Introduction
Osteosarcoma (OS) represents a type of highly
aggressive bone tumor prevalent in adolescents and is characterized
by composite genetic defects (1–4). The
failure in osteosarcoma therapy is mainly due to tumor recurrence
or lung metastasis (5–7). Therefore, identification of new
molecular targets and relative mechanism of metastasis is urgently
needed for the effective management of OS.
In clinical specimens from different stages of human
tumors, the early precursor lesions commonly express markers of an
activated DNA damage response (8).
DNA helicases have crucial roles in maintaining genome stability
and stable DNA replication in all organisms (9). Mammalian HELQ is a 3′-5′ DNA helicase
with strand displacement activity (10). It opens up the parental strands at a
blocked DNA replication fork and remodels nascent lagging strand
intermediates to facilitate the loading of subsequent factors
required for DNA damage processing or restart of DNA replication
(11). Adelman et al
uncovered a critical role for HELQ in germ cell maintenance and
tumor suppression in mammals, which attributed to its role in
replication-coupled DNA repair by interacting with RAD51 paralogue
(12). Recent studies have
identified single nucleotide polymorphisms at loci within or near
HELQ that are associated with increased risks for several different
cancers including upper aerodigestive tract cancers, ovarian
cancers, and head and neck cancers (13). However, whether HELQ is involved in
OS tumorigenesis and the molecular mechanisms of its tumorigenesis
have yet to be defined.
Chk1 is the principal direct effector of the DNA
damage and replication checkpoints (14). Inhibition of Chk1 promoted genomic
DNA damage and decreased homologous recombination repair. Studies
showed that defects in CHK1-RAD51 signaling result in defective
homologous recombination and chromosome instabilities, and Rad51
inactivation induced aberrant replication dynamics, consequently
leading to tumorigenesis (15,16).
HELQ has a role in promoting CHK1 activation and HELQ colocalizes
with Rad51 involved in the repair of damaged forks by homologous
recombination (11). It was also
reported that the Caenorhabditis elegans ortholog helq-1
plays a role in meiotic DSB repair by promoting postsynaptic RAD-51
filament disassembly (17). All
these findings suggested the association between HELQ, CHK1-RAD51
pathway in DNA repairing which may affect OS phenotype. In the
present study, we tested this hypothesis and found that HELQ may be
involved in OS cell malignant phenotype via activating the
CHK1-RAD51 signaling pathway.
Materials and methods
Cell lines
The human osteosarcoma cell lines U2-OS and 143B
were purchased from the cell bank of Type Culture Collection of
Chinese Academy of Sciences (Shanghai, China). The U2-OS cells were
cultured in 1640 medium (Gibco) and 143B maintained in Dulbecco's
modified Eagle's medium (DMEM) (Gibco), both supplemented with 10%
fetal bovine serum (FBS) with 100 U/ml penicillin and 100 U/ml
streptomycin. Cells were cultured at 37°C in 5% CO2.
RNA isolation and qPCR
Total RNA from U2-OS and 143B cells was extracted
using TRIzol (Invitrogen) method. HELQ expression level was
evaluated by quantificational real time-PCR, and GAPDH was used as
the endogenous reference genes. All amplifications were performed
in the final reaction mixture (20 µl). Primer sequences used were:
HELQ forward primer 5′-GAAGGTGTCACTATTGAACCTGG-3′, HELQ reverse
primer 5′-GAGGATGACTTCCAATCCCTTTC-3′; GAPDH forward primer
5′-GGAGCGAGATCCCTCCAAAAT-3′, GAPDH reverse primer
5′-GGCTGTTGTCATACTTCTCATGG-3′. The amplification reaction was
performed using StepOne Real-Time PCR System for 40 cycles.
Relative expression was calculated using the 2−∆∆Ct
method.
Lentivirus-Vector construction and
cell transfection
The Lentivirus-Vectors were prepared by Yingqi
Biotechnology Company (Wuhan, Hubei, China). The cells were
transfected with Lentivirus-Vectors of upregulating HELQ (Lv-HELQ),
Lentivirus-Vectors of downregulating HELQ (Lv-shHELQ), and negative
Lentivirus-Vectors (NC), respectively.
Migration assays
In brief, cells were grown to confluence in 6-well
tissue culture plastic dishes to a density of approximately
5×106 cells/well. The cells were wounded by drawing a
line with a rubber policeman (Fisher Scientific, Hampton, NH, USA)
through the center of the plate. Cultures were rinsed with PBS and
replaced with fresh quiescent medium alone or containing 10% FBS,
following which the cells were incubated at 37°C for 24 h.
Photographs were taken at 0 and 24 h, and the migrated distance was
measured by ImageJ (NCBI). The cell migration rate was obtained by
counting three fields per area and represented as the average of
six independent experiments done over multiple days.
Transwell invasion assays
Invasion of U2-OS and 143B cells was measured using
the BD BioCoat™ BD Matrigel™ Invasion Chamber (BD Biosciences,
Franklin Lakes, NJ, USA) according to the manufacturer's protocol.
The medium in the lower chamber contained 5% fetal calf serum which
acts as a source of chemoattractants (in the absence of FCS in the
upper chamber). Cells were suspended in serum-free medium and added
to the upper chambers at the same time. Cells that passed through
the Matrigel-coated membrane were stained with Diff-Quik (Sysmex,
Kobe, Japan) and photographed (x400). Photographs were taken at 24
h, and cell counting was measured by ImageJ (NCBI). The values for
invasion were obtained by counting three fields per membrane and
represented as the average of six independent experiments done over
multiple days.
CCK8 assay
Cell viability was evaluated by a non-radioactive
cell counting kit (CCK-8, TransGen) assay. U2-OS and 143B cells
were seeded out in 96-well (4000/well) tissue culture plates and
cultured for 24, 48 and 72 h, respectively. Then, 10 µl of CCK8
solution was added to each well, and the plates were incubated for
an additional 2 h at 37°C. Cell viability was measured as the
absorbance at 450 nm with a microplate reader. The mean optical
density (OD) values from triplicate wells for each treatment were
used as the index of cell viability.
Comet assay
The level of DNA damage was evaluated by Comet
assay. This assay was performed using a Comet assay kit (Trevigen,
Gaithersburg, MD, USA). All steps were processed according to the
described procedures (18).
Western blot analysis
Total protein from the OS cells was extracted using
RIPA lysis buffer containing 6 µg/ml PMSF. Protein concentration
was determined by Bradford assay. Equal amounts of protein were
electrophoresed by 8% SDS-PAGE and transferred onto a pure
Nitrocellulose blotting membrane (0.22 ml). Membranes were blocked
with 5% skim milk for 1 h at room temperature, then blocked with
primary antibodies (mouse anti-HELQ IgG, 1:200, Santa Cruz
Biotechnology, Inc., sc-81095, Shanghai, China; rabbit anti-CHK1
IgG, 1:1000, Abcam, ab47574, UK; rabbit anti-RAD51 IgG, 1:800;
Abcam, ab63801, UK) overnight at 4°C. Membranes were washed before
incubated with appropriate peroxidase-conjugated secondary
antibodies (anti-rabbit and anti-mouse, 1:2000). The immune
complexes were detected with pro-light HRP kit (Tiangen Biotech
Co., Ltd., Beijing, China). GAPDH (1:2000, Santa Cruz
Biotechnology, Inc.) protein expression was used as a normalization
control for protein loading. All experiments were repeated by six
times over multiple days.
Statistical analysis
Data are expressed as mean ± SD. One-way ANOVA was
used in multiple-sample analysis. A value of p<0.05 was
considered to indicate a statistically significant difference. All
analyses were performed with SPSS Version 13.0 (SPSS Inc, Chicago,
IL, USA).
Results
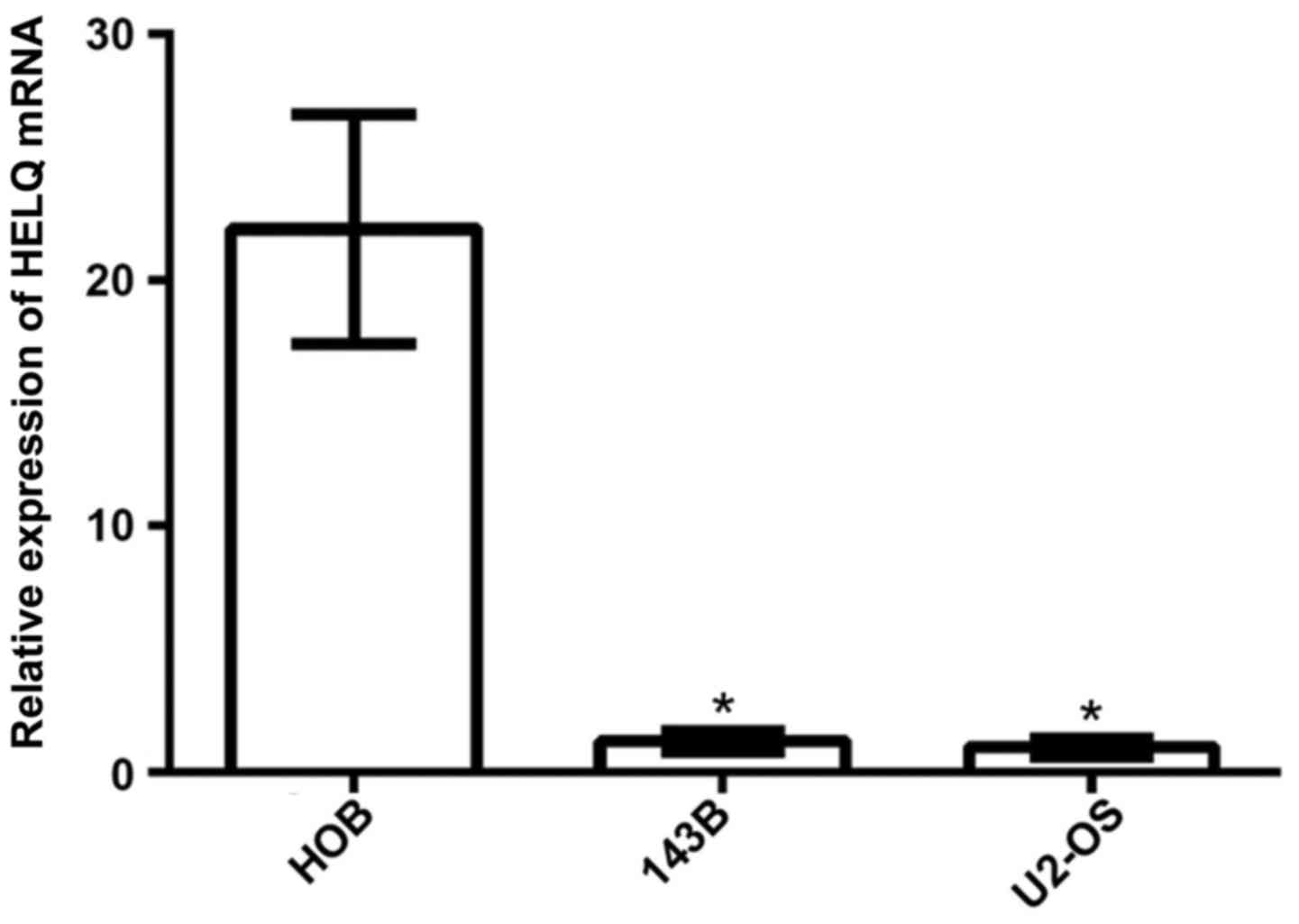
Expression of HELQ mRNA is lower in OS
cell lines than in osteoblast cell line
HELQ mRNA expression in OS cell lines (U2-OS and
143B cells) and osteoblast cell line (HOB cells) were detected by
real-time PCR analysis. The results showed that expression of HELQ
mRNA was significantly lower in U2-OS and 143B cells compared to
HOB cells (Fig. 1).
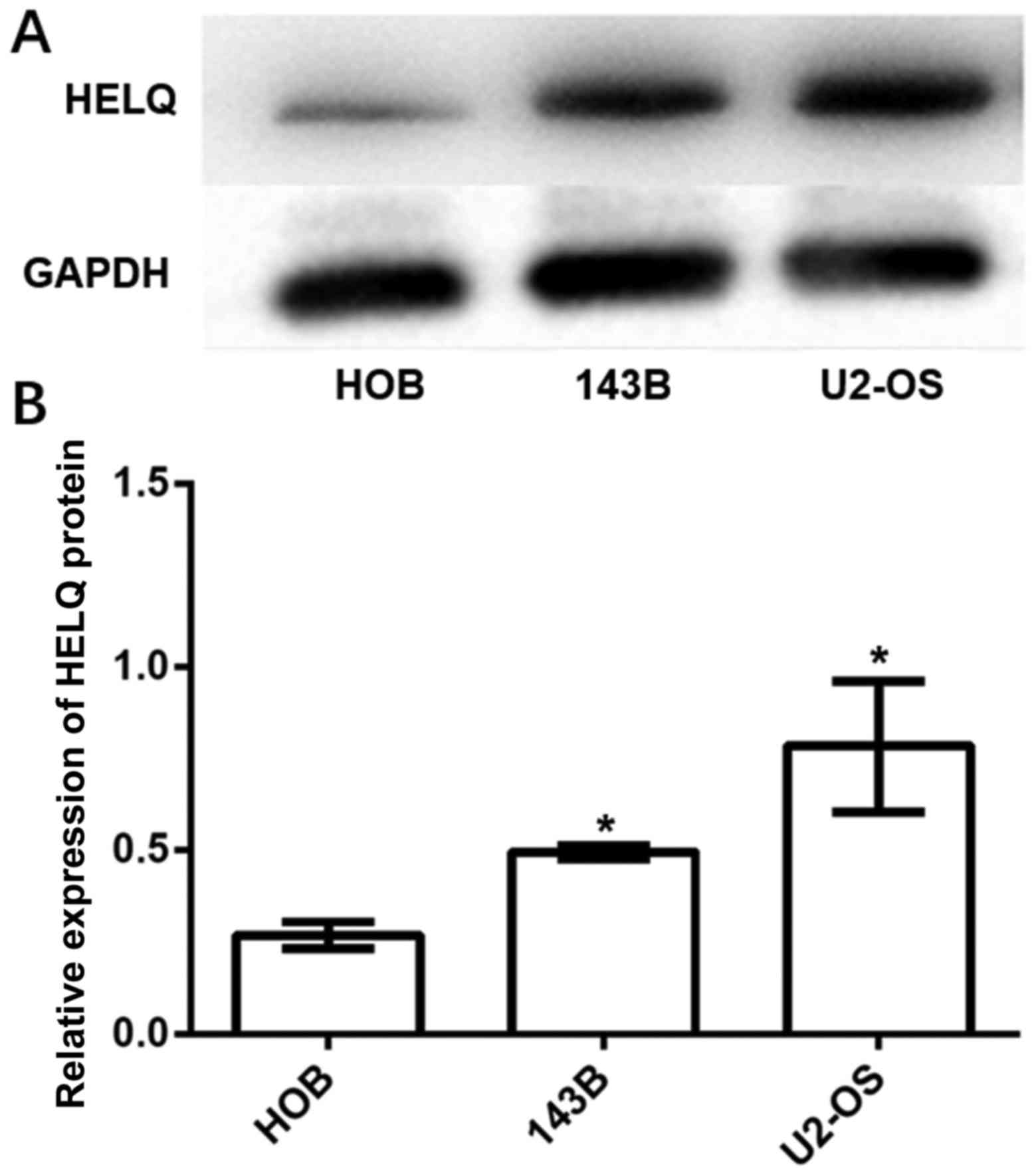
Expression of HELQ protein is higher
in OS cell lines than in osteoblast cell line
HELQ protein expression in OS cell lines (U2-OS and
143B cells) and osteoblast cell line (HOB cells) were detected by
western blot analysis. The results showed that expression of HELQ
protein was significantly higher in U2-OS and 143B cells compared
to HOB cells (Fig. 2).
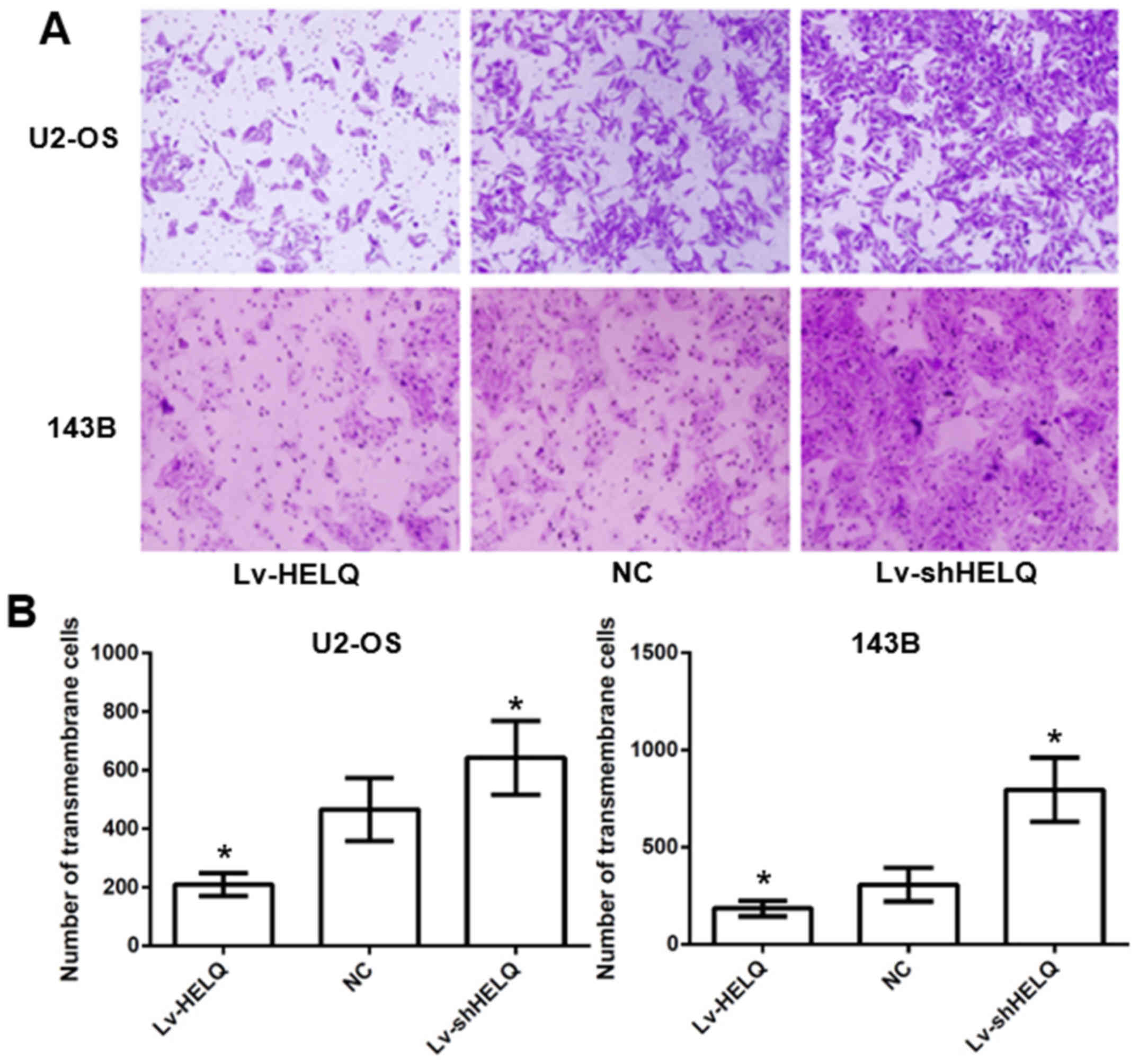
HELQ retrograde expression regulates
U2-OS and 143B cell invasion ability in vitro
Transwell invasion assays were performed to measure
the invasion of U2-OS and 143B cells. Fig. 3 showed decreased transmembrane cells
(U2-OS: 209.2±38.6 cells/membrane; 143B: 185.3±40.4 cells/membrane)
in HELQ-lentivirus-transfected cells and increased transmembrane
cells (U2-OS: 642.3±125.6 cells/membrane; 143B: 795.7±164.4
cells/membrane) in shRNA-HELQ-lentivirus-transfected cells,
compared with the negative control (U2-OS: 465.4±107.8
cells/membrane; 143B: 307.1±86.9 cells/membrane), indicating that
HELQ could inhibit OS cells invasion in vitro.
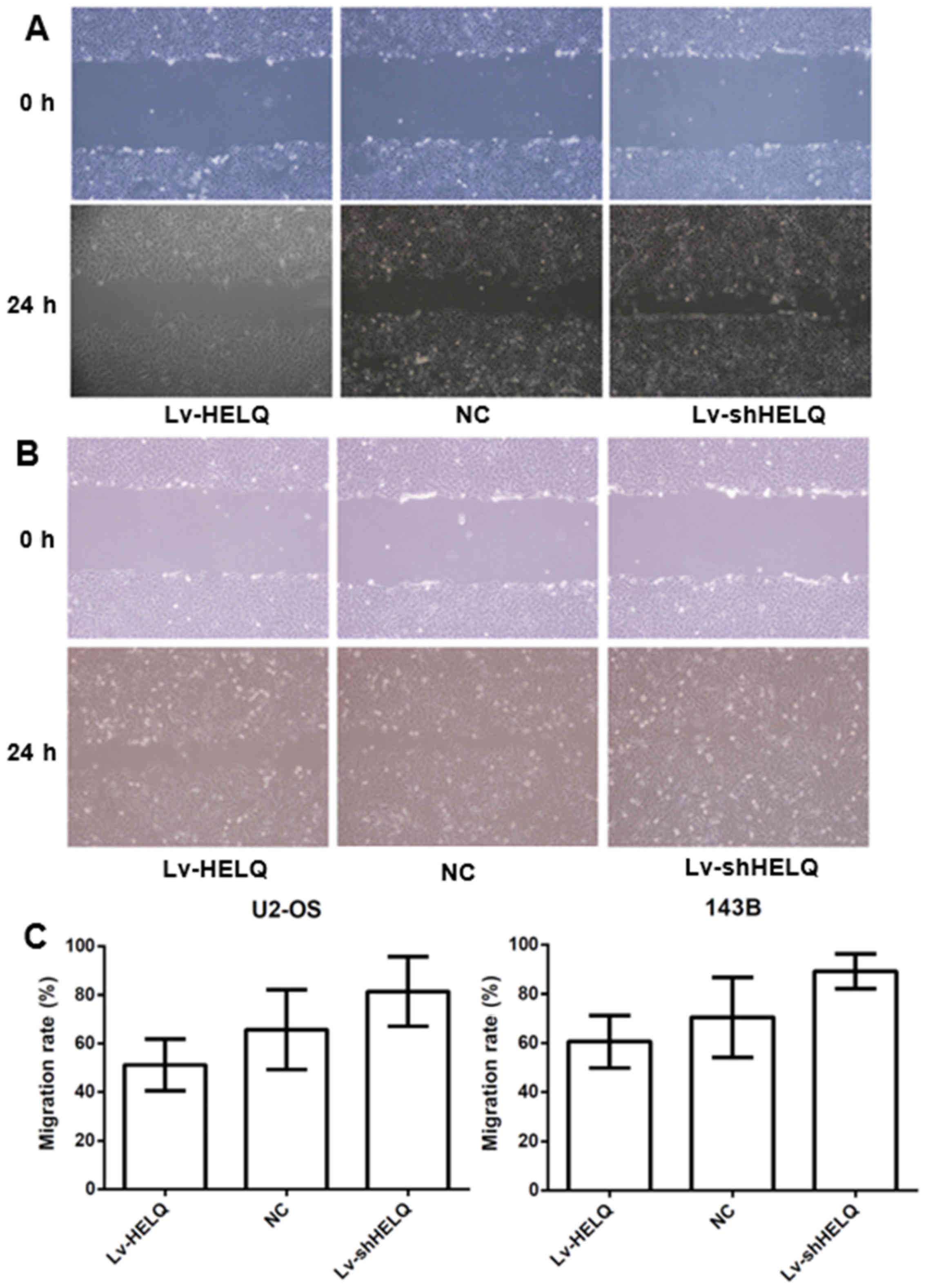
HELQ retrograde expression regulates
U2-OS and 143B cell migration ability in vitro
Wound healing assay revealed the inhibited migration
in HELQ-lentivirus-transfected cells (U2-OS: 51.2±10.6%, 143B:
60.6±10.7%) and enhanced migration in
shRNA-HELQ-lentivirus-transfected cells (U2-OS: 81.4±14.3%, 143B:
89.2±7.1%), compared with the negative control (U2-OS: 65.7±16.4%,
143B: 70.5±16.3%) (Fig. 4). The
results indicated that HELQ could inhibit OS cells migration.
HELQ retrograde expression regulates
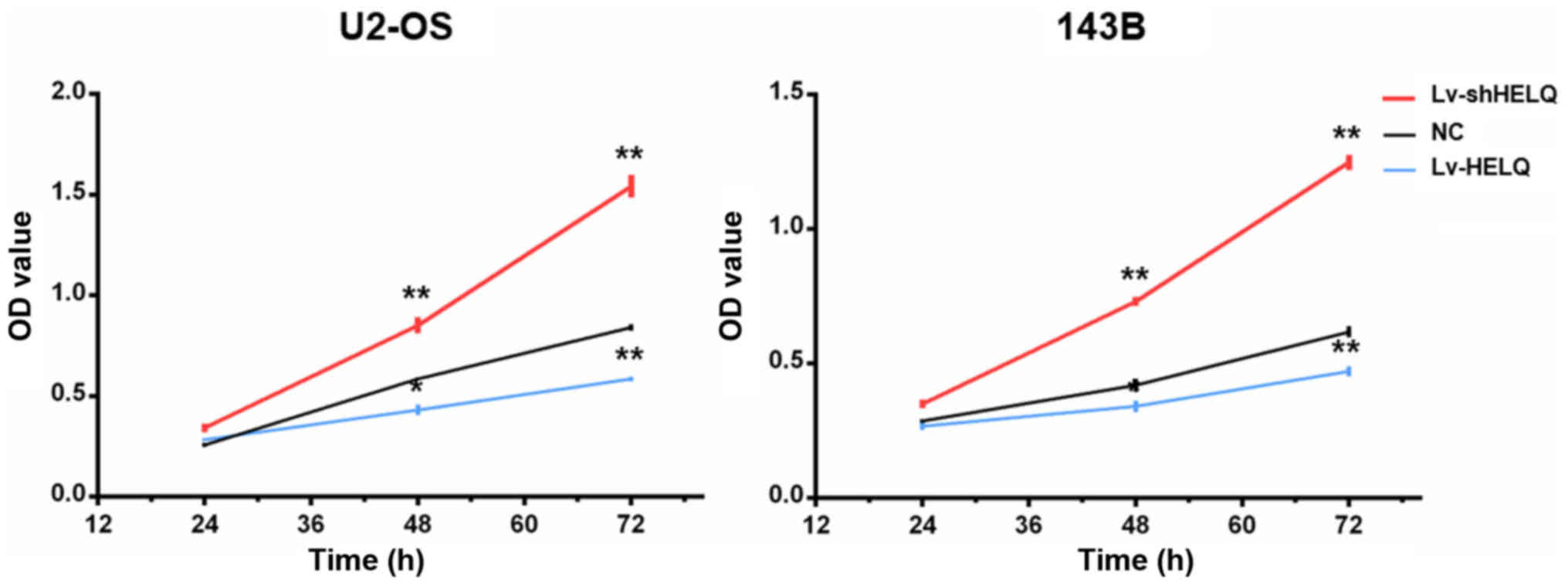
U2-OS and 143B cell proliferation
The CCK8 assay was used to assessed the roles of
HELQ in OS cell proliferation. Downregulating HELQ expression by
shRNA-lentivirus enhanced the proliferation abilities of U2-OS and
143B cells. Moreover, overexpression of HELQ with transfection of
HELQ-lentivirus significantly suppressed the proliferation of U2-OS
and 143B cells (Fig. 5). The
results suggested that activation of HELQ signaling inhibits OS
cell proliferation.
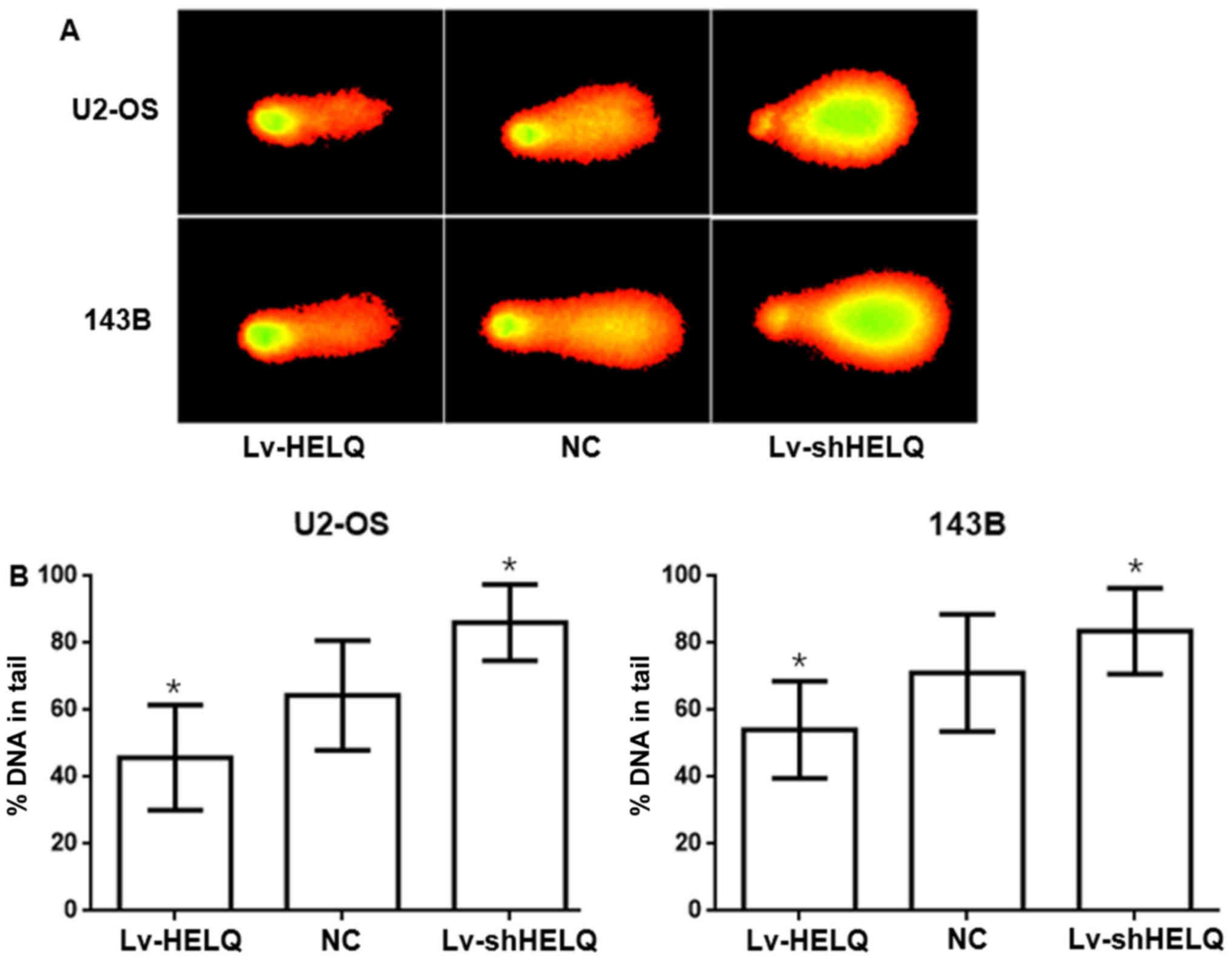
HELQ is critical to DNA repair
The comet assay, also known as the single cell gel
electrophoresis assay, is a very sensitive and rapid quantitative
technique used to detect DNA damage at the individual cell level
(19,20). As illustrated in Fig. 6, the DNA damage levels, measured by
%DNA in tail, were higher in the shRNA-HELQ-lentivirus group
(U2-OS: 86.1632±11.3856%, 143B: 83.5961±12.7842%) but lower in the
HELQ-lentivirus group (U2-OS: 45.8098±15.7096%, 143B:
54.1610±14.4469%) as compared to negative control (U2-OS: U2-OS:
64.4241±16.3866%, 143B: 71.0989±17.4967%). The results demonstrated
that HELQ promoted DNA repair after damage in OS cells.
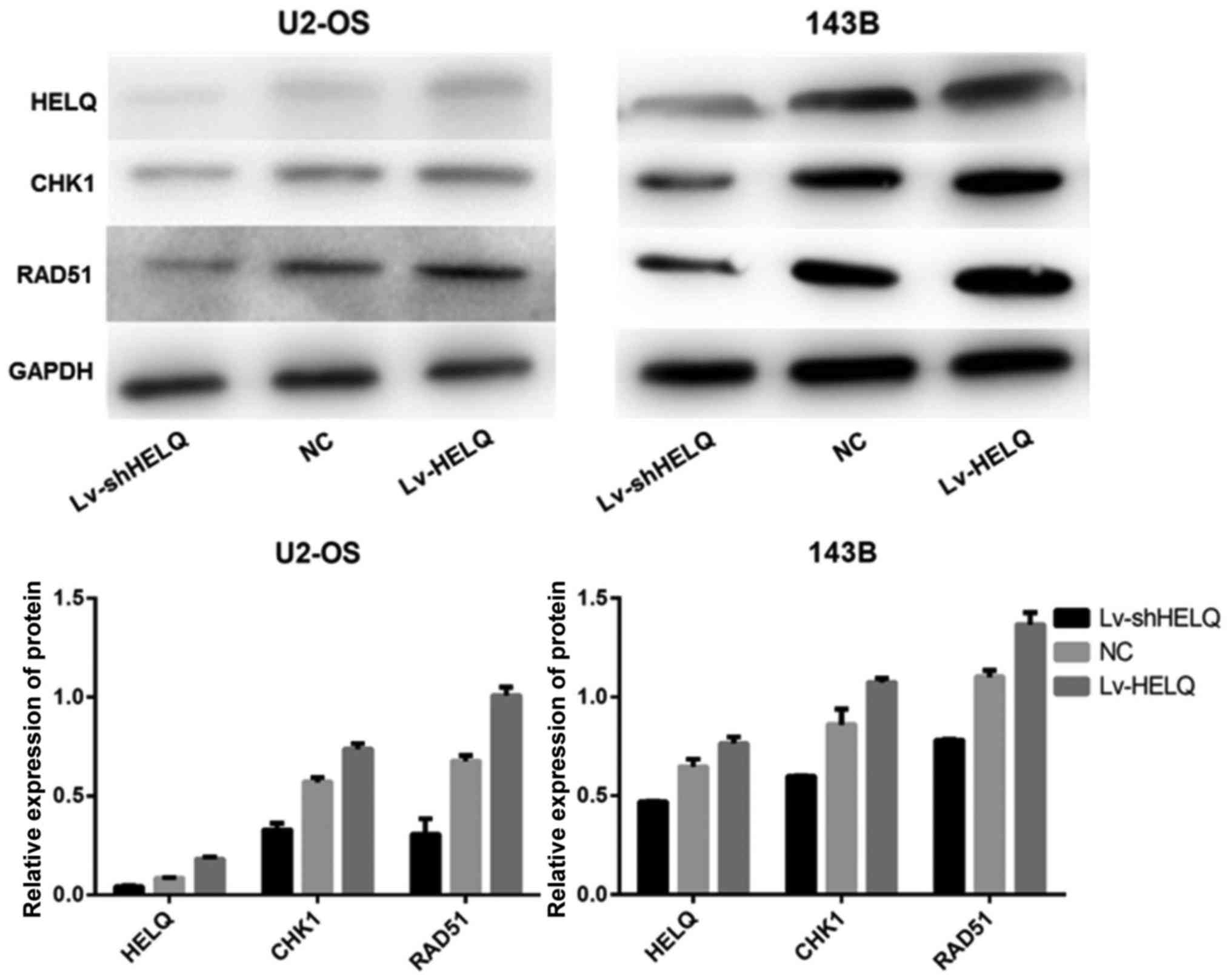
HELQ regulates the CHK1-RAD51
signaling pathway
To investigate the effect of HELQ on CHK1-RAD51
signaling pathway, the expression levels of HELQ, CHK1 and RAD51
proteins were measured with western blot analysis. Results indicate
that HELQ, CHK1 and RAD51 protein expression levels were higher in
cells transfected with HELQ-lentivirus and lower in cells
transfected with shRNA-HELQ-lentivirus, compare to the negative
control (Fig. 7). These
observations demonstrate that altered HELQ expression could impact
CHK1 and RAD51 protein expression in U2-OS and 143B cells,
suggesting that HELQ may regulate the CHK1-RAD51 pathway.
Discussion
HELQ family members contribute to the repair of
replication-blocking lesions such as DNA interstrand cross-links
(11). HELQ is a superfamily II DNA
helicase, conserved from archaea through to humans (21,22).
Studies have established that human HELQ is an ATP-dependent enzyme
that unwinds DNA with a 3′ to 5′ polarity (23,24).
HELQ helicase-deficient mice exhibit subfertility, germ cell
attrition, ICL sensitivity and tumor predisposition (12). Previous study reported that single
nucleotide polymorphisms at loci within or near HELQ that are
associated with increased risks for several different cancers
including upper aerodigestive tract cancers, ovarian cancers, head
and neck cancers (25–28). In the present study, our results
showed that the migration, invasion and proliferation were
significantly suppressed by activation of HELQ signaling in OS
cells, indicating that modulating HELQ could effectively reverse
the malignant phenotype of OS cells.
Modulation of CHK1 and RAD51 is also tightly
regulated in DNA repair and HELQ function. HELQ can promote CHK1
activation, CHK1 phosphorylation was significantly reduced in
HELQ−/− cells (10). The
Chk1 protein kinase is activated in response to damaged DNA and
stalled replication forks and acts as a central effector of the DNA
damage and replication checkpoint responses in vertebrate cells
(29). Supernumerary centrosomes in
human lymphoblastoid cells exposed to ionizing radiation were
eradicated by treatment with 2 mM caffeine or by the depletion of
Chk1, suggesting that Chk1 may be involved in promoting centrosome
amplification induced by DNA damage (30). CHK1 physically interacts with Rad51
to regulate homologous recombination and defects in Chk1-Rad51
signaling result in HR defective and chromosome instabilities,
consequently leading to tumorigenesis (14). Parplys et al have described a
role for RAD51 in driving genomic instability caused by impaired
replication and intra-S-phase mediated CHK1 signaling by studying
an inducible RAD51 overexpression model as well as ten breast
cancer cell lines (31). Our
results revealed that HELQ is involved in the DNA damage of
osteosarcoma via the CHK1-RAD51 signaling pathway.
In conclusion, our present study demonstrates a role
of HELQ signaling in repairing DNA damage and modulating OS cell
phenotype via targeting the CHK1-RAD51 pathway after tumorigenesis.
Targeting HELQ and CHK1-RAD51 pathway may be a potential strategy
for treating OS metastases. Further research is required to
identify the detailed roles of HELQ in osteosarcoma.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81260400), the
Science and Technology Program of Shenzhen (JCYJ20150402152130176),
the Technology Research Development and Creative Design Program of
Shenzhen, Nanshan District (2015002) and the Natural Science
Foundation of Jiangxi Province (no. 20142BAB205064).
References
|
1
|
Meyers PA, Schwartz CL, Krailo M,
Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W,
Gebhardt M, Goorin AM, et al: Osteosarcoma: A randomized,
prospective trial of the addition of ifosfamide and/or muramyl
tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J
Clin Oncol. 23:2004–2011. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bacci G, Forni C, Longhi A, Ferrari S,
Mercuri M, Bertoni F, Serra M, Briccoli A, Balladelli A and Picci
P: Local recurrence and local control of non-metastatic
osteosarcoma of the extremities: A 27-year experience in a single
institution. J Surg Oncol. 96:118–123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jawad MU, Cheung MC, Clarke J, Koniaris LG
and Scully SP: Osteosarcoma: Improvement in survival limited to
high-grade patients only. J Cancer Res Clin Oncol. 137:597–607.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohata N, Ito S, Yoshida A, Kunisada T,
Numoto K, Jitsumori Y, Kanzaki H, Ozaki T, Shimizu K and Ouchida M:
Highly frequent allelic loss of chromosome 6q16-23 in osteosarcoma:
Involvement of cyclin C in osteosarcoma. Int J Mol Med.
18:1153–1158. 2006.PubMed/NCBI
|
|
5
|
Mialou V, Philip T, Kalifa C, Perol D,
Gentet JC, Marec-Berard P, Pacquement H, Chastagner P, Defaschelles
AS and Hartmann O: Metastatic osteosarcoma at diagnosis: Prognostic
factors and long-term outcome - the French pediatric experience.
Cancer. 104:1100–1109. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hegyi M, Semsei AF, Jakab Z, Antal I, Kiss
J, Szendroi M, Csoka M and Kovacs G: Good prognosis of localized
osteosarcoma in young patients treated with limb-salvage surgery
and chemotherapy. Pediatr Blood Cancer. 57:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stokkel MP, Linthorst MF, Borm JJ,
Taminiau AH and Pauwels EK: A reassessment of bone scintigraphy and
commonly tested pretreatment biochemical parameters in newly
diagnosed osteosarcoma. J Cancer Res Clin Oncol. 128:393–399. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartkova J, Horejsí Z, Koed K, Krämer A,
Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et
al: DNA damage response as a candidate anti-cancer barrier in early
human tumorigenesis. Nature. 434:864–870. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu L and Hickson ID: DNA helicases
required for homologous recombination and repair of damaged
replication forks. Annu Rev Genet. 40:279–306. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takata K, Reh S, Tomida J, Person MD and
Wood RD: Human DNA helicase HELQ participates in DNA interstrand
crosslink tolerance with ATR and RAD51 paralogs. Nat Commun.
4:23382013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tafel AA, Wu L and McHugh PJ: Human HEL308
localizes to damaged replication forks and unwinds lagging strand
structures. J Biol Chem. 286:15832–15840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Adelman CA, Lolo RL, Birkbak NJ, Murina O,
Matsuzaki K, Horejsi Z, Parmar K, Borel V, Skehel JM, Stamp G, et
al: HELQ promotes RAD51 paralogue-dependent repair to avert germ
cell loss and tumorigenesis. Nature. 502:381–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stolk L, Perry JR, Chasman DI, He C,
Mangino M, Sulem P, Barbalic M, Broer L, Byrne EM, Ernst F, et al:
LifeLines Cohort Study: Meta-analyses identify 13 loci associated
with age at menopause and highlight DNA repair and immune pathways.
Nat Genet. 44:260–268. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smith J, Tho LM, Xu N and Gillespie DA:
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and
cancer. Adv Cancer Res. 108:73–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krajewska M, Fehrmann RS, Schoonen PM,
Labib S, de Vries EG, Franke L and van Vugt MA: ATR inhibition
preferentially targets homologous recombination-deficient tumor
cells. Oncogene. 34:3474–3481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jia Y, Song W, Zhang F, Yan J and Yang Q:
Akt1 inhibits homologous recombination in Brca1-deficient cells by
blocking the Chk1-Rad51 pathway. Oncogene. 32:1943–1949. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ward JD, Muzzini DM, Petalcorin MI,
Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F and
Boulton SJ: Overlapping mechanisms promote postsynaptic RAD-51
filament disassembly during meiotic double-strand break repair. Mol
Cell. 37:259–272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Neri M, Milazzo D, Ugolini D, Milic M,
Campolongo A, Pasqualetti P and Bonassi S: Worldwide interest in
the comet assay: A bibliometric study. Mutagenesis. 30:155–163.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashby J, Tinwell H, Lefevre PA and Browne
MA: The single cell gel electrophoresis assay for induced DNA
damage (comet assay): Measurement of tail length and moment.
Mutagenesis. 10:85–90. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu HF, Lai TY, Hsia TC, Tang YJ, Yang JS,
Chiang JH, Lu CC, Liu CM, Wang HL and Chung JG: Danthron induces
DNA damage and inhibits DNA repair gene expressions in GBM 8401
human brain glioblastoma multiforms cells. Neurochem Res.
35:1105–1110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Woodman IL and Bolt EL: Molecular biology
of Hel308 helicase in archaea. Biochem Soc Trans. 37:74–78. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marini F and Wood RD: A human DNA helicase
homologous to the DNA cross-link sensitivity protein Mus308. J Biol
Chem. 277:8716–8723. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Muzzini DM, Plevani P, Boulton SJ, Cassata
G and Marini F: Caenorhabditis elegans POLQ-1 and HEL-308 function
in two distinct DNA interstrand cross-link repair pathways. DNA
Repair (Amst). 7:941–950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guy CP and Bolt EL: Archaeal Hel308
helicase targets replication forks in vivo and in vitro and unwinds
lagging strands. Nucleic Acids Res. 33:3678–3690. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li WQ, Hu N, Hyland PL, Gao Y, Wang ZM, Yu
K, Su H, Wang CY, Wang LM, Chanock SJ, et al: Genetic variants in
DNA repair pathway genes and risk of esophageal squamous cell
carcinoma and gastric adenocarcinoma in a Chinese population.
Carcinogenesis. 34:1536–1542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao Y, He Y, Xu J, Xu L, Du J, Zhu C, Gu
H, Ma H, Hu Z, Jin G, et al: Genetic variants at 4q21, 4q23 and
12q24 are associated with esophageal squamous cell carcinoma risk
in a Chinese population. Hum Genet. 132:649–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liang C, Marsit CJ, Houseman EA, Butler R,
Nelson HH, McClean MD and Kelsey KT: Gene-environment interactions
of novel variants associated with head and neck cancer. Head Neck.
34:1111–1118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McKay JD, Truong T, Gaborieau V, Chabrier
A, Chuang SC, Byrnes G, Zaridze D, Shangina O, Szeszenia-Dabrowska
N, Lissowska J, et al: A genome-wide association study of upper
aerodigestive tract cancers conducted within the INHANCE
consortium. PLoS Genet. 7:e10013332011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Montano R, Thompson R, Chung I, Hou H,
Khan N and Eastman A: Sensitization of human cancer cells to
gemcitabine by the Chk1 inhibitor MK-8776: Cell cycle perturbation
and impact of administration schedule in vitro and in vivo. BMC
Cancer. 13:6042013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bourke E, Dodson H, Merdes A, Cuffe L,
Zachos G, Walker M, Gillespie D and Morrison CG: DNA damage induces
Chk1-dependent centrosome amplification. EMBO Rep. 8:603–609. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Parplys AC, Seelbach JI, Becker S, Behr M,
Wrona A, Jend C, Mansour WY, Joosse SA, Stuerzbecher HW, Pospiech
H, et al: High levels of RAD51 perturb DNA replication elongation
and cause unscheduled origin firing due to impaired CHK1
activation. Cell Cycle. 14:3190–3202. 2015. View Article : Google Scholar : PubMed/NCBI
|