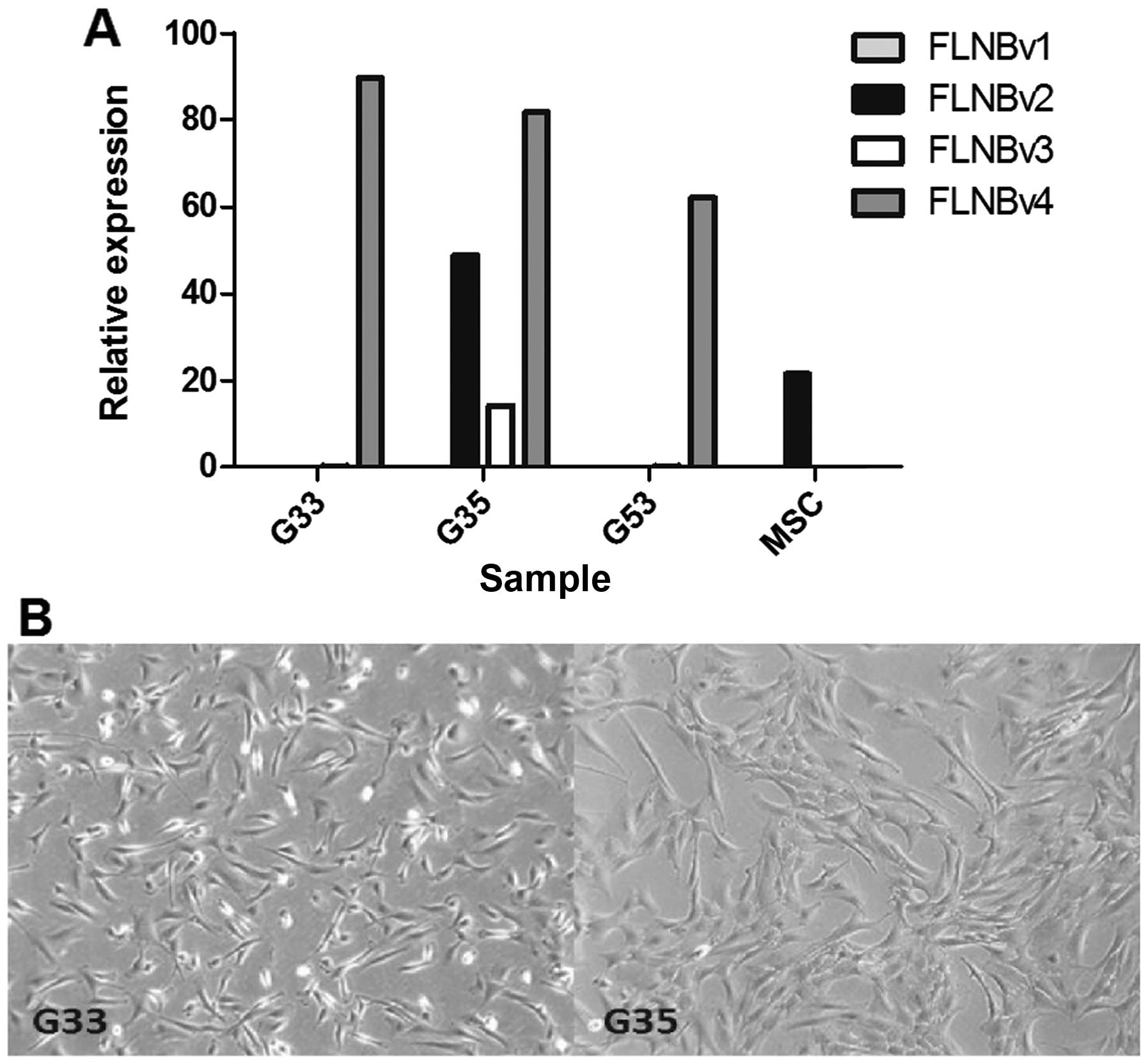
different (Fig. 1A). G33 and G53
are FLNBv4 dominant while MSC are FLNBv2 dominant. It is
interesting that G35 contains significant amounts of FLNBv2 and
FLNBv3, although FLNBv4 is still the major transcript variant. In
terms of cell morphology, we found that G33 is more polygonal and
fan-shaped while G35 is more fibroblast-like (Fig. 1B). Moreover, according to our
clinical record, the patient contributing the G35 cell line has had
two recurrences. This may explain why the expression pattern of G35
is so different from the other two cell lines. To see whether this
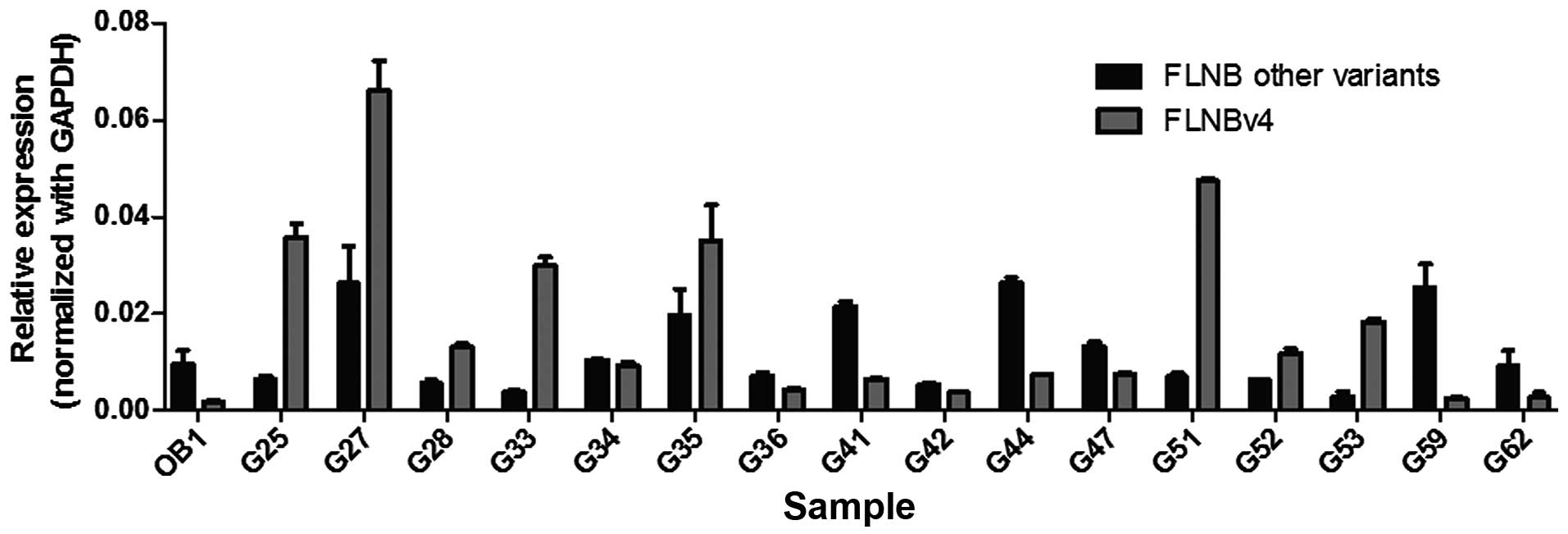
is a general phenomenon in GCT, real-time PCR was performed using
16 more GCT stromal cell lines (Fig.
2). Primers were designed specific to the sequence encoding the
hinge 1 region and to the FLNBv4 transcript so as to differentiate
FLNBv4 from other variants. Results show that the normal
osteoblasts (OB) expressed a low level of FLNBv4. This is
consistent with the observation of MSC results in the transcriptome
analysis. In the GCT stromal cell lines, 14 have increased
proportion of FLNBv4 relative to other variants. Of importance,
eight of them have more FLNBv4 transcripts than the overall amount
of other transcripts. Taken together, the amount of FLNBv4
transcripts is significantly elevated in GCT stromal cells.
Differential expression of FLNBv4 in
liver cancer and adjacent normal tissues
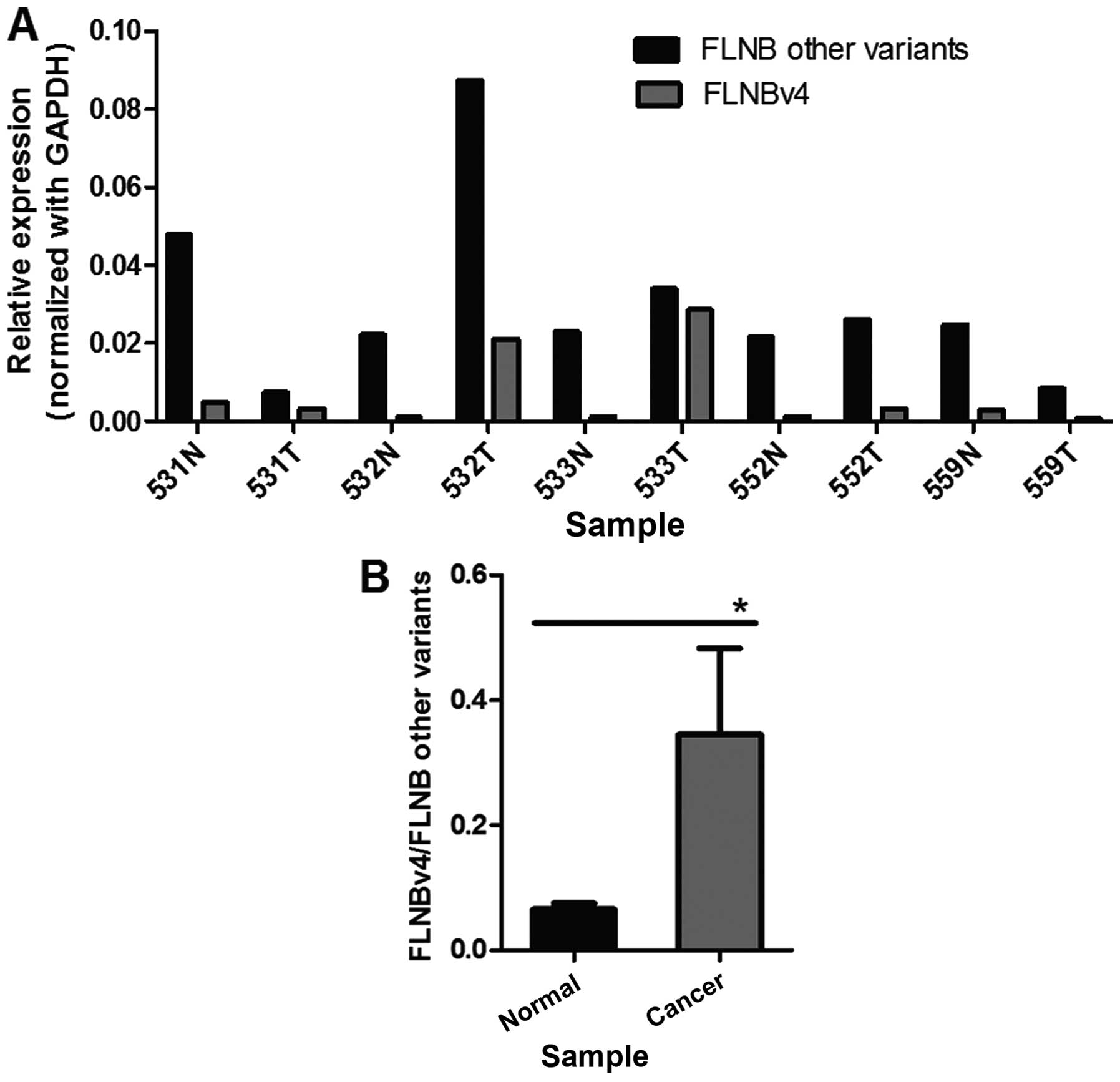
To explore whether the increase in the transcript
level of FLNBv4 could be observed in other cancer types, five liver
cancer and adjacent normal tissue pairs were screened for the
expression of FLNBv4 and other variants using real-time PCR
(Fig. 3A). Statistical analysis
revealed that the proportion of FLNBv4 transcripts relative to
other variants was significantly increased by ~5-fold in liver
cancer tissues (Fig. 3B). These
results were consistent with our observation in GCT cell lines.
Changes in gene expression triggered
by FLNBv2 and FLNBv4
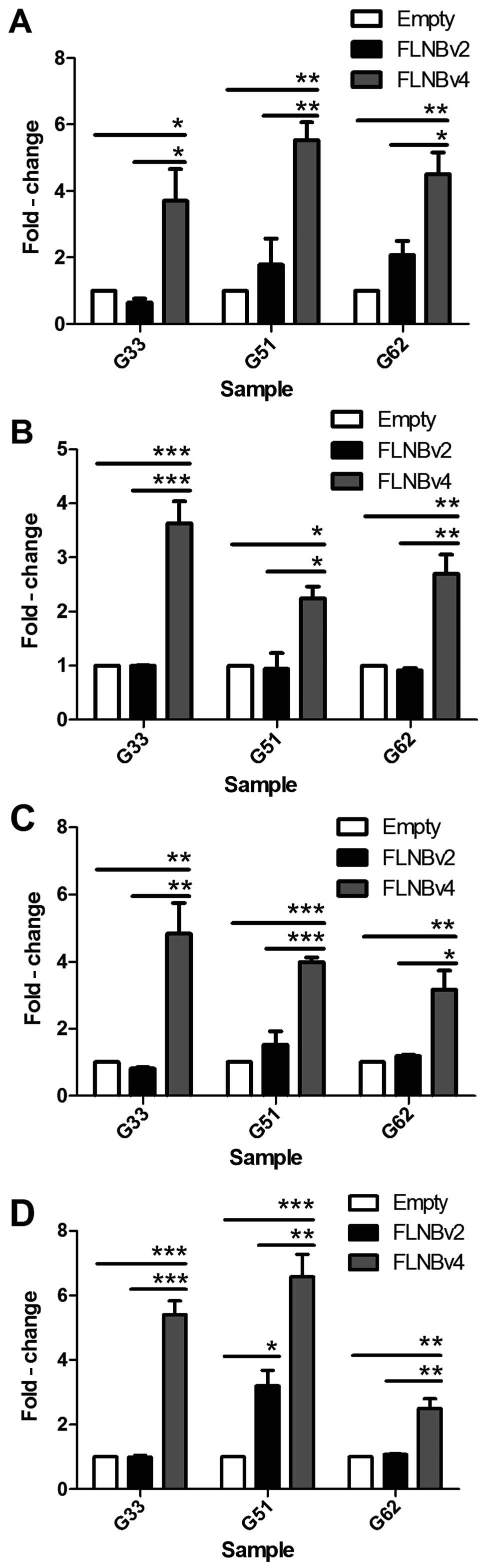
G33, G51 and G62 cells were transfected with FLNBv2,
FLNBv4 and empty vector. After 72 h, RNA was extracted and
converted to cDNA. Real-time PCR was then used to examine the
expression level of RANKL, OPG, OCN and RUNX2 in the transfected
cells. We found that RANKL was significantly upregulated by
2–4-fold in FLNBv4 transfected GCT cell lines, whereas, FLNBv2 also
induced a mild upregulation in G51 and G62 cells when compared with
the empty vector transfected cells (Fig. 4A). Moreover, FLNBv4 could
significantly and specifically upregulate OPG, OCN and RUNX2 by
>2-fold in all three GCT cell lines (Fig. 4B-D).
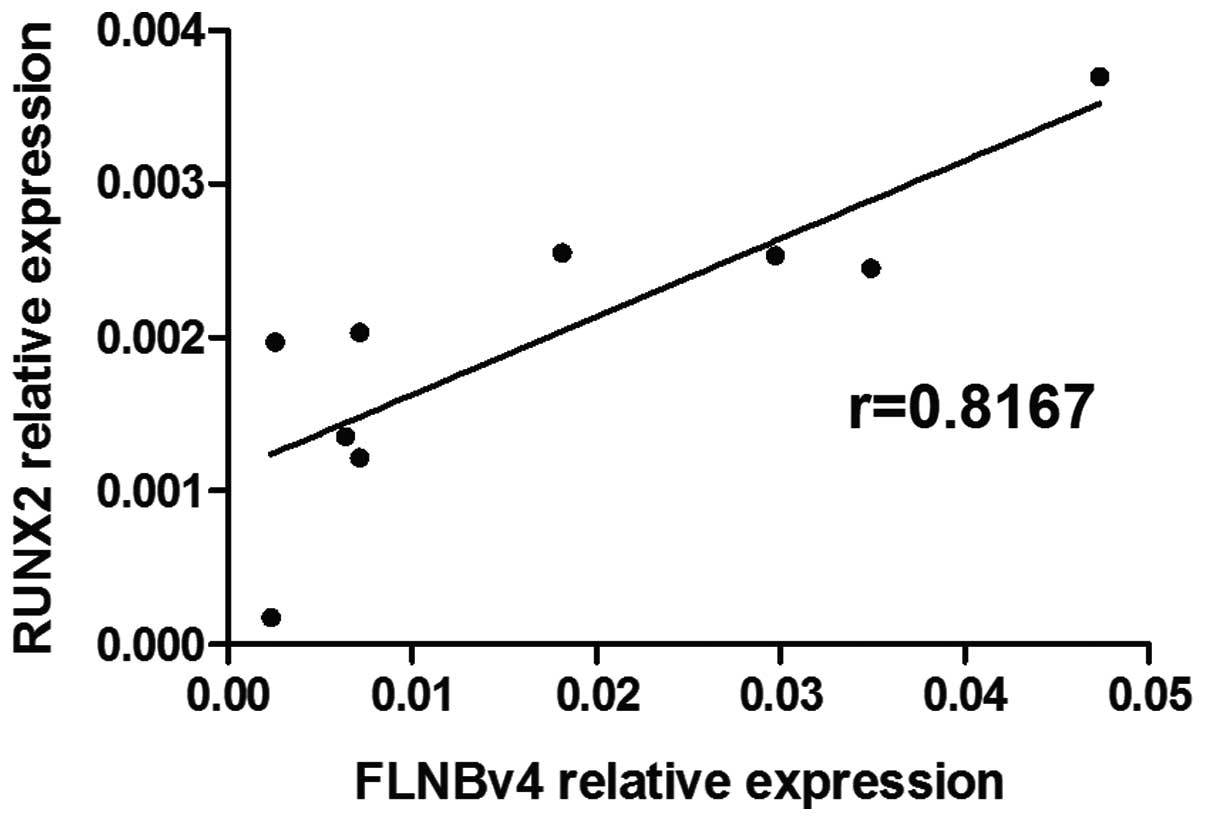
Correlation of expression levels of
FLNBv4 and RUNX2 in GCT cells
To study whether the endogenous expression level of
FLNBv4 could be correlated with the expression of RANKL, OPG, OCN
and RUNX2 in GCT cells, the expression of these four genes and the
FLNBv4 was examined in 9 GCT cell lines by real-time PCR. A
positive and high correlation (P=0.0174 and r=0.8167) between
FLNBv4 and RUNX2 was identified (Fig.
5) while no significant correlation was found between FLNBv4
and the other three genes.
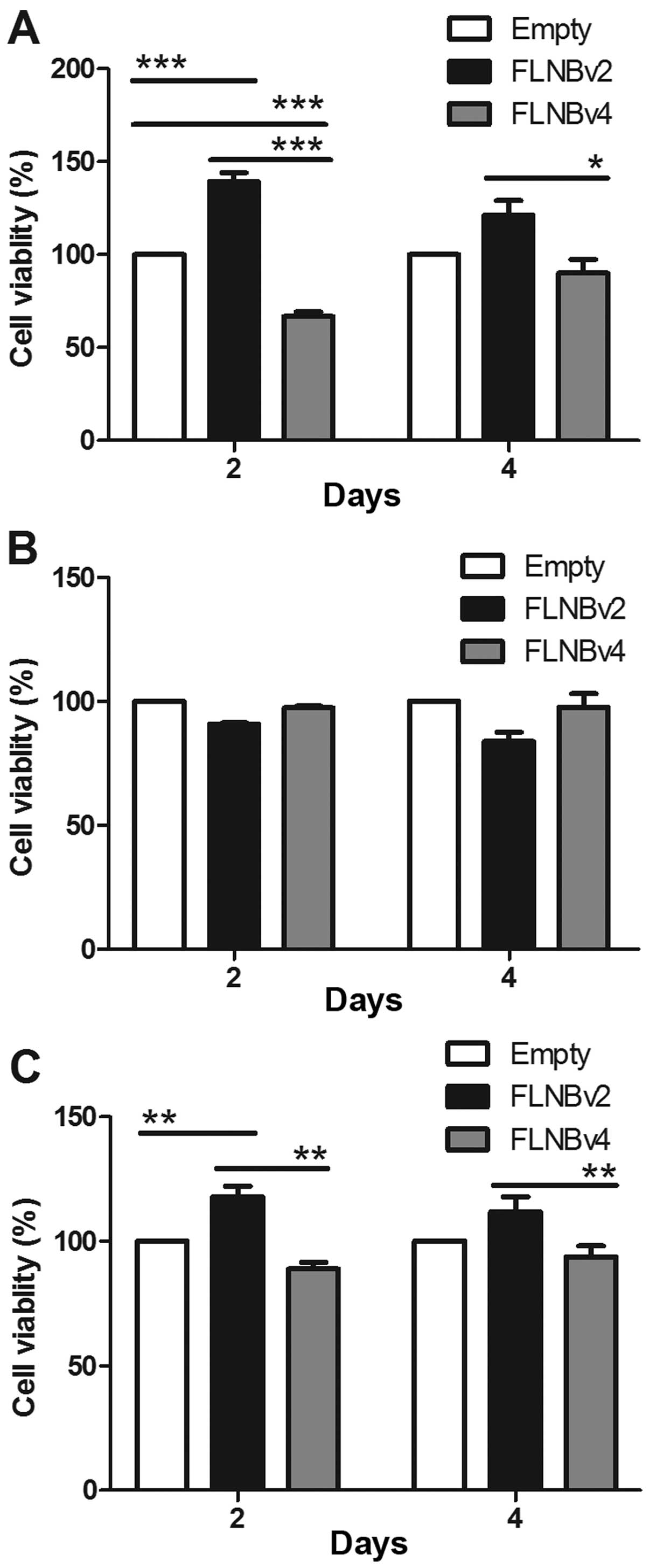
Effects of FLNBv2 and FLNBv4 on the
proliferation of GCT cells
To investigate whether FLNBv2 and FLNBv4 could
affect the proliferation of GCT cells, we transfected the G33, G51
and G62 cell lines with the recombinant plasmids expressing the
FLNBv2 and FLNBv4 and the cell proliferation was subsequently
determined by the CCK-8 cell viability assay at 2 and 4 days
post-transfection. In G33 cells, FLNBv2 transfected cells have a
significantly higher cell viability when compared with FLNBv4
transfected cells in 2 and 4 days post-transfection (Fig. 6A). This phenomenon could be also
observed in G62 cells in 2 days after post-transfection. However,
the situation in G51 cells was totally different. The proliferation
of FLNBv4 transfected cells was higher than that of FLNBv2
transfected cells, although the difference was not significant. It
is notable that among the three cell lines G51 was the only cell
line derived from a recurrent GCT.
Discussion
In our data, GCT cells express FLNBv4 which do not
contain FLNB hinge 1 region. The expression level of FLNBv4 in
normal osteoblast OB1 was undetectable. This phenomenon indicated
that FLNBv4 is commonly found in GCT cells but not normal
osteoblast cells. In our transcriptome analysis data, G33 and G53
are FLNBv4 dominant and G35 has a comparable amount of FLNBv2 and
FLNBv4. The difference between G33, G53 and G35 may be explained by
difference in morphology and tumor environment. Liver cancer
tissues also have a higher FLNBv4 to other FLNB variants ratio
compared with their own adjacent normal counterparts. Based on this
observation, we suspected that high expression level of FLNBv4 is
correlated with tumor formation.
In this study, we found that FLNBv4 can specifically
upregulated RANKL in GCT cells. RANKL is well known to be highly
expressed in GCT compared with normal osteoblasts or MSCs, and
contributing for the accumulation of giant cells in GCT.
Overexpression of RANKL in GCT microenvironment stimulates the
secretion of tumor necrosis factor-α (TNF-α) and interleukin-1β
from giant cells, which in turn induces GCT stromal cell
proliferation. RANKL is also an important signaling pathway which
plays a vital role in osteoclastogenesis and bone resorption
(22,23). Bone resorption is necessary for the
establishment and promotion of tumor metastasis in the bone
(24,25). In this pathway, RANKL can interact
with its receptor RANK, which is on the surface of osteoclast
precursors, to enhance activation, formation and survival of
osteoclasts in normal stage or tumor stage (26). Therefore, the effect of FLNBv4 on
RANKL can promote the tumor formation and bone resorption in
GCT.
Moreover, our results suggest that
FLNBv4-transfected cells have a higher level of RUNX2 when compared
with FLNBv2-transfected cells. There is also a positive correlation
between the expression level of FLNBv4 and RUNX2. RUNX2 is a
regulatory gene of matrix metalloproteinase 13 (MMP13) and its
encoded protein, Runx2, can activate the expression of MMP13 and
hence promote the osteoclast differentiation and osteolysis
(27). Moreover, Runx2 contributes
to the progression of cell cycle exit through binding with the
hypophosphorylated form of pRb. Our results suggest that FLNBv4 may
promote GCT stromal cell differentiation through Runx2. It is
notable that OCN, which is a non-collagenous, vitamin K-dependent
protein secreted in the late stage of osteoblasts differentiation
(28), was also upregulated by
FLNBv4, the expression level of OPG was increased by FLNBv4. OPG is
an osteoclastogenesis inhibitory factor of the TNF family. It can
inhibit the formation of osteoclast-like giant cell and prevent
bone resorption (29). All the
above results imply that the effect of FLNBv4 on Runx2, OCN and OPG
is able to promote the differentiation and prevent bone resorption
in GCT.
Taken together, FLNBv4 can produce opposite effects
in GCT stromal cells through the transcriptional regulation of
different genes. The balance between the promotion of proliferation
and differentiation therefore determines the overall effect of
FLNBv4. This is consistent with the fact that the expression level
of FLNBv4 is highly variable in different GCT stromal cell lines.
In addition, FLNBv4 can only inhibit the proliferation of two out
of the three cell lines tested. The exact role of this splicing
variant may depend on the genetic background and microenvironment
of GCT stromal cells.
Acknowledgements
This study is supported by the Health and Medical
Research Fund from the Food and Health Bureau, Hong Kong Special
Administrative Region (reference no. 14130312).
References
|
1
|
Yip KM, Leung PC and Kumta SM: Giant cell
tumor of bone. Clin Orthop Relat Res. 323:60–64. 1996. View Article : Google Scholar
|
|
2
|
Leung KH, Lam AY, Ho KW and Shek TW: Giant
cell tumor of the humeral head treated by denosumab: Implication to
shoulder surgeons. Int J Shoulder Surg. 9:135–138. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng MH, Robbins P, Xu J, Huang L, Wood D
and Papadimitriou J: The histogenesis of giant cell tumour of bone:
a model of interaction between neoplastic cells and osteoclasts.
Histol Histopathol. 16:297–307. 2001.PubMed/NCBI
|
|
4
|
Sobti A, Agrawal P, Agarwala S and Agarwal
M: Giant cell tumor of bone - An overview. Arch Bone Jt Surg.
4:2–9. 2016.PubMed/NCBI
|
|
5
|
James IE, Walsh S, Dodds RA and Gowen M:
Production and characterization of osteoclast-selective monoclonal
antibodies that distinguish between multinucleated cells derived
from different human tissues. J Histochem Cytochem. 39:905–914.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang L, Teng XY, Cheng YY, Lee KM and
Kumta SM: Expression of preosteoblast markers and Cbfa-1 and
Osterix gene transcripts in stromal tumour cells of giant cell
tumour of bone. Bone. 34:393–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Razinia Z, Mäkelä T, Ylänne J and
Calderwood DA: Filamins in mechanosensing and signaling. Annu Rev
Biophys. 41:227–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeon GW, Lee MN, Jung JM, Hong SY, Kim YN,
Sin JB and Ki CS: Identification of a de novo heterozygous missense
FLNB mutation in lethal atelosteogenesis type I by exome
sequencing. Ann Lab Med. 34:134–138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakamura F, Stossel TP and Hartwig JH: The
filamins: Organizers of cell structure and function. Cell Adhes
Migr. 5:160–169. 2011. View Article : Google Scholar
|
|
10
|
Popowicz GM, Schleicher M, Noegel AA and
Holak TA: Filamins: Promiscuous organizers of the cytoskeleton.
Trends Biochem Sci. 31:411–419. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stossel TP: Filamins and the potential of
complexity. Cell Cycle. 9:14632010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gorlin JB, Yamin R, Egan S, Stewart M,
Stossel TP, Kwiatkowski DJ and Hartwig JH: Human endothelial
actin-binding protein (ABP-280, nonmuscle filamin): A molecular
leaf spring. J Cell Biol. 111:1089–1105. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pudas R, Kiema TR, Butler PJ, Stewart M
and Ylänne J: Structural basis for vertebrate filamin dimerization.
Structure. 13:111–119. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sawyer GM and Sutherland-Smith AJ: Crystal
structure of the filamin N-terminal region reveals a hinge between
the actin binding and first repeat domains. J Mol Biol.
424:240–247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakamura F, Osborn TM, Hartemink CA,
Hartwig JH and Stossel TP: Structural basis of filamin A functions.
J Cell Biol. 179:1011–1025. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu W, Xie Z, Chung DW and Davie EW: A
novel human actin-binding protein homologue that binds to platelet
glycoprotein Ibalpha. Blood. 92:1268–1276. 1998.PubMed/NCBI
|
|
17
|
Dalkilic I, Schienda J, Thompson TG and
Kunkel LM: Loss of FilaminC (FLNc) results in severe defects in
myogenesis and myotube structure. Mol Cell Biol. 26:6522–6534.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van der Flier A, Kuikman I, Kramer D,
Geerts D, Kreft M, Takafuta T, Shapiro SS and Sonnenberg A:
Different splice variants of filamin-B affect myogenesis,
subcellular distribution, and determine binding to integrin [beta]
subunits. J Cell Biol. 156:361–376. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giancotti FG and Ruoslahti E: Integrin
signaling. Science. 285:1028–1032. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lau CP, Ng PK, Li MS, Tsui SK, Huang L and
Kumta SM: p63 regulates cell proliferation and cell cycle
progression associated genes in stromal cells of giant cell tumor
of the bone. Int J Oncol. 42:437–443. 2013.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu PF, Tang JY and Li KH: RANK pathway in
giant cell tumor of bone: Pathogenesis and therapeutic aspects.
Tumour Biol. 36:495–501. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang T, Yin H, Wang J, Li Z, Wei H, Liu Z,
Wu Z, Yan W, Liu T, Song D, et al: MicroRNA-106b inhibits
osteoclastogenesis and osteolysis by targeting RANKL in giant cell
tumor of bone. Oncotarget. 6:18980–18996. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lau CP, Huang L, Tsui SK, Ng PK, Leung PY
and Kumta SM: Pamidronate, farnesyl transferase, and geranylgeranyl
transferase-I inhibitors affects cell proliferation, apoptosis, and
OPG/RANKL mRNA expression in stromal cells of giant cell tumor of
bone. J Orthop Res. 29:403–413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang L, Cheng YY, Chow LT, Zheng MH and
Kumta SM: Tumour cells produce receptor activator of NF-kappaB
ligand (RANKL) in skeletal metastases. J Clin Pathol. 55:877–878.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong SS, Liu XG, Chen Y, Guo Y, Wang L,
Zhao J, Xiong DH, Xu XH, Recker RR and Deng HW: Association
analyses of RANKL/RANK/OPG gene polymorphisms with femoral neck
compression strength index variation in Caucasians. Calcif Tissue
Int. 85:104–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang Q, Jiang Z, Meng T, Yin H, Wang J,
Wan W, Cheng M, Yan W, Liu T, Song D, et al: MiR-30a inhibits
osteolysis by targeting RunX2 in giant cell tumor of bone. Biochem
Biophys Res Commun. 453:160–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Neve A, Corrado A and Cantatore FP:
Osteocalcin: Skeletal and extra-skeletal effects. J Cell Physiol.
228:1149–1153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang L, Xu J, Wood DJ and Zheng MH: Gene
expression of osteoprotegerin ligand, osteoprotegerin, and receptor
activator of NF-kappaB in giant cell tumor of bone: Possible
involvement in tumor cell-induced osteoclast-like cell formation.
Am J Pathol. 156:761–767. 2000. View Article : Google Scholar : PubMed/NCBI
|