Introduction
Gliomas are the most malignant and common type of
brain tumors (1). Despite
considerable advances in surgery, radiation, and chemotherapy
(temozolomide, TMZ), the prognosis for glioma has not significantly
improved (2). With a median
survival time of 12–15 months, fewer than 3% of glioma patients
live longer than 5 years after diagnosis (3–5).
Because of the restricted anatomical location and absence of
metastases outside the central nervous system (CNS), targeted gene
therapy may provide a promising strategy to treat gliomas (6). Therefore, identification of target
genes that drive tumorigenicity is essential for developing
targeted therapy for glioma (7–10). One
promising family for gene targeting is the Forkhead-box (FOX)
family. The FOX family consists of at least 43 transcription
factors that regulate the expression of genes that support cell
growth, proliferation and differentiation (11). Many FOX genes function as oncogenes.
For example, FOXA1 is amplified and overexpressed in esophageal and
lung adenocarcinomas (12). FOXO1
fused to PAX3 or PAX7 serve as prognostic indicators in alveolar
rhabdomyosarcoma (13–15).
FOXD1 (also known as FREAC-4) is highly expressed in
the kidney and controls cellularity in the renal capsule, a
structure required for normal renal development (16–19).
FOXD1 is also expressed in the brain and the retina and is
necessary for normal development of the retina and optic chiasm
(20,21). FOXD1 has also been implicated as an
oncogene, as it promotes breast cancer proliferation and
chemotherapeutic drug resistance by targeting p27 (22). However, the role of FOXD1 in
tumorigenesis and progression, particularly in glioma, is still
limited.
In the present study, we first revealed the role of
FOXD1 in glioma tumorigenesis and progression. We found that FOXD1
expression is upregulated and directly correlated with the glioma
grade. Using FOXD1-siRNA, we determined that silencing FOXD1
expression inhibits cell proliferation and migration. Our results
suggest that FOXD1 may be a potential therapy target for patients
with glioma.
Materials and methods
Cell culture and transfection
Human glioma cell lines U87 and U251 were cultured
in DMEM (C11995500BT, Gibco) supplemented with 10% fetal bovine
serum (FBS, 10099-141, Gibco). Cells were cultured at 37°C in a
humidified atmosphere of 5% CO2. Trypsin-EDTA (0.25%)
(25200072, Gibco) was used to detach the cells from the flask. The
FOXD1-siRNA and negative control siRNA (NC-siRNA) were purchased
from Santa-Cruz Biotechnology (sc-60649; sc-637007) and Sigma.
Cells were transfected using Lipofectamine® RNAiMAX
reagent (13778150, Invitrogen) according to the protocol.
Glioma tumor specimens
All patients undergoing surgical treatment at the
Hunan Cancer Hospital (Changsha, Hunan, China) for primary brain
cancers between 2007 and 2013 were invited to participate in this
institutional review board-approved study. Seventy patients
diagnosed with low grade glioma (LGG, n=36) or high grade glioma
(HGG, n=34) tumors were included in the study. Tissue samples were
collected from glioma patient tumors. Normal brain tissue (n=20)
was also obtained from patient with brain injury. The tissue
samples were flash frozen in liquid nitrogen immediately after
resection and stored at −80°C for future processing.
RNA extraction and quantitative
real-time PCR
Total RNA was extracted by TRIzol reagent according
to the manufacturer's protocol. Two micrograms of RNA was
reverse-transcribed into cDNAs using the Primescript RT reagent kit
with gDNA Eraser (Takara Bio Inc., Japan). The PCR system was
performed using SYBR Premix DimerEraser kit (Takara Bio Inc). The
reactions were cycled 40 times [95°C, 30 sec, (95°C, 5 sec; 55°C,
30 sec; and 72°C, 30 sec)] with fluorescence measurements. A
melting curve was performed at the end of amplification cycles to
verify the specificity of the PCR products. Primers used for
real-time PCR were as follows: FOXD1, F: AAGAACCCGCTGGTGAAG; R:
GTCCAGTAGTTGCCCTTGC. GAPDH, F: GAGTCAACGGATTTGGTCGT; R:
TTGATTTTGGAGGGATCTCG. All of the determinations were performed in
duplicate. The relative expression of FOXD1 mRNA was normalized to
the expression level of GAPDH mRNA using the 2−∆Ct
method.
Western blotting
Total cellular protein was extracted with RIPA.
SDS-PAGE was performed with 50 µg total cellular proteins using 10%
gradient Tris-glycine gels. Primary antibodies used included
anti-FOXD1 (WH0002297M1, Sigma-Aldrich LLC, USA), and anti-GAPDH
(G8795, Sigma-Aldrich LLC). The bound antibodies were visualized
using an enhanced chemiluminescence reagent (RPN2232, GE
Healthcare, UK) and quantified by densitometry using ChemiDoc XRS+
image analyzer (Bio-Rad, USA). Densitometric analyses of bands were
adjusted with GAPDH as loading control. Triplicate experiments with
triplicate samples were performed.
MTS assay
Cells were collected in the logarithmic phase of
growth and seeded into 96-well culture plates (1,000 cells/well).
The cells were incubated in 100 µl MTS reagent (1:9) for 1 h. The
absorbance values were measured at 490 nm by the
BioTek®Eon (Synergy™, HT, USA). In the absence of cells,
background absorbance of the medium was subtracted. Each assay was
performed in triplicate.
Colony formation assay
U251 cells were collected in the logarithmic phase
of growth and seeded in triplicate into 6-well plates at a density
of 1,000 cells/well. Cells were cultured for 12 days at 37°C in an
incubator with a 5% CO2 atmosphere. The cells were then
fixed with 4% paraformaldehyde for 30 min and stained with Giemsa
(C0121, Beyotime, China) for 20 min. After washing with PBS several
times, the cells were photographed with a camera (Canon, Japan) and
the cell colonies were counted.
Wound healing assay
Cells (2×105 cells/well.) were seeded
onto a 6-well plate overnight. The confluent monolayers were
scratched using sterile pipette tips and washed with
phosphate-buffered saline (PBS) 3 times to remove detached cells.
The cells were then transfected with siRNA for 36 h. Photographs of
the wounded areas were obtained using a Leica DMI3000 B inverted
microscope (Leica, German). The migration rate was calculated as
the ratio of the width of the scratch that remained cell-free after
migration to the initial average width of the scratch. The size of
the wounded areas was quantified using ImageJ Version 1.41o
software (National Institutes of Health).
Immunofluorescent staining
Glioma cells were seeded into 6-well plates and
cultured with siRNA for 48 h. The cells were then fixed with 4%
paraformaldehyde for 10 min at room temperature, washed and stained
with Hoechst 33258 (C0003, Beyotime) for 5 min. Cells were observed
under a Leica DMI3000 B inverted microscope equipped with ebq100-04
(Leica, Germany).
Statistical analysis
SPSS16.0 software was used for general statistics
analyses. Comparisons between two experimental groups were
performed using Student's t-test. Survival rate was calculated
using the Kaplan-Meier method with the log-rank test applied for
comparison. All tests performed were two sided and the criterion
for statistical significance was p<0.05.
Results
High FOXD1 expression in glioma
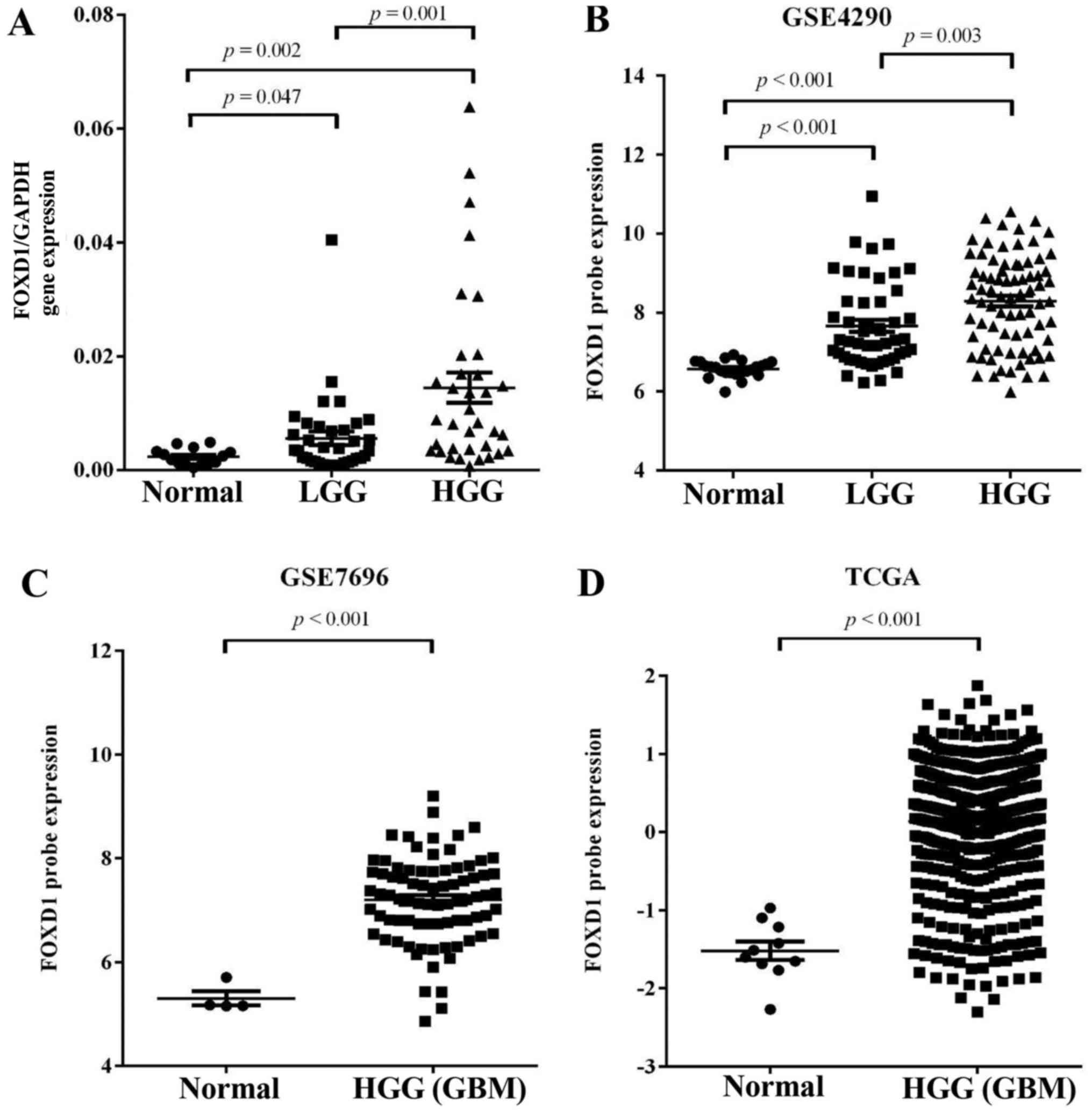
The expression pattern of FOXD1 was evaluated in 70
glioma tissues and 20 normal brain tissues using quantitative
real-time PCR (Q-PCR). FOXD1 expression was significantly higher in
glioma tissues (LGG, p=0.047; HGG, p=0.002) compared with normal
brain samples. FOXD1 expression was also higher in high grade
glioma compared with low grade glioma (p=0.001, Fig. 1A). Furthermore, the association
between FOXD1 expression and clinicopathological parameters was
analyzed in the present study. No significant association was
observed between FOXD1 expression and patient age or gender
(Table I). To further validate
these results, we analyzed expression of FOXD1 in expression
profiles (GSE4290 and GSE7696) acquired from the Gene Expression
Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database, and glioma
datasets acquired from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/). We found that
expression of upregulated FOXD1 (>2-fold upregulated) expression
was significantly higher in malignant gliomas compared to lower
grade gliomas and non-tumor brain tissue and directly correlated
with the glioma grade in GSE4290 dataset (Fig. 1B). Analysis of the GSE7696 data and
TCGA revealed that FOXD1 have >2-fold upregulation at the
transcription level and were greatly increased in malignant gliomas
when compared to non-tumor brain tissue (Fig. 1C and D). These results suggest that
increased FOXD1 expression correlates with glioma tumor grade.
 | Table I.Correlation between FOXD1 expression
and glioma clinicopathological features in 70 patients. |
Table I.
Correlation between FOXD1 expression
and glioma clinicopathological features in 70 patients.
|
|
|
| FOXD1 expression
levels |
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | N % | High expression | Low expression | Ratio High/low | p-value |
|---|
| Gender |
|
|
|
|
|
| Male | 48 (68.57) | 14 | 35 | 0.4 | 0.092 |
|
Female | 22 (31.43) | 10 | 12 | 0.833 |
|
| Age years |
|
|
|
|
|
|
<45 | 44 (62.85) | 13 | 31 | 0.419 | 0.056 |
| ≥45 | 26 (37.14) | 11 | 15 | 0.733 |
|
| Grade |
|
|
|
|
|
| Low
(I+II) | 36 (51.42) | 6 | 30 | 0.2 | 0.002 |
| High
(III+IV) | 34 (48.58) | 18 | 16 | 1.125 |
|
Prognostic value of FOXD1 expression
in gliomas
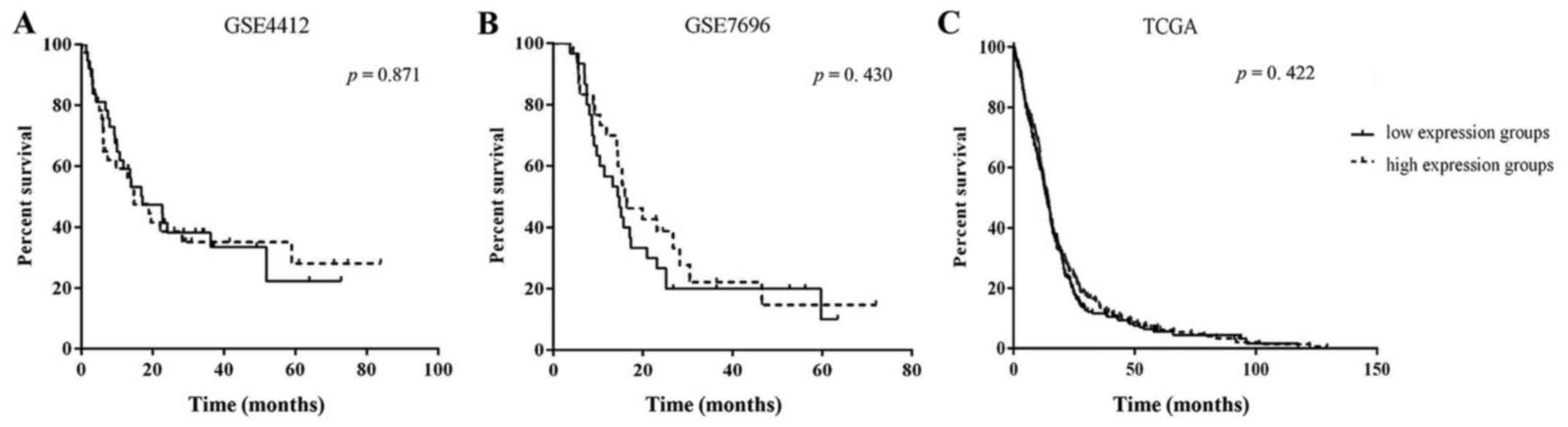
To determine the association between FOXD1
expression and clinical outcome, Kaplan-Meier analysis was
conducted for expression profiles (GSE4290 and GSE7696) and the
TCGA data. The analysis showed no significant difference in outcome
between patients with high FOXD1 expressing and low FOXD1
expressing gliomas for both gene expression profiles and the TCGA
datasets (Fig. 2). The clinical
outcome of those patients with high FOXD1 expression was observed
to be better than those with a low FOXD1 expression. Even so, FOXD1
expression may not serve as a prognostic factor of glioma
patients.
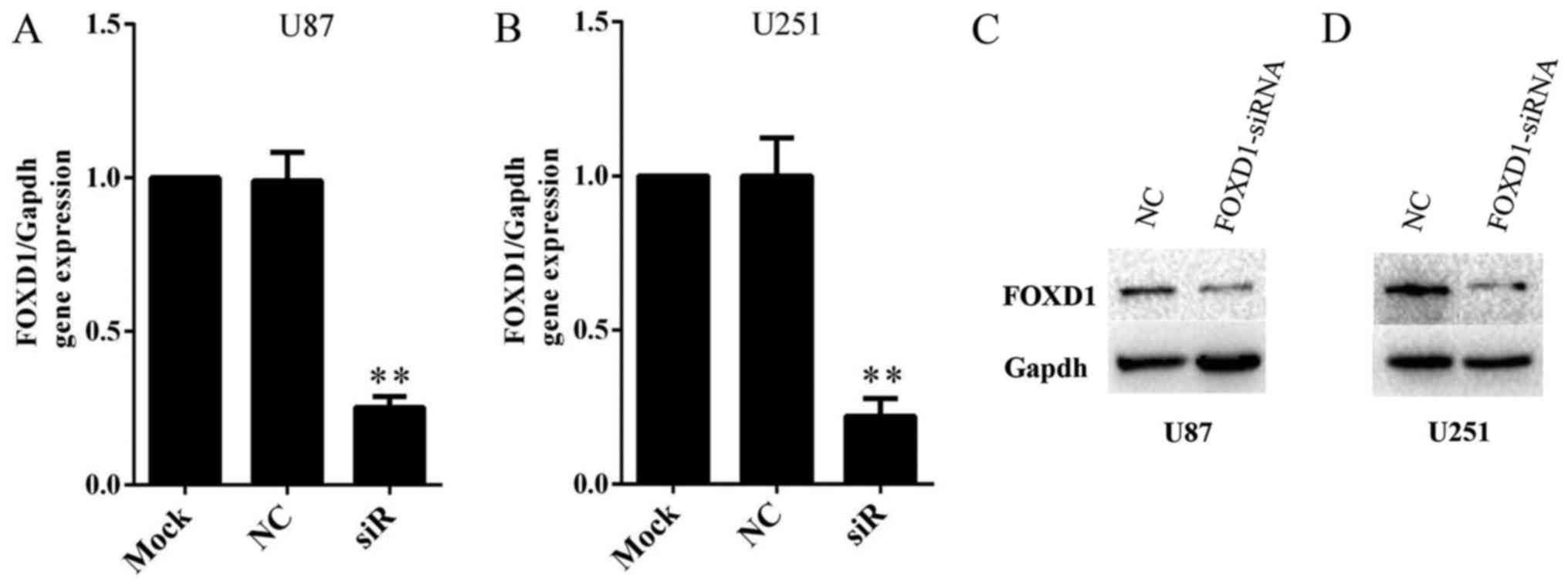
SiRNA inhibits FOXD1 expression
efficiently in human glioma cell lines
In order to investigate the role of FOXD1 in glioma,
we knockdown expression of FOXD1 using a FOXD1-siRNA. Malignant
human glioma cell lines U87 and U251 were treated with FOXD1-siRNA
or NC-siRNA. Treatment with FOXD1-siRNA significantly reduced FOXD1
mRNA expression by ~80% and nearly depleted protein expression in
U87 (Fig. 3A and C) and U251
(Fig. 3B and D) cells. Therefore,
the present results demonstrated that a highly efficient knockdown
of FOXD1 expression at the mRNA and protein levels was achieved
after transfection.
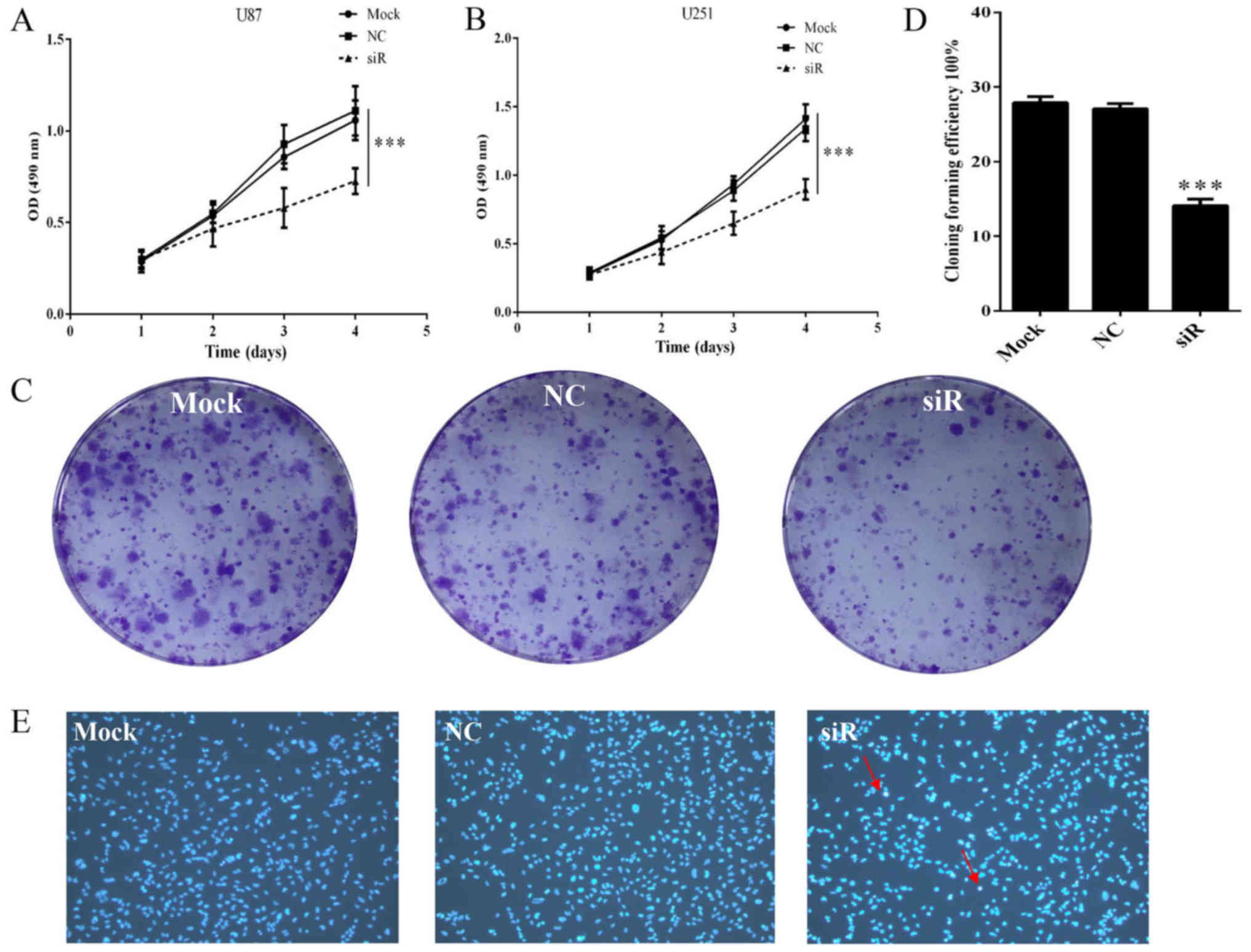
Knockdown of FOXD1 suppresses
proliferation and promotes apoptosis in glioma cells
To investigate the effect of FOXD1 silencing on
glioma cell proliferation, U87 and U251 cells transfected with
FOXD1-siRNA or NC-siRNA were analyzed by MTS assay. Proliferation
of U87 cells treated with FOXD1-siRNA began to decrease 72 h after
treatment. On day 4, proliferation of cells treated with
FOXD1-siRNA was significantly reduced compared to those treated
with NC-siRNA or non-treated cells (p<0.001; Fig. 4A). These results are consistent with
U251 cells (p<0.001; Fig. 4B).
Moreover, no difference was seen between NC-siRNA treated cells and
Mock cells. The relatively long-term effects of FOXD1 knockdown on
glioma cell proliferation were also examined using a colony
formation assay. As shown in Fig. 4C
and D, silencing of FOXD1 expression in U251 cells
substantially reduced colony formation (p<0.001).
In addition to unlimited proliferation and elevated
clonogenic capacity, evading apoptosis is also essential for tumor
initiation and development. To determine whether the reduction in
cell number following FOXD1 knockdown is due in part to cell
apoptosis, we treated U251 cells with FOXD1-siRNA or NC-siRNA and
stained them with Hoechst 33258 to evaluate cell viability.
Apoptotic bodies containing nuclear fragments were generated in
transfected FOXD1-siRNA group (Fig.
4E), suggesting that FOXD1 knockdown may induce cell apoptosis
in glioma cells.
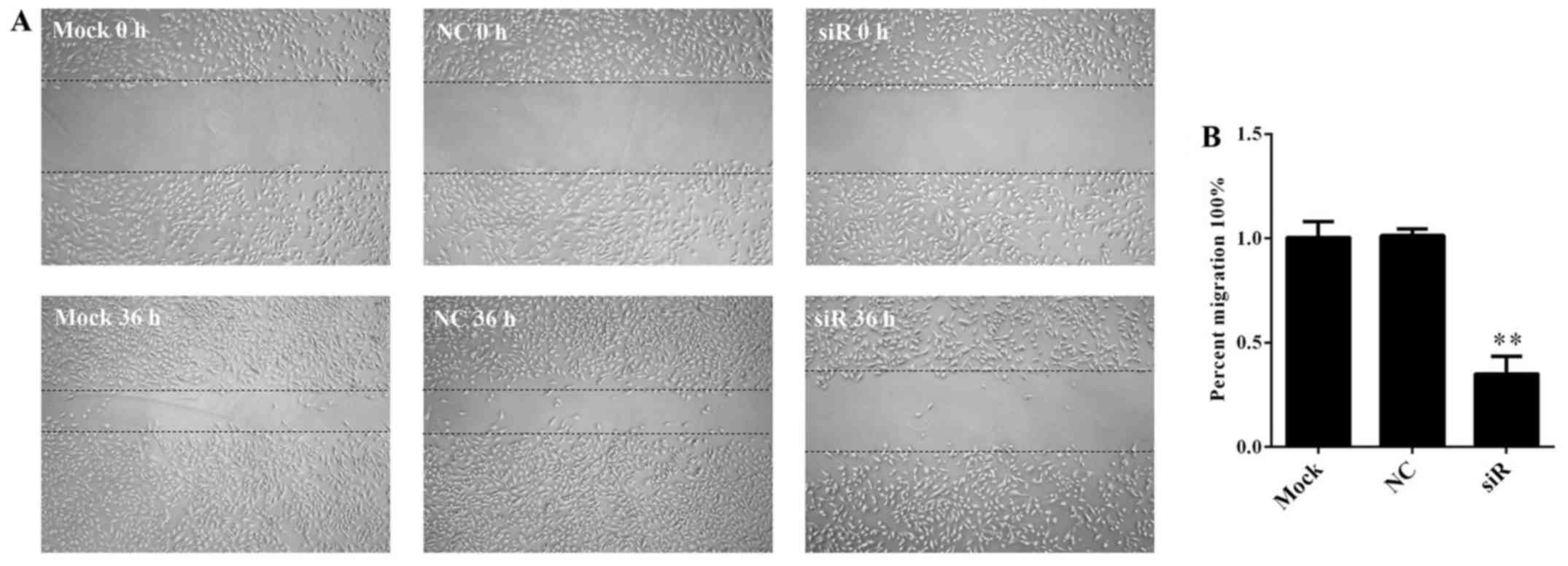
Silencing effect of FOXD1 on migration
in U251 cells
We investigated the effect of FOXD1 silencing on
cell migration. FOXD1 knockdown markedly reduced glioma cell
migration 36 h after transfection with the FOXD1-siRNA (Fig. 5A). Migration rates were
significantly lower for FOXD1-siRNA treated cells compared to
non-treated and NC-siRNA treated cells (p=0.002) (Fig. 5B). No significant differences were
found between Mock and NC-siRNA treated cells. These results
suggest that FOXD1 contributes to glioma cell migration.
Discussion
Despite advances in standard treatments of glioma,
little progress has been made in patient outcome. This is due in
part to the non-specific nature of these treatments. Thus,
alternative therapies that specifically target tumor-specific genes
may provide a promising alternative treatment to the current
standards of care. This study identified FOXD1 as a potential
target for the development of tumor targeted therapies.
We show that FOXD1 is upregulated in glioma tissues
compared to normal brain tissues and that silencing FOXD1
expression inhibits glioma cell proliferation and migration rates.
Blocking FOXD1 expression also generated apoptotic bodies
containing nuclear fragments suggesting that FOXD1 promotes cell
death. Taken together, the results suggest that FOXD1 functions as
an oncogene in glioma.
Forkhead box (FOX) proteins, an evolutionarily
conserved family of transcriptional regulators, mediate a wide
range of biological processes, such as proliferation, metabolism,
differentiation, apoptosis, and migration (23–27),
and take part in the onset and progression of tumors (28). Some studies show that FOXD1 is
related to organization development (19,29),
cell reprogramming (30), cell
differentiation (31). These
results are consistent with previous studies that have shown that
FOX transcription factors promote cancer cell migration and
proliferation.
Our study also parallels the previous studies that
found FOXD1 promotes cell growth. In 2015, Zhao et al
(22) showed that FOXD1 promotes
the cell cycle progression by inducing G1→S transition, suggesting
that FOXD1 induces G1→S transition by down-regulation of p27
expression. Further investigation into the mechanisms underlying
FOXD1 is needed to determine how FOXD1 supports cell proliferation
and other oncogenic activities in glioma.
In order to confirm the relationship between FOXD1
and its prognosis, we collected the datasets GSE4412, GSE7696 and
TCGA. Kaplan-Meier analysis showed no significant differences in
GSE4412 or GSE7696 between the high FOXD1 expression and low FOXD1
expression groups in patients with glioma, and it is the same as in
TCGA data. FOXD1 expression may not serve as a prognostic factor of
glioma patients.
In conclusion, our results show that FOXD1
expression is highly expressed in malignant gliomas and directly
correlates with glioma grade. Kaplan-Meier analysis showed FOXD1
expression may not serve as a prognostic factor of glioma patients.
However, reduced expression of FOXD1 inhibits glioma cell
proliferation, cell survival and migration. These findings
implicate FOXD1 as an oncogene in glioma that could potentially
serve as a therapeutic target for glioma treatment.
Acknowledgements
This study was supported by the National High-tech
R&D Program of China (863 Program) (2012AA02A517), National
Natural Science Foundation of China (81373490, 81573508, 81573463),
and Hunan Provincial Science and Technology Plan of China
(2015TP1043), and Open Foundation of Innovative Platform in
University of Hunan Province of China (2015–14).
References
|
1
|
Schucht P, Beck J, Seidel K and Raabe A:
Extending resection and preserving function: Modern concepts of
glioma surgery. Swiss Med Wkly. 145:w140822015.PubMed/NCBI
|
|
2
|
Haque A, Banik NL and Ray SK: Emerging
role of combination of all-trans retinoic acid and interferon-gamma
as chemoimmunotherapy in the management of human glioblastoma.
Neurochem Res. 32:2203–2209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohgaki H: Epidemiology of brain tumors.
Methods Mol Biol. 472:323–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: European Organisation for Research and Treatment of
Cancer Brain Tumor and Radiotherapy Groups; National Cancer
Institute of Canada Clinical Trials Group: Radiotherapy plus
concomitant and adjuvant temozolomide for glioblastoma. N Engl J
Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pulkkanen KJ and Yla-Herttuala S: Gene
therapy for malignant glioma: Current clinical status. Mol Ther.
12:585–598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weigel D, Jürgens G, Küttner F, Seifert E
and Jäckle H: The homeotic gene fork head encodes a nuclear protein
and is expressed in the terminal regions of the Drosophila embryo.
Cell. 57:645–658. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lai E, Prezioso VR, Smith E, Litvin O,
Costa RH and Darnell JE Jr: HNF-3A, a hepatocyte-enriched
transcription factor of novel structure is regulated
transcriptionally. Genes Dev. 4:1427–1436. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lai E, Prezioso VR, Tao WF, Chen WS and
Darnell JE Jr: Hepatocyte nuclear factor 3 alpha belongs to a gene
family in mammals that is homologous to the Drosophila homeotic
gene fork head. Genes Dev. 5:416–427. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaestner KH, Knochel W and Martinez DE:
Unified nomenclature for the winged helix/forkhead transcription
factors. Genes Dev. 14:142–146. 2000.PubMed/NCBI
|
|
11
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
12
|
Lin L, Miller CT, Contreras JI, Prescott
MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et
al: The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1),
on chromosome band 14q13 is amplified and overexpressed in
esophageal and lung adenocarcinomas. Cancer Res. 62:5273–5279.
2002.PubMed/NCBI
|
|
13
|
Sorensen PH, Lynch JC, Qualman SJ,
Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ and
Barr FG: PAX3-FKHR and PAX7-FKHR gene fusions are prognostic
indicators in alveolar rhabdomyosarcoma: A report from the
children's oncology group. J Clin Oncol. 20:2672–2679. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hillion J, Le Coniat M, Jonveaux P, Berger
R and Bernard OA: AF6q21, a novel partner of the MLL gene in
t(6;11)(q21;q23), defines a forkhead transcriptional factor
subfamily. Blood. 90:3714–3719. 1997.PubMed/NCBI
|
|
15
|
Parry P, Wei Y and Evans G: Cloning and
characterization of the t(X;11) breakpoint from a leukemic cell
line identify a new member of the forkhead gene family. Genes
Chromosomes Cancer. 11:79–84. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hatini V, Tao W and Lai E: Expression of
winged helix genes, BF-1 and BF-2, define adjacent domains within
the developing forebrain and retina. J Neurobiol. 25:1293–1309.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pierrou S, Hellqvist M, Samuelsson L,
Enerbäck S and Carlsson P: Cloning and characterization of seven
human forkhead proteins: Binding site specificity and DNA bending.
EMBO J. 13:5002–5012. 1994.PubMed/NCBI
|
|
18
|
Hatini V, Huh SO, Herzlinger D, Soares VC
and Lai E: Essential role of stromal mesenchyme in kidney
morphogenesis revealed by targeted disruption of Winged Helix
transcription factor BF-2. Genes Dev. 10:1467–1478. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levinson RS, Batourina E, Choi C,
Vorontchikhina M, Kitajewski J and Mendelsohn CL: Foxd1-dependent
signals control cellularity in the renal capsule, a structure
required for normal renal development. Development. 132:529–539.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Herrera E, Marcus R, Li S, Williams SE,
Erskine L, Lai E and Mason C: Foxd1 is required for proper
formation of the optic chiasm. Development. 131:5727–5739. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gumbel JH, Patterson EM, Owusu SA, Kabat
BE, Jung DO, Simmons J, Hopkins T and Ellsworth BS: The forkhead
transcription factor, Foxd1, is necessary for pituitary luteinizing
hormone expression in mice. PLoS One. 7:e521562012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao YF, Zhao JY, Yue H, Hu KS, Shen H,
Guo ZG and Su XJ: FOXD1 promotes breast cancer proliferation and
chemotherapeutic drug resistance by targeting p27. Biochem Biophys
Res Commun. 456:232–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Banerjee S, Kong D, Li Y and
Sarkar FH: Down-regulation of Forkhead Box M1 transcription factor
leads to the inhibition of invasion and angiogenesis of pancreatic
cancer cells. Cancer Res. 67:8293–8300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ray PS, Wang J, Qu Y, Sim MS, Shamonki J,
Bagaria SP, Ye X, Liu B, Elashoff D, Hoon DS, et al: FOXC1 is a
potential prognostic biomarker with functional significance in
basal-like breast cancer. Cancer Res. 70:3870–3876. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paik JH, Kollipara R, Chu G, Ji H, Xiao Y,
Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al: FoxOs are
lineage-restricted redundant tumor suppressors and regulate
endothelial cell homeostasis. Cell. 128:309–323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Balli D, Ustiyan V, Zhang Y, Wang IC,
Masino AJ, Ren X, Whitsett JA, Kalinichenko VV and Kalin TV: Foxm1
transcription factor is required for lung fibrosis and
epithelial-to-mesenchymal transition. EMBO J. 32:231–244. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Humphreys BD, Lin SL, Kobayashi A, Hudson
TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP and Duffield
JS: Fate tracing reveals the pericyte and not epithelial origin of
myofibroblasts in kidney fibrosis. Am J Pathol. 176:85–97. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koga M, Matsuda M, Kawamura T, Sogo T,
Shigeno A, Nishida E and Ebisuya M: Foxd1 is a mediator and
indicator of the cell reprogramming process. Nat Commun.
5:31972014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fetting JL, Guay JA, Karolak MJ, Iozzo RV,
Adams DC, Maridas DE, Brown AC and Oxburgh L: FOXD1 promotes
nephron progenitor differentiation by repressing decorin in the
embryonic kidney. Development. 141:17–27. 2014. View Article : Google Scholar : PubMed/NCBI
|