Introduction
Colon cancer (CC) is a common neoplasm and presents
a considerable disease burden worldwide. The incidence and
mortality of CC in China has increased in recent decades (1). 5-Fluorouracil (5-FU) is a mainstream
chemotherapeutic drug that is widely used in CC treatment (2,3). The
pharmacological effects of 5-FU arise from the blocking of DNA
replication by interrupting the synthesis of pyrimidine thymidine.
Furthermore, 5-FU induces cell cycle arrest and apoptosis (4). However, many CC patients display
resistance and a high recurrence rate following chemotherapy with
5-FU. Therefore, drug resistance has been considered as one of the
biggest obstacles in chemotherapy, and new strategies for enhancing
the sensitivity of chemotherapy and preventing drug resistance are
urgently needed.
Epidermal growth factor receptor (EGFR) is a member
of tyrosine kinase receptors and has been reported to show a robust
increase in levels in many types of cancer, such as non-small cell
lung cancer (NSCLS), gastric cancer and CC (5,6). The
phosphorylation of EGFR leads to the activation of extracellular
signal-regulated kinase (ERK) and the phosphatidylinositol 3-kinase
(PI3K)/Ser/Thr kinase (AKT) pathways (7). These two pathways are correlated with
cell migration and apoptosis. Meanwhile, EGFR contributes to drug
resistance in tumor cells (8,9). The
human a disintegrin and metalloprotease 9 (ADAM9) is a
membrane-anchored metalloprotease that is often markedly
upregulated in several human carcinomas and accelerates the
adherence and migration of CC and other tumor cells (10,11). A
recent study found that ADAM9 is critical for promoting cell
proliferation in esophageal squamous cell carcinoma by targeting
EGFR-AKT signaling (12). In fact,
ADAM9 is considered as a key component in EGFR signaling and
development (13,14). Moreover, ADAMs have been suggested
as the novel mechanisms involved in the drug resistance in human
breast carcinoma MCF-7 and other cells (15,16).
Thus, we can speculate that ADAM9 may be able to increase drug
resistance by regulating EGFR.
MicroRNAs (miRNAs) are small non-coding regulatory
RNAs that are involved in temporal and tissue-specific eukaryotic
gene regulation. These miRNAs pair with either full or partial
complementary sequences in the 3′ untranslated regions (3UTRs) of
target mRNAs, leading to sequence-specific mRNA cleavage and/or
translational repression (17).
miRNAs have been virtually linked to the pathogenesis of many
diseases and pharmacological mechanisms of drug therapy. Previous
studies have indicated that specific targeting of miRNAs can open
new avenues for cancer treatment by decreasing drug resistance and
thereby improving the response to chemotherapy (18). In recent years, miR-20b has been
found to be downregulated in many types of cancer tissues. In
addition, miR-20b inhibited the proliferation, migration and
invasion of bladder cancer EJ cells via targeting of cell cycle
regulation and Sp-1-mediated MMP-2 expression (19). Moreover, miR-20b showed decreased
expression in colon tumors relative to normal colon tissue
(20). However, the role and
mechanism of miR-20b in the drug resistance of CC cells remain
unknown.
Given that a better understanding of the underlying
mechanism of drug resistance could provide fresh insight into the
treatment of CC, we examined the involvement of miR-20b in
chemoresistance to 5-FU in CC cells using two isogenic HCT116 cell
lines that are sensitive or resistant to 5-FU. Our results provide
evidence that miR-20b can directly modulate chemoresistance in
these cells by regulating its downstream target, the ADAM9/EGFR
signaling pathway.
Materials and methods
Tissue samples and ethics
statement
A total of 26 pairs of CC and adjacent non-cancerous
tissues were collected from CC patients who were sensitive or
resistant to 5-FU. All subjects were enrolled at the Henan Cancer
Hospital and their ages ranged from 35 to 60 years. The CC patients
were diagnosed according to the modified World Health Organization
diagnostic criteria for CC by a pathologist. All tissue samples
were obtained with informed consent, and the present study was
approved by the Ethics Committee of Henan Cancer Hospital and was
carried out in accordance with The Code of Ethics of the World
Medical Association.
Cell culture
CC cell line HCT116 is sensitive to 5-FU, and was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The HCT116-R cell line is resistant to 5-FU and
was generated by incubating HCT116 cells with increasing levels of
5-FU (Sigma-Aldrich, St. Louis, MO, USA) as previously described
(21). Normal colon and CC cells
were cultured in McCoy's 5A serum-free medium (Sigma, St. Louis,
MO, USA) supplemented with 20 µg/ml insulin, 10 ng/ml epidermal
growth factor and 4 µg/ml transferrin at 37°C and 5%
CO2. Resistant cell lines were maintained under constant
treatment with the drug.
Cell transfection
The cells were seeded in 6-well plates at
1.0×105/ml/well and transfected in logarithmic growth
phase with 25 nmol/l miR-20b mimic, miR-20b inhibitor or its
non-specific control (NC). All transfection reagents were purchased
from GenePharma Co., Ltd. (Shanghai, China). Transfection was
performed according to the manual provided with the siPOR™ NeoFX™
Transfection Agent (Ambion, Grand Island, NE, USA) and the
transfection efficiency was determined by RT-PCR. Aliquots of 0.5
µg pcDNA3.1/ADAM9 and pcDNA3.1 control vector were obtained from
OriGene and transfected using Lipofectamine™ 2000 (Life
Technologies, Carlsbad, CA, USA) as per the manufacturer's
protocol.
Cell proliferation and cell viability
assays
Cells were plated in 96-well plates at a
concentration of 2×104 cells/100 µl and treated with
5-FU (0, 5 and 10 µg/ml, respectively) for the indicated time to
assess proliferation. The surviving fractions were stained for 2 h
at 1–5 days with thiazolyl blue tetrazolium bromide (MTT) (Sigma).
The absorbance at 570 nm was measured by a spectrophotometer after
dissolving in dimethyl sulfoxide (DMSO) (Sigma). Cell viability was
calculated as the ratio of optical density (OD) values according to
the manufacturer's instructions using a CellTiter 96 AQueous One
Solution Cell Proliferation Assay kit (Promega Corporation,
Madison, WI, USA).
Cell cycle analysis
Cells were seeded into 6-well plates at
5×105 cells/well. After pretreatment with serum-free
medium for 48 h, the cells were harvested and fixed overnight with
ice-cold 70% ethanol. The fixed cells were quantified using a Cell
Cycle Analysis kit (KeyGen Biotech, Nanjing, China) according to
the manufacturer's instructions. Each experiment was performed in
triplicate.
Apoptosis assay
After transfection for 72 h, the cells were
collected and incubated in phosphate-buffered saline (PBS) for 10
min. Analysis of apoptosis was performed using an Annexin V
apoptosis detection kit (BD Pharmingen™, San Diego, CA, USA)
according to the manufacturer's instructions. Cells were washed and
suspended in 100 ml binding buffer, at a density of
1×106 cells/ml, and then labeled with 5 µl of Annexin
V-FITC and PI for 15 min. Thereafter, flow cytometric analysis was
performed on a FACSAria flow cytometry system (BD Biosciences).
Data were analyzed using BD FACSDiva software.
Luciferase reporter assay
The pGL3 control vector (Promega) carrying the 3′UTR
sequence of ADAM9, which was predicted to interact with miR-20b or
a mutated sequence within the predicted target sites, was
transfected into HCT116-R cells. Before luciferase activity
analysis, cells were further transfected with the miR-20b mimic.
Cells were divided into five groups: control, miR-20b mimic,
miR-20b + pcDNA3.1, miR-20b + pcDNA3.1/ADAM9 and mimic NC. Firefly
luciferase activity was measured using the Dual-Luciferase Assay
kit (Promega) according to the manufacturer's instructions, and the
data were then normalized using a Renilla luciferase
reference plasmid.
Real-time quantitative polymerase
chain reaction (RT-qPCR) assay
Total RNA was extracted using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) from cells or
tissues. The concentration and integrity of the RNA were determined
by electrophoresis, and the RNA was reverse transcribed using a
RevertAid First Strand cDNA Synthesis kit (Invitrogen). For
transfection with ADAM9, the cDNA of ADAM9 was synthesized using a
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems,
Foster City, CA, USA). The Maxima SYBR-Green Master Mix (2X) was
obtained from Applied Biosystems according to the manufacturer's
instructions. The PCR involved an initial step at 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min and 72°C
for 45 sec. Primer pairs for miR-20b were designed and synthesized
by RiboBio Inc. (Guangzhou, China). Data were analyzed using the
comparative 2−ΔΔCt method.
Western blotting
Cells were homogenized with lysate (Beyotime)
supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF)
(Sigma), and then centrifuged. The supernatants were collected and
the contents were quantified using the BCA kit (Pierce, Rockford,
IL, USA). Samples of 20 µg protein were separated by SDS-PAGE using
a 12% polyacrylamide gel and transferred to a polyvinylidene
difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After
blockage of non-specific binding sites with 5% non-fat milk, the
membrane was incubated overnight at 4°C with various primary
antibodies, including antibodies against ADAM9, EGFR and β-actin
[Cell Signaling Technology (CST); Danvers, MA, USA)] (1:500). Next,
the nitrocellulose membrane was washed three times with PBS
containing 0.1% Tween-20 and further incubated with the secondary
antibody (1:800) at room temperature. The immunoblots were
visualized using an ECL Western Blotting Detection kit (Millipore)
and analysis software Quantity One (Bio-Rad, Hercules, CA,
USA).
Statistical analysis
Quantitative data are expressed as mean ± standard
deviation (SD). Measurement data were analyzed using Student's
t-test when there were only two groups. For other situations,
statistical analyses were performed using one-way or two-way ANOVA
in SPSS 19.0. The level of significance was set at P<0.01.
Results
miR-20b is downregulated, while ADAM9
and EGFR are upregulated in tissues of the 5-FU-resistant CC
patients
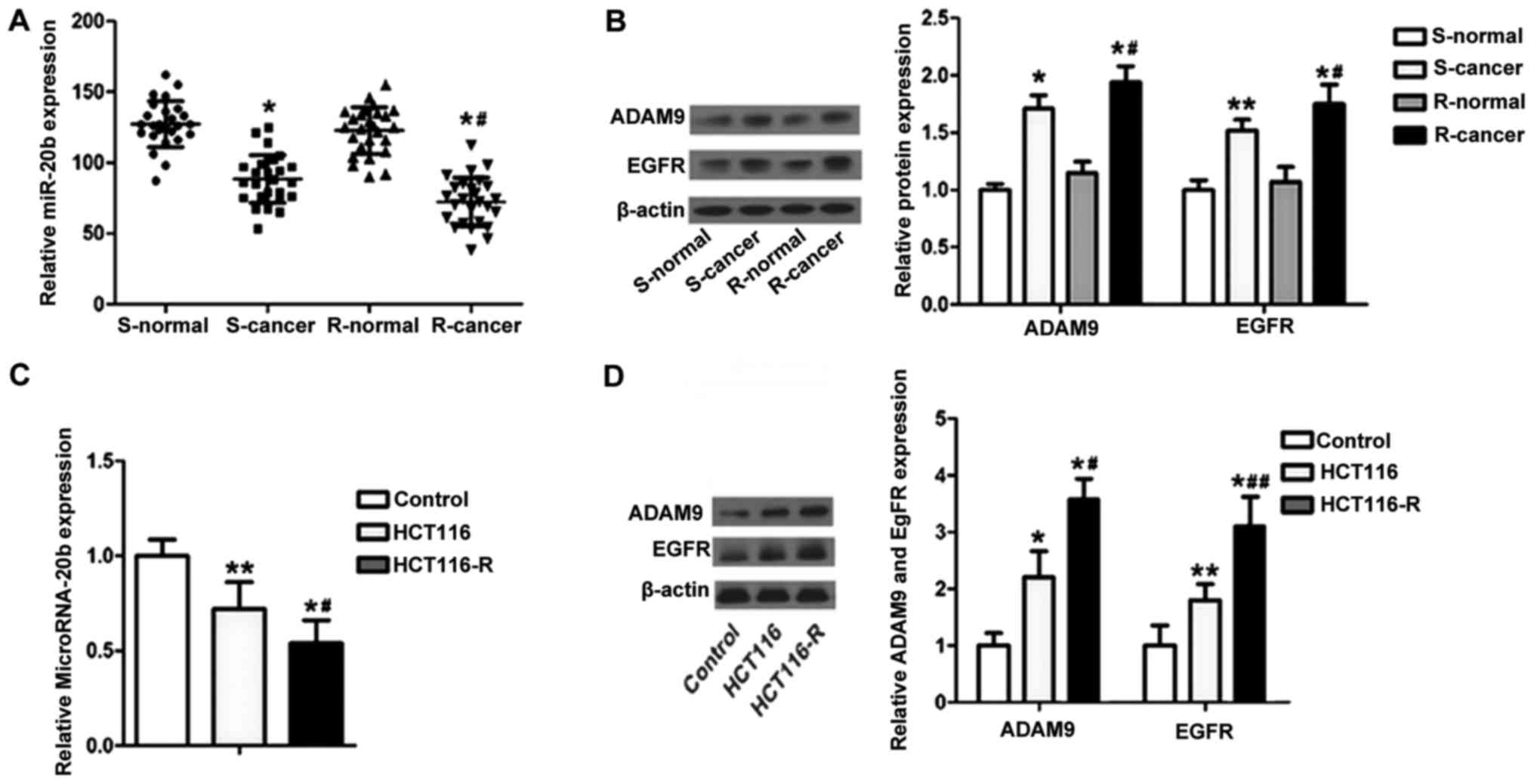
To investigate the potential roles of miR-20b in CC,
we first examined the expression of miR-20b in an expanded cohort
of 26 CC patients. Our results showed that miR-20b expression was
markedly downregulated in the CC tissues compared to the level in
the corresponding adjacent non-cancerous tissues (Fig. 1A). Meanwhile, miR-20b expression was
lower in the CC patients who were sensitive to 5-FU than the level
in those who were resistant. In order to identify the role of ADAM9
and EGFR in 5-FU resistance, the levels of ADAM9 and EGFR were
detected by western blotting. As shown in Fig. 1B, the expression levels of ADAM9 and
EGFR were significantly higher in the cancer cells than levels in
the normal control cells. Similarly, levels of ADAM9 and EGFR
expression were higher in CC patients who were sensitive to 5-FU
than these levels noted in the patients who were resistant.
To further explore the mechanisms of 5-FU resistance
in the two isogenic CC cell lines (HCT116 and HCT116-R), HCT116,
HCT116-R and normal CC cells were cultured and the levels of
miR-20b (Fig. 1C), ADAM9 and EGFR
(Fig. 1D) were measured. The
results for miR-20b, ADAM9 and EGFR expression in the CC tissue and
CC cell lines were consistent. These data showed that the
expression of miR-20b was universally lower whereas levels of ADAM9
and EGFR were higher in the 5-FU resistant CC patients and cells
than in the resistant patients and cells, indicating that miR-20b,
ADAM9 and EGFR may be involved in 5-FU resistance in CC cells.
HCT116-R cell line is resistant to
5-FU
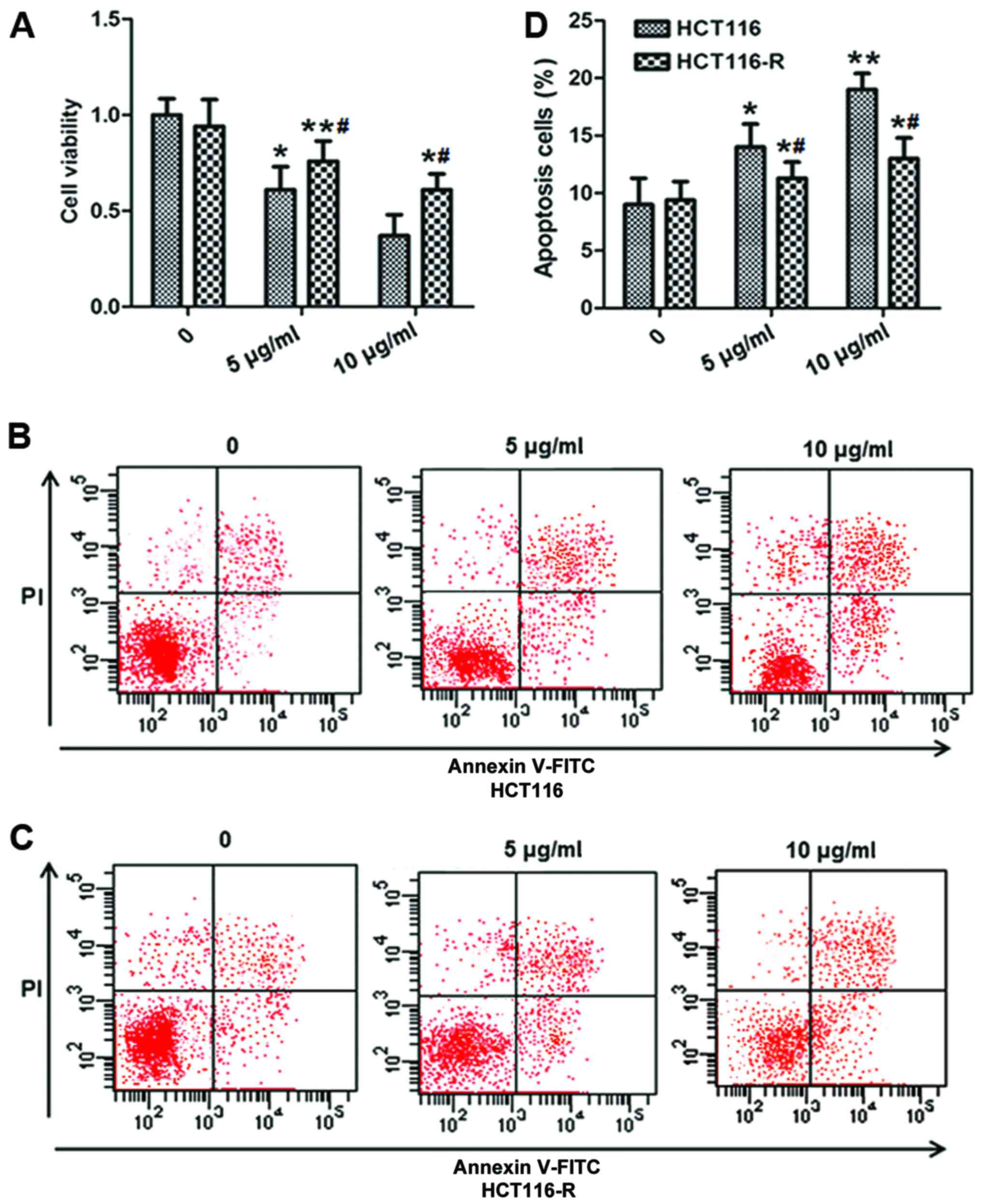
To understand the mechanisms of 5-FU resistance, we
determined the effect of 5-FU on the growth of HCT116 and HCT116-R
cell lines, and found that 5-FU decreased cell viability in a
dose-dependent manner, with a more obvious effect on HCT116 cells
than HCT116-R cells (Fig. 2A).
Fig. 2B and C respectively show the
percentage of apoptosis in the HCT116 and HCT116-R cells after 5-FU
treatment. The statistical data on the percentage of apoptotic
cells is shown in Fig. 2D.
Similarly, HCT116 cells showed higher sensitivity to 5-FU-induced
apoptosis than HCT116-R cells, as demonstrated by cell
viability.
miR-20b mediates proliferation and
apoptosis in HCT116-R cells in vitro
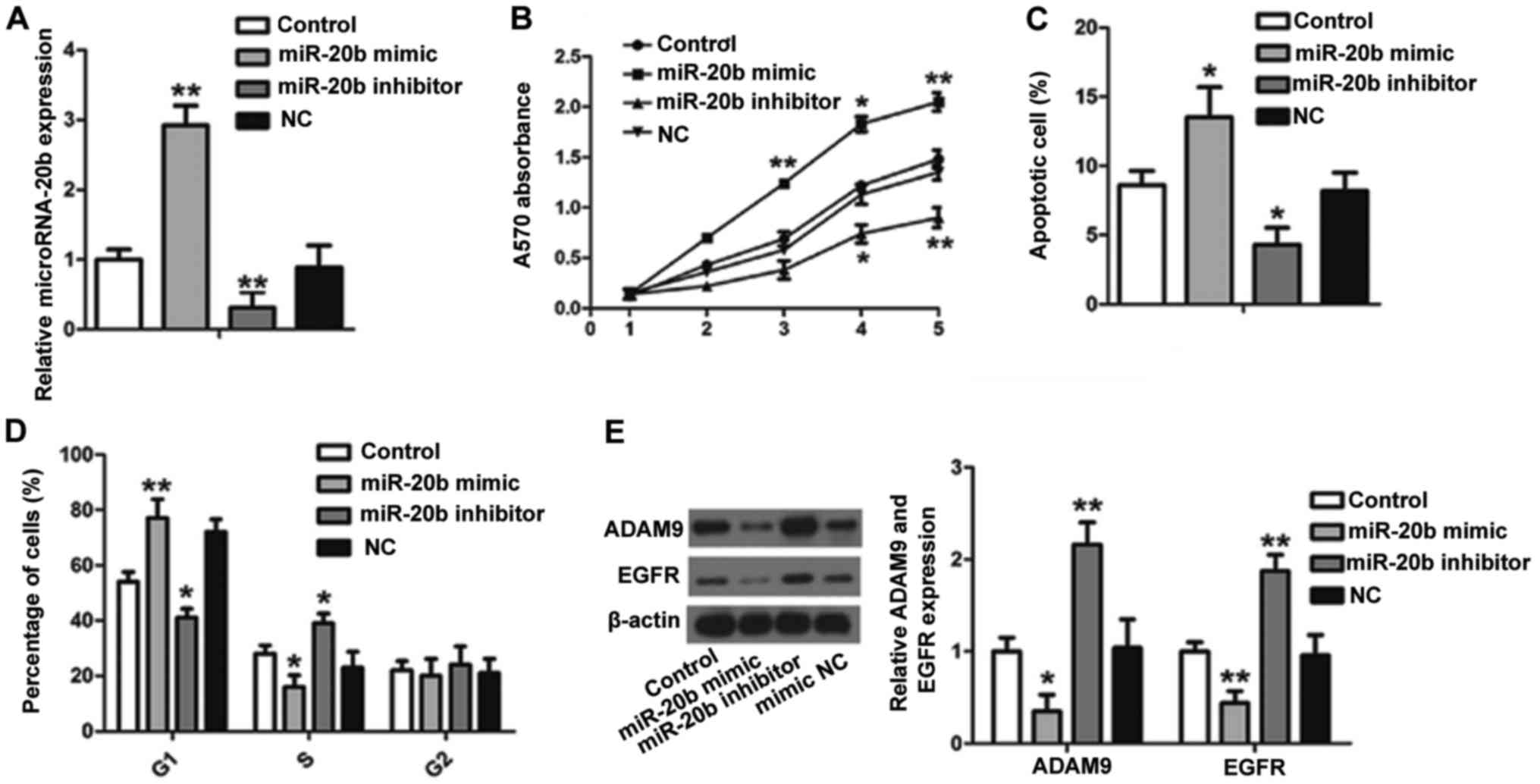
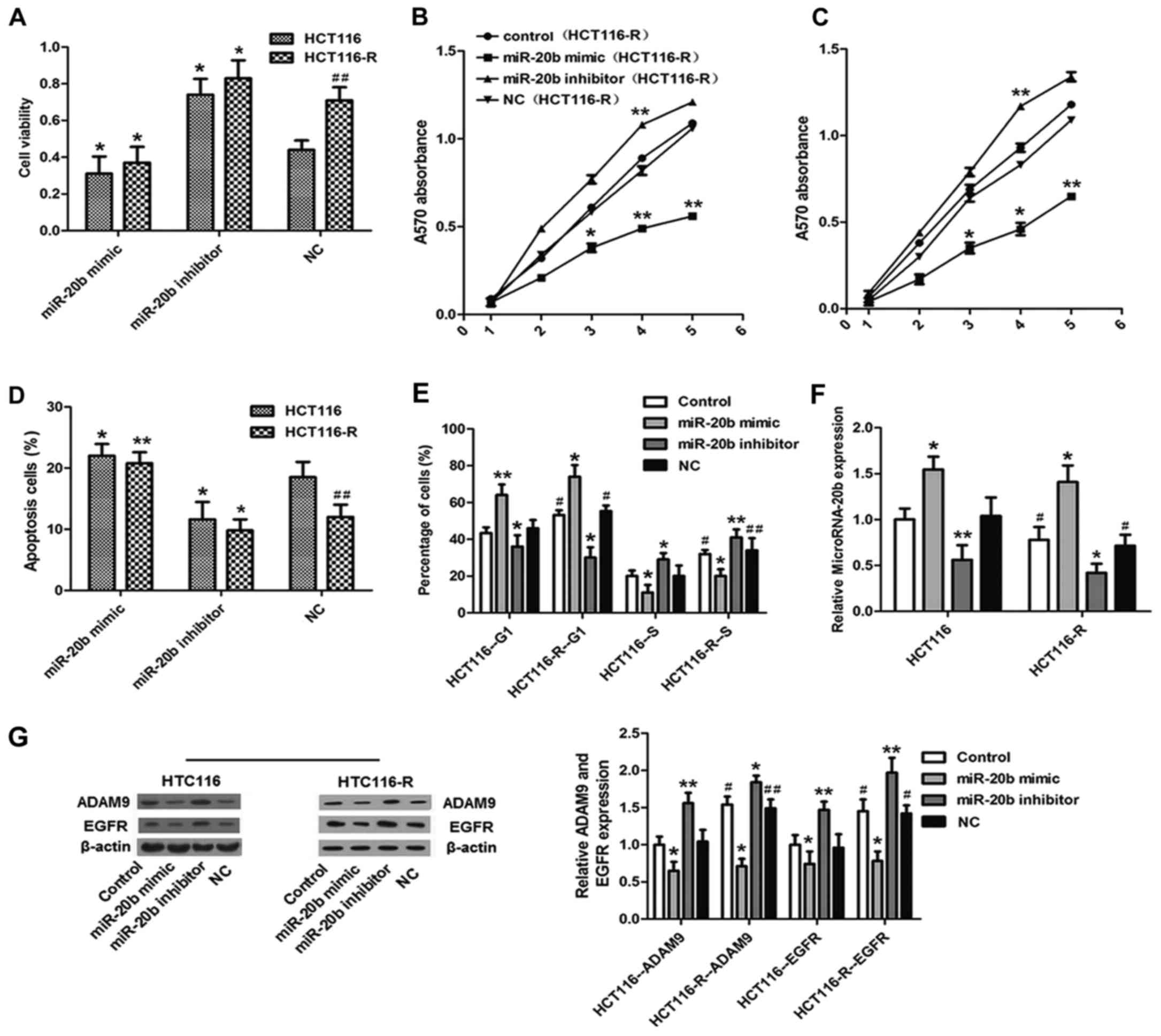
To identify the role of miR-20b in CC cells, miR-20b
was overexpressed or suppressed in the HCT116-R cells by
transfection with miR-20b mimic or miR-20b inhibitor (Fig. 3A). As shown in Fig. 3B, cell proliferation was decreased
in the miR-20b-overexpressed group in comparison with the control
group according to the MTT assay. Conversely, in the
miR-20b-suppressed group, cell proliferation was accelerated.
Moreover, the percentage of apoptotic cells increased to 13.5%
after transfection with the miR-20b mimic (Fig. 3C). Inhibition of the expression of
miR-20b decreased the apoptosis of the HCT116-R cells.
Next, we sought to determine whether miR-20b has any
impact on cell cycle progression in the HCT116-R cells (Fig. 3D). We found that the number of cells
in the G1 phase was increased in the miR-20b mimic group compared
with this percentage in the control, but this percentage was
decreased in the S phase. Meanwhile, the miR-20b mimic and miR-20b
inhibitor groups showed contrasting results. The percentage of
cells in G2 did not differ significantly between groups. In
addition, the protein expression of ADAM9 and EGFR was
downregulated in the miR-20b mimic group and upregulated in the
miR-20b inhibitor group compared with the control (Fig. 3E). Overall, these findings suggest
that miR-20b inhibits the proliferation and induces the apoptosis
of CC by suppressing cell cycle progression at the G1/S transition
in CC cells. Moreover, ADAM9 and EGFR may be involved in this
process.
miR-20b suppresses cell proliferation
and apoptosis and regulates cell cycle progression by targeting
ADAM9 in HCT116-R cells
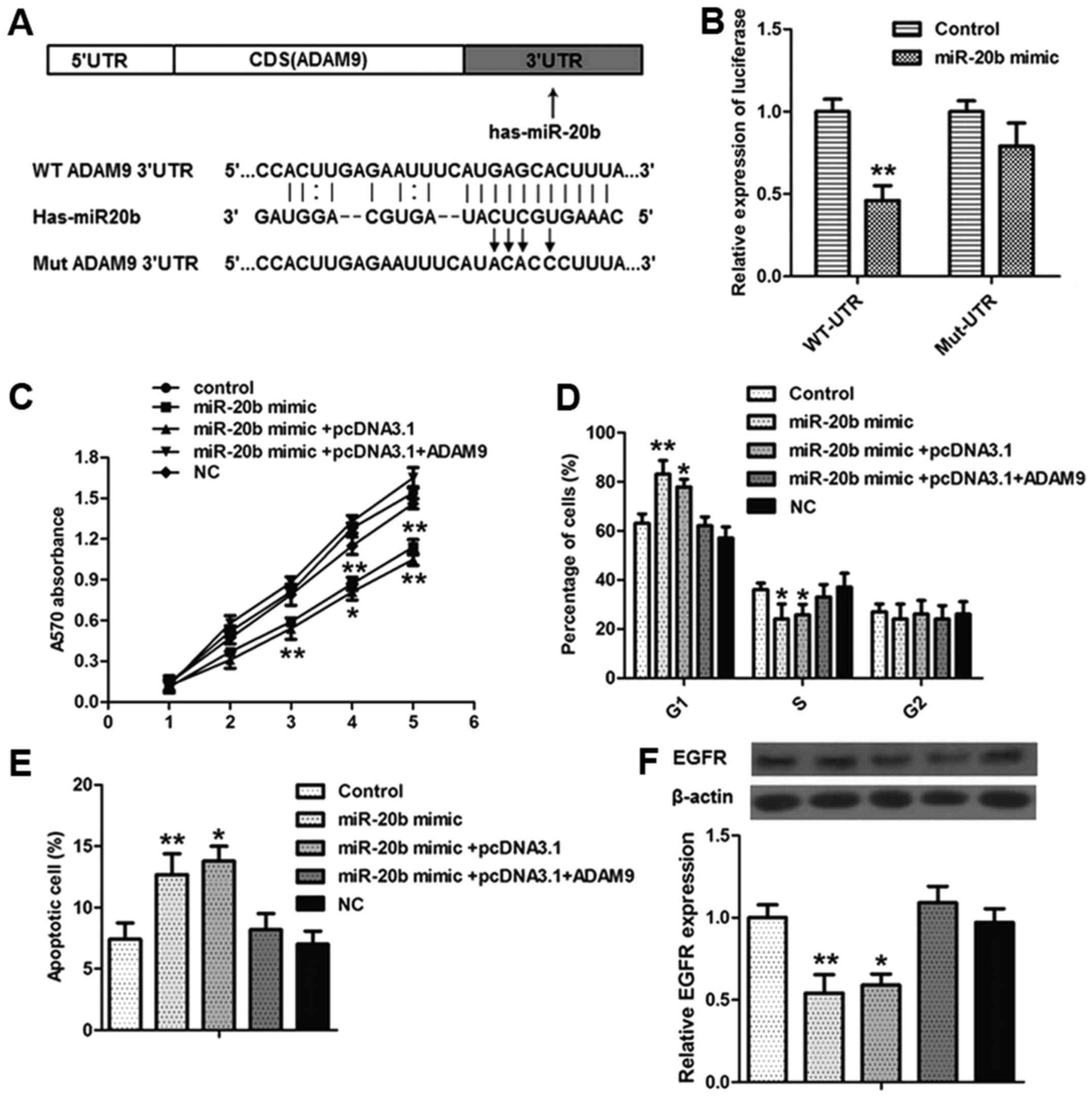
In order to investigate the relationship between
miR-20b and ADAM9/EGFR in CC, ADAM9 was identified as one of the
putative targets of miR-20b. We constructed the wild-type and
mutant ADAM9 and cloned it downstream of a luciferase reporter gene
(Fig. 4A). We found that miR-20b
decreased the luciferase intensity by ~63% in the wild-type ADAM9
3′UTR of HCT116-R cells compared with the normal cells. However,
luciferase activity did not differ from that in the ADAM9 3′UTR-mut
groups (Fig. 4B). These results
suggest that ADAM9 is a direct target of miR-20b in CC.
To further confirm that ADAM9 is a functional target
of miR-20b in CC cells, we co-transfected HCT116-R cells with
pcDNA3.1-ADAM9 and miR-20b mimic to construct a ADAM9
overexpression model. Our results showed an increase in cell
proliferation, a reduction in cell cycle arrest at G1/S and a
reduction in apoptosis rate in the ADAM9-overexpressed group
(Fig. 4C-E). Accordingly, ADAM9
overexpression elevated the expression of EGFR (Fig. 4F). All these data indicate that
miR-20b suppresses CC cell proliferation, apoptosis and cell cycle
progression by directly targeting ADAM9.
miR-20b reduces 5-FU resistance in the
HCT116-R cells
To determine whether miR-20b reduces the 5-FU
resistance of CC cells, HCT116 and HCT116-R cells were transfected
with miR-20b mimic. Next, these cells were treated with 10 µg/ml
5-FU for 72 h. We then assessed the cell viability, proliferation,
apoptosis, cell cycle progression and expression of ADAM9 and EGFR
as mentioned above. Our results showed that miR-20b suppressed cell
viability and proliferation in both the HCT116 and HCT116-R cells
(Fig. 5A-C). In contrast,
apoptosis, cell cycle progression and ADAM9 and EGFR expression
were promoted by miR-20b in the HCT116 and HCT116-R cells (Fig. 5D-G). Meanwhile, none of the measured
parameters differed between HCT116 and HCT116-R cells in the mimic
and miR-20b inhibitor groups. These results indicate that miR-20b
attenuated the 5-FU resistance of HCT116-R cells.
Taken together, the present study demonstrated that
miR-20b reduced 5-FU resistance by suppressing the ADAM9/EGFR
signaling pathway in CC tissues and in vitro.
Discussion
5-FU is widely considered to be the standard therapy
for CC and is usually combined with leucovorin. Although 5-FU can
improve the median survival of CC patients by almost two years
(22), drug resistance to 5-FU is a
serious issue. In the present study, in order to investigate the
potential mechanism of chemoresistance to 5-FU in CC cells, we
established a 5-FU-resistant cell line from human CC cell line
HCT116 and constructed an ADAM9 overexpression model by
co-transfecting HCT116-R cells with pcDNA3.1-ADAM9 and miR-20b
mimic. The major finding was that miR-20b was positively associated
with 5-FU resistance but inversely associated with the expression
of ADAM9/EGFR in CC cells. Moreover, miR-20b reduced the 5-FU
resistance of CC cells by inhibiting ADAM9 expression, and EGFR
expression was inhibited as well. These results suggest that
targeting miR-20b may represent a novel therapeutic approach for
CC.
Increasing evidence suggests that miRNAs play
important roles in cancers and many are aberrantly expressed in CC
cells (23–25). Ahmed proposed that miRNAs could be
markers for the diagnostic screening of CC (26). In the present study, we found that
miR-20b was downregulated in CC patients and significantly
inhibited the proliferation of CC cells in vitro, which is
consistent with a previous study that found that miR-20b was
downregulated in colon tumors. Furthermore, miR-20b was reported to
inhibit the proliferation of melanoma cells by regulating the
proteinase-activated receptor-1 (PAR-1), thrombin receptor
(27), and to inhibit cell cycle
progression by targeting MMP-2 in bladder cancer EJ cells (19).
Recent evidence has shown that miRNAs attenuate or
contribute to 5-FU resistance in human CC cells (28). MicroRNA array showed that
microRNA-195 chemosensitizes CC cells to the chemotherapeutic drug
doxorubicin by targeting the first binding site of B-cell
CLL/lymphoma 2-like protein 2 (BCL2L2) mRNA (29). Mussnich et al reported that
miR-199a-5p and miR-375 affect CC cell sensitivity to cetuximab by
targeting the PH domain and leucine rich repeat protein phosphatase
1 (PHLPP1) (30). MicroRNA-425-5p
was also reported to reduce 5-FU resistance in colorectal cancer
cells via the regulation of programmed cell death 10 (21). Our present study found that miR-20b
reduced 5-FU resistance in CC cells, which may be a novel potential
mechanism for suppressing 5-FU resistance in CC cells.
EGFR has been identified as a tumor-promoting gene
in many types of cancers. EGFR-targeted therapy has recently been
implemented as a new therapeutic strategy in various malignancies
(31). The anti-EGFR monoclonal
antibodies cetuximab and panitumumab showed good clinical activity
in ~10% of patients with metastatic colorectal cancer that was
resistant to chemotherapy (32).
Patel et al demonstrated that curcumin enhanced the effects
of 5-FU and oxaliplatin in mediating growth inhibition of CC cells
by regulating EGFR (33), which
indicates that EGFR is closely linked to 5-FU resistance in CC
cells. Indeed, EGFR is widely considered as a drug resistance gene
in NSCLS, glioblastoma, colorectal cancer and other cancers, and
could be modulated by many genes (9,34). It
was also found that overexpression of ADAM9 enhanced growth
factor-mediated recycling of E-cadherin in the human CC cell line
HT29 (35). In the present study,
we demonstrated that EGFR was not only upregulated with ADAM9
synchronously in CC, but was also positively regulated by ADAM9.
Furthermore, cell viability, cell cycle progression and cell
proliferation were inhibited when ADAM9/EGFR were elevated, whereas
cell apoptosis was accelerated. Additionally, the expression of
ADAM9 and EGFR was higher in HCT116 cells than that in HCT11-R
cells. Thus ADAM9/EGFR showed a positive correlation with the 5-FU
resistance of CC cells. These results are consistent with previous
studies that found that ADAM9 is correlated with cell adherence and
migration in pancreatic cancer cells (36), and that the expression of EGFR and
ADAM9 was increased and that EGFR is the downstream target gene of
ADAM9 in esophageal squamous cell carcinoma cells (12). Our results thus confirm that the
ADAM9/EGFR pathway plays an important role in the 5-FU resistance
of CC cells.
Although ADAM9 has been reported to be upregulated
in CC, there is little research on the mechanism of action of ADAM9
in CC. The present study found that ADAM9/EGFR could be
downregulated by miR-20b mimic and upregulated by miR-20b inhibitor
in CC cells, supporting the hypothesis that ADAM9/EGFR is the
functional downstream target of miR-20b in vitro. A
luciferase reporter assay further confirmed that ADAM9 functions as
a direct target of miR-20b and prevented the cell apoptosis induced
by miR-20b in CC. Notably, we observed that the level of ADAM9/EGFR
and 5-FU resistance did not differ between HCT11-R and HCT116 cells
after transfection with miR-20b in the presence of 5-FU. These
results indicate that miR-20b attenuates 5-FU resistance in CC
cells by depressing ADAM9/EGFR expression.
In summary, the present data suggest that miR-20b is
downregulated in 5-FU-resistant CC patients and cells compared with
sensitive patients and cells. 5-FU sensitivity could be increased
by miR-20b through regulation of the ADAM9/EGFR pathway. Our study
broadens our understanding of the contribution of miR-20b to 5-FU
sensitivity in CC cells via the targeting of several other genes,
and indicates that it could be a powerful target to reduce 5-FU
resistance in CC cells.
References
|
1
|
Hou L, Ji BT, Blair A, Dai Q, Gao YT and
Chow WH: Commuting physical activity and risk of colon cancer in
Shanghai, China. Am J Epidemiol. 160:860–867. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li J, Hou N, Faried A, Tsutsumi S,
Takeuchi T and Kuwano H: Inhibition of autophagy by 3-MA enhances
the effect of 5-FU-induced apoptosis in colon cancer cells. Ann
Surg Oncol. 16:761–771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Talmon G and Wang J: MicroRNA-587
antagonizes 5-FU-induced apoptosis and confers drug resistance by
regulating PPP2R1B expression in colorectal cancer. Cell Death Dis.
6:e18452015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Laurent-Puig P, Cayre A, Manceau G, Buc E,
Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, et al:
Analysis of PTEN, BRAF, and EGFR status in determining benefit from
cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin
Oncol. 27:5924–5930. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Noordhuis MG, Eijsink JJH, Ten Hoor KA,
Roossink F, Hollema H, Arts HJ, Pras E, Maduro JH, Reyners AK, de
Bock GH, et al: Expression of epidermal growth factor receptor
(EGFR) and activated EGFR predict poor response to (chemo)radiation
and survival in cervical cancer. Clin Cancer Res. 15:7389–7397.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao Y, Tan B, Kuo MS, Liu L and Breyer
MD: Abstract 5439: Metabolomic study of EGFR drug resistance
mechanisms. Cancer Res. 74:(Suppl 19). S5439. 2014. View Article : Google Scholar
|
|
9
|
Taylor TE, Furnari FB and Cavenee WK:
Targeting EGFR for treatment of glioblastoma: Molecular basis to
overcome resistance. Curr Cancer Drug Targets. 12:197–209. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Ji Z, Qiao C, Qi Y and Shi W:
Overexpression of ADAM9 promotes colon cancer cells invasion. J
Invest Surg. 26:127–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jia AY, Castillo-Martin M, Bonal DM,
Sánchez-Carbayo M, Silva JM and Cordon-Cardo C: MicroRNA-126
inhibits invasion in bladder cancer via regulation of ADAM9. Br J
Cancer. 110:2945–2954. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu R, Gu J, Jiang P, Zheng Y, Liu X,
Jiang X, Huang E, Xiong S, Xu F, Liu G, et al: DNMT1-microRNA126
epigenetic circuit contributes to esophageal squamous cell
carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res.
21:854–863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Blobel CP: ADAMs: Key components in EGFR
signalling and development. Nat Rev Mol Cell Biol. 6:32–43. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pham NA, Schwock J, Iakovlev V, Pond G,
Hedley DW and Tsao MS: Immunohistochemical analysis of changes in
signaling pathway activation downstream of growth factor receptors
in pancreatic duct cell carcinogenesis. BMC Cancer. 8:432008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Işeri OD, Kars MD, Arpaci F and Gündüz U:
Gene expression analysis of drug-resistant MCF-7 cells:
Implications for relation to extracellular matrix proteins. Cancer
Chemother Pharmacol. 65:447–455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kyula JN, Van Schaeybroeck S, Doherty J,
Fenning CS, Longley DB and Johnston PG: Chemotherapy-induced
activation of ADAM-17: A novel mechanism of drug resistance in
colorectal cancer. Clin Cancer Res. 16:3378–3389. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park SL, Cho TM, Won SY, Song JH, Noh DH,
Kim WJ and Moon SK: MicroRNA-20b inhibits the proliferation,
migration and invasion of bladder cancer EJ cells via the targeting
of cell cycle regulation and Sp-1-mediated MMP-2 expression. Oncol
Rep. 34:1605–1612. 2015.PubMed/NCBI
|
|
20
|
Sarver AL, French AJ, Borralho PM,
Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM,
Boardman LA, Cunningham JM, et al: Human colon cancer profiles show
differential microRNA expression depending on mismatch repair
status and are characteristic of undifferentiated proliferative
states. BMC Cancer. 9:401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Hu X, Miao X, Zhu K, Cui S, Meng
Q, Sun J and Wang T: MicroRNA-425-5p regulates chemoresistance in
colorectal cancer cells via regulation of Programmed Cell Death 10.
J Cell Mol Med. 20:360–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Giacchetti S, Itzhaki M, Gruia G, Adam R,
Zidani R, Kunstlinger F, Brienza S, Alafaci E, Bertheault-Cvitkovic
F, Jasmin C, et al: Long-term survival of patients with
unresectable colorectal cancer liver metastases following
infusional chemotherapy with 5-fluorouracil, leucovorin,
oxaliplatin and surgery. Ann Oncol. 10:663–669. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Linda Y, Narasimhaswamy B and David HB:
MicroRNA and colorectal cancer. Dig Liver Dis Off J Ital Soc
Gastroenterol Ital Assoc Study Liver. 44:66–70. 2012.
|
|
24
|
Gregersen LH, Jacobsen AB, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-145 targets YES and STAT1 in colon
cancer cells. PLoS One. 5:e88362010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Valeri N, Braconi C, Gasparini P, Murgia
C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat
F, et al: MicroRNA-135b promotes cancer progression by acting as a
downstream effector of oncogenic pathways in colon cancer. Cancer
Cell. 25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ahmed FE: miRNA as markers for the
diagnostic screening of colon cancer. Expert Rev Anticancer Ther.
14:463–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saleiban A, Faxälv L, Claesson K, Jönsson
JI and Osman A: miR-20b regulates expression of
proteinase-activated receptor-1 (PAR-1) thrombin receptor in
melanoma cells. Pigment Cell Melanoma Res. 27:431–441. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh
HJ, Lee JH, Nam SW and Lee EK: A long non-coding RNA snaR
contributes to 5-fluorouracil resistance in human colon cancer
cells. Mol Cells. 37:540–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W,
Jiang X, Zhang C and Qu J: MicroRNA-195 chemosensitizes colon
cancer cells to the chemotherapeutic drug doxorubicin by targeting
the first binding site of BCL2L2 mRNA. J Cell Physiol. 230:535–545.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mussnich P, Rosa R, Bianco R, Fusco A and
D'Angelo D: MiR-199a-5p and miR-375 affect colon cancer cell
sensitivity to cetuximab by targeting PHLPP1. Expert Opin Ther
Targets. 19:1017–1026. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
West CM, Joseph L and Bhana S: Epidermal
growth factor receptor-targeted therapy. Br J Radiol. 81:S36–S44.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moroni M, Veronese S, Benvenuti S,
Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, Gambacorta M,
Siena S and Bardelli A: Gene copy number for epidermal growth
factor receptor (EGFR) and clinical response to antiEGFR treatment
in colorectal cancer: A cohort study. Lancet Oncol. 6:279–286.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Patel BB, Sengupta R, Qazi S, Vachhani H,
Yu Y, Rishi AK and Majumdar AP: Curcumin enhances the effects of
5-fluorouracil and oxaliplatin in mediating growth inhibition of
colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer.
122:267–273. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Engel J, Richters A, Getlik M, Tomassi S,
Keul M, Termathe M, Lategahn J, Becker C, Mayer-Wrangowski S,
Grütter C, et al: Targeting drug resistance in EGFR with covalent
inhibitors: A structure-based design approach. J Med Chem.
58:6844–6863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hirao T, Nanba D, Tanaka M, Ishiguro H,
Kinugasa Y, Doki Y, Yano M, Matsuura N, Monden M and Higashiyama S:
Overexpression of ADAM9 enhances growth factor-mediated recycling
of E-cadherin in human colon cancer cell line HT29 cells. Exp Cell
Res. 312:331–339. 2006.PubMed/NCBI
|
|
36
|
Hamada S, Satoh K, Fujibuchi W, Hirota M,
Kanno A, Unno J, Masamune A, Kikuta K, Kume K and Shimosegawa T:
MiR-126 acts as a tumor suppressor in pancreatic cancer cells via
the regulation of ADAM9. Mol Cancer Res. 10:3–10. 2012. View Article : Google Scholar : PubMed/NCBI
|