Introduction
Nasopharyngeal carcinoma (NPC) is a common head and
neck cancer in southern China and Southeast Asia (1). At present, radiotherapy is the major
treatment for patients with undifferentiated NPC. Thus, the
responsiveness of NPC cells to radiotherapy will have a direct
impact on the treatment outcomes (2). The reasons underlying the presentation
of radiation resistant phenotype in a subgroup of patients remain
poorly understood. In case of radiation treatment failure, patients
will have high chance to develop distant metastasis and recurrent
disease (3–5). Although the use of molecular markers
in predicting treatment outcome shows promising results in multiple
solid tumors, the use of it in monitoring radiation response or
prognostication remains limited in NPC (6). Thus, there is a need to identify key
molecular markers with predictive value in the NPC patients.
MicroRNA is a group of non-coding RNA which
functions as post-transcriptional regulator in controlling specific
gene expression (7). Mature
microRNA could form a thermodynamically stable duplex with the
target mRNA by binding to the 3′ untranslated region (UTR) or open
reading frame (8). Binding to
microRNA was able to hinder the translation process and suppress
the corresponding protein level. In addition, the microRNA/mRNA
duplex recruited Argonaute 2 (Ago2) forming the RNA-induced
silencing complex (RISC) which subsequently promotes mRNA cleavage
and degradation (8). MicroRNA
dysregulation has been reported in nearly all the human
malignancies. Cancers had differential microRNA expression profiles
depending on the cellular context. The differentially expressed or
suppressed microRNA could contribute to the pathological
development of specific cancer phenotype by regulating
characteristic gene expression.
MicroRNA-138-5p expression was significant reduced
in NPC tissues and NPC cell lines (9). However, little is know about the
functions of miR-138-5p in NPC. Liu et al performed a
functional study on the role of miR-138-5p in NPC cell lines. They
showed that re-introduction of miR-138-5p into the NPC cells could
suppress the tumorigenicity of NPC cells in vivo (9). In this study, we addressed the
functional role of miR-138-5p in the responsiveness of NPC cells to
radiation treatment. Computational analysis showed that miR-138-5p
could potentially target the transcript of eukaryotic initiation
factor 4E binding protein 1 (EIF4EBP1). EIF4EBP1 is a translational
regulator (10). EIF4EBP1 represses
eukaryotic translation initiation factor 4E (eIF4E), the rate
limiting cap-binding protein which regulates cap-dependent mRNA
translation by binding to the 7-methyl-GTP 5′ capped end of the
mRNA (11). A recent study showed
that increased EIF4EBP1 stability could promote therapeutic
resistance in prostate cancer cell (12). Therefore, we suggest that miR-138-5p
could modulate the responsiveness to radiation treatment by
targeting EIF4EBP1 in NPC.
Materials and methods
Cell culture
Human NPC cell lines namely HONE1 and HK1, were
derived from poorly and well differentiated NPC, respectively
(13,14). HONE1 and HK1 cells were maintained
in RPMI-1640 medium additionally supplemented with 10% fetal bovine
serum, 200 U/ml penicillin G sodium, 200 µg/ml streptomycin
sulfate, and 0.5 µg/ml amphotericin B in a 37°C humidified
incubator with 5% CO2. Cell irradiation was performed by
Gammacell® 3000 Elan system (Best Theratronics, Ottawa,
ON, Canada).
miR-138 precursor transfection
HONE1 and HK1 cells were transiently transfected
with miR-138 precursor and negative control (Applied Biosystems,
Carlsbad, CA, USA) by HiPerFect reagent (Qiagen, Valencia, CA, USA)
according to the manufacturer's protocol. After 72 h of
transfection, cells were collected and the efficiency of miR-138
precursor transfection was determined.
Real-time quantitative RT-PCR
(QPCR)
Total RNA was extracted by TRIzol (Life
Technologies, Grand Island, NY, USA) according to the protocol of
the manufacturer. High-Capacity cDNA Reverse Transcription kit
(Applied Biosystems) was used for reverse transcription. Transcript
levels of miR-138-5p and U6 control snRNA were measured by TaqMan
Gene Expression assays (Applied Biosystems). The PCR primer-probe
pairs for EIF4EBP1 were as follows: forward,
5′-AGCCCTTCCAGTGATGAGC-3′; reverse, 5′-TGTCCATCTCAAACTGTGACTCTT-3′;
probe no. 21 of Universal ProbeLibrary (Roche Applied Science,
Indianapolis, IN, USA). Primers were synthesized by Integrated DNA
Technologies (Coralville, IA, USA). Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was used as an endogenous control and
quantified by Universal ProbeLibrary Human GAPD Gene assay (Roche
Applied Science). The fold changes were calculated using
2−∆∆Ct method.
Colony formation assay
Colony formation assay was carried out in 6-well
cell culture plate with 600 cells per well. Cells were irradiated
at a single dose of 2, 4, or 6 Gy and allowed to grow for two
weeks. Then, colonies were stained with 0.5% crystal violet after
fixation with 70% ethanol and colonies with >50 cells were
counted.
Proliferation assay
Proliferation assay was performed by using RTCA DP
instrument of xCELLigence Real-Time Cell Analyzer (Roche Applied
Science). Cells were inoculated into E-Plate 16 and were irradiated
by Gammacell 3000 Elan system (Best Theratronics). The
proliferation rates of HONE1 and HK1 cells were examined
constantly.
γH2AX phosphorylation detection
HONE1 and HK1 cells were plated on chamber slides
and irradiated at a single dose of 2, 4, or 6 Gy. Then, cells were
washed, fixed and incubated with rabbit polyclonal anti-γH2AX
antibodies (Abcam, Cambridge, UK) and CF™488A Secondary Antibody
Conjugates (Biotium, Hayward, CA, USA). Nucleus and F-actin were
counterstained with DAPI (Life Technologies) and Alexa
Fluor® 635 phalloidin (Life Technologies) respectively.
Images were taken under a fluorescence microscope (Nikon, Tokyo,
Japan) and the number of γH2AX foci in the cells was counted.
Acridine orange (AO) staining
AO (Sigma-Aldrich, St. Louis, MO, USA) staining was
performed to detect the formation of acidic vesicular organelles
(AVOs) during autophagy process. Cells were stained with AO at the
concentration of 4 µg/ml for 15 min at 37°C. Stained cells were
observed by a fluorescent microscope (Nikon).
Microarray
Affymetrix HG-U133 Plus 2 array (Affymetrix, Santa
Clara, CA, USA) was used to detect global gene expression
profiling. The quality of total RNA was evaluated using Agilent
2100 Bioanalyzer (Agilent Technologies). Data analysis was
performed using GeneSpring version 13 (Agilent Technologies).
Microarray was conducted by the Centre for Genomic Sciences, the
University of Hong Kong.
Luciferase reporter assay
The sense and antisense strands of wild-type and
mutant 3′-UTR of EIF4EBP1 were synthesized. The oligonucleotides
were as follows: wild-type sense strand:
5′-CCAGGGCACCTGCCCCCTCCTCTTCGTGAACACCAGCAGATACCTCCTTGTGA-3′;
wild-type antisense strand:
5′-AGCTTCACAAGGAGGTATCTGCTGGTGTTCACGAAGAGGAGGGGGCAGGTGCCCTGGAGCT-3′;
mutant sense strand:
5′-CCAGGGCACCTGCCCCCTCCTCTTCGTGAAAGTACTAAGATACCTCCTTGTGA-3′; mutant
antisense strand:
5′-AGCTTCACAAGGAGGTATCTTAGTACTTTCACGAAGAGGAGGGGGCAGGTGCCCTGGAGCT-3′.
The annealed sense and antisense strands of wild-type or mutant
3′-UTR of EIF4EBP1 were cloned into the SacI and
HindIII sites of pMIR-Report Luciferase vector (Applied
Biosystems) to produce Luc-wild-type or Luc-mutant construct,
respectively. HONE1 cells were co-transfected with 200 ng
Luc-wild-type vector or Luc-mutant vector, 50 nM miR-138 precursor
or negative control (Applied Biosystems), together with 200 ng
pMIR-Report β-galactosidase control vector (Applied Biosystems)
using Lipofectamine 2000 (Life Technologies). After 48 h, firefly
luciferase and β-galactosidase activities were determined using
Dual-Light luminescent reporter gene assay kit (Applied Biosystems)
on an LB 96V microplate luminometer (EG&G Berthold, Bad
Wildbad, Germany).
siRNA transfection
EIF4EBP1 siRNA-1, EIF4EBP1 siRNA-2 and negative
control siRNA (Qiagen) were transfected into HONE1 and HK1 cells
using HiPerFect transfection reagent (Qiagen) in accordance with
the protocol from the manufacturer. After 72 h, cells were
collected and the efficiency of EIF4EBP1 silencing was determined
by QPCR.
Statistical analysis
Student's t-test was employed to compare
quantitative variables between two groups. All the tests were
2-sided. Results were only considered statistically significant if
p<0.05.
Results
Overexpression of miR-138-5p enhanced
the sensitivity of NPC cells to radiation
To evaluate the effect of miR-138-5p on
radiosensitivity of NPC cells, we overexpressed miR-138-5p by
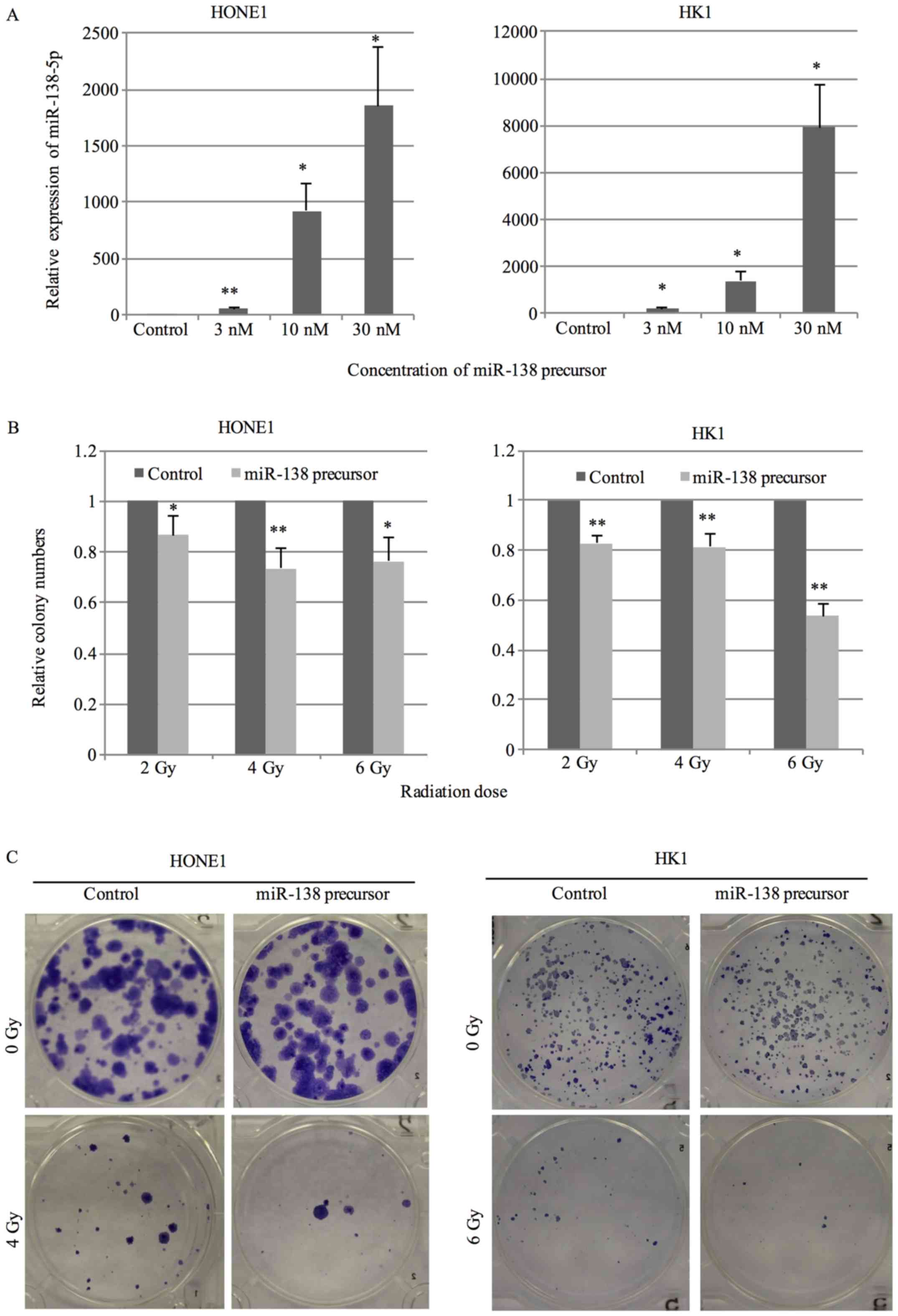
transfection of miR-138 precursor. A dose-dependent increase in
miR-138-5p expression was observed (Fig. 1A). Colony formation assay was
performed to assess the sensitivity to radiation treatment. Cells
transfected with 10 nM miR-138 precursor or negative control were
exposed to radiation at the dose of 2, 4 or 6 Gy and the survival
fraction was counted after 14 days. An apparent decrease in colony
number was observed in cells transfected with miR-138 precursor in
comparison with negative control under radiation treatment
(Fig. 1B and C).
Overexpression of miR-138-5p inhibited
proliferation and enhanced the γH2AX foci formation and autophagy
of NPC cells under radiation treatment
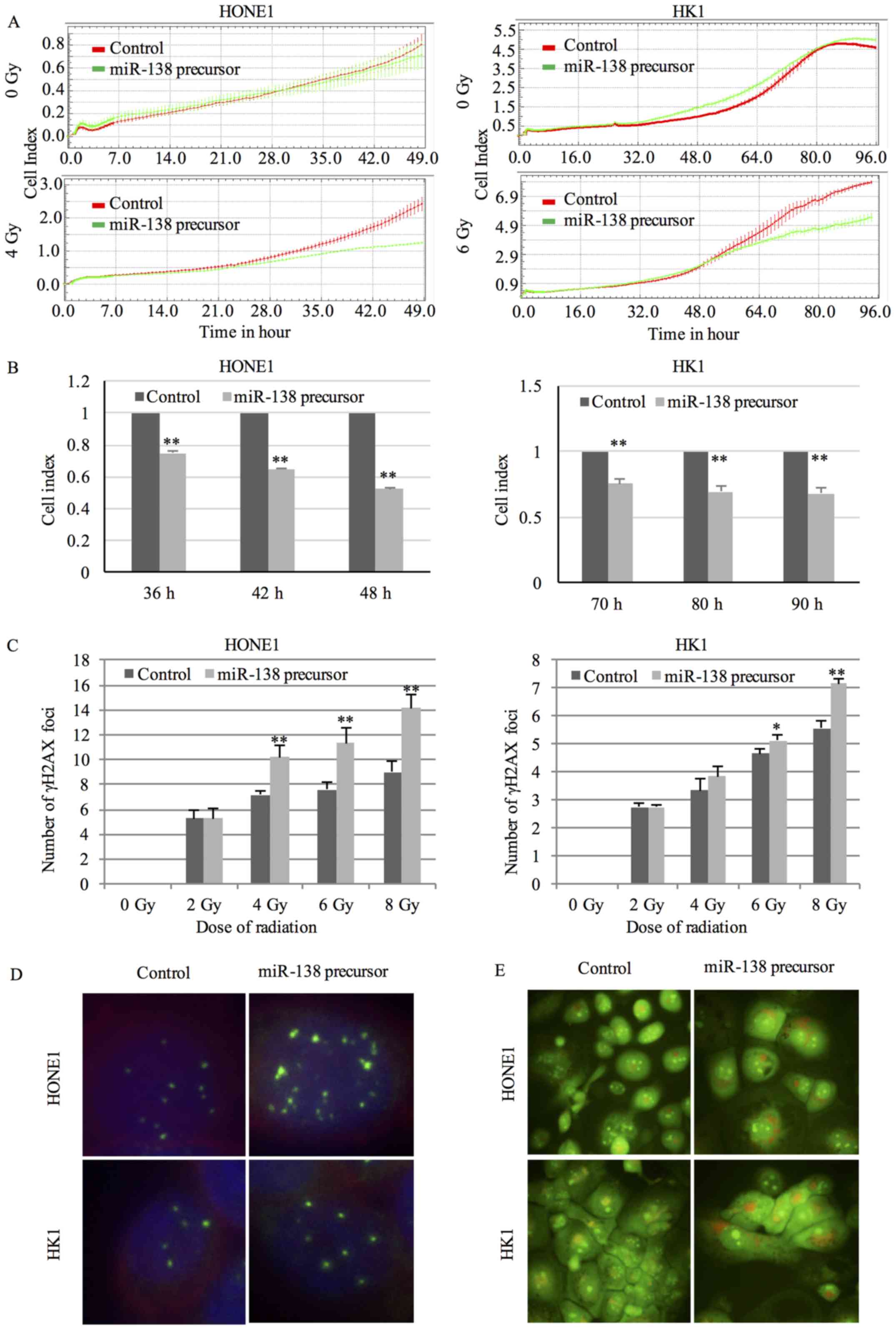
The proliferation rate after radiation treatment was
used to assess the radiosensitivity. Cells transfected with 10 nM
miR-138 precursor were exposed to radiation at 24 hours after
transfection and cell proliferation was continuously monitored.
Although miR-138 precursor itself did not affect the proliferation
rate of cells without radiation exposure, cells transfected with
miR-138 precursor exhibited a significant reduction in
proliferation rate in a time-dependent manner under radiation
treatment (Fig. 2A and B).
Radiation killed cells by inducing DNA double-strand breaks (DSBs).
DSBs were recognized by varied protein complexes including γH2AX.
Staining of cells with anti-γH2AX antibody generated foci
corresponding to DSBs in nuclei (15). Overexpression of miR-138-5p resulted
in an obvious increase in the number of γH2AX foci after radiation
treatment (Fig. 2C and D),
indicating a higher degree of radiation-induced DNA damage.
Radiation could induce autophagy in cancer cells. Formation of AVOs
was a characteristic feature of autophagy. In AO-stained cells,
cytoplasm and nucleolus displayed green fluorescence. In contrast,
AVOs fluoresced bright red (16).
An increase in red signal was observed in NPC cells transfected
with miR-138 precursor compared to negative control after exposure
to radiation (Fig. 2E), implicating
a higher level of radiation-induced autophagy.
miR138-5p directly targets
EIF4EBP1
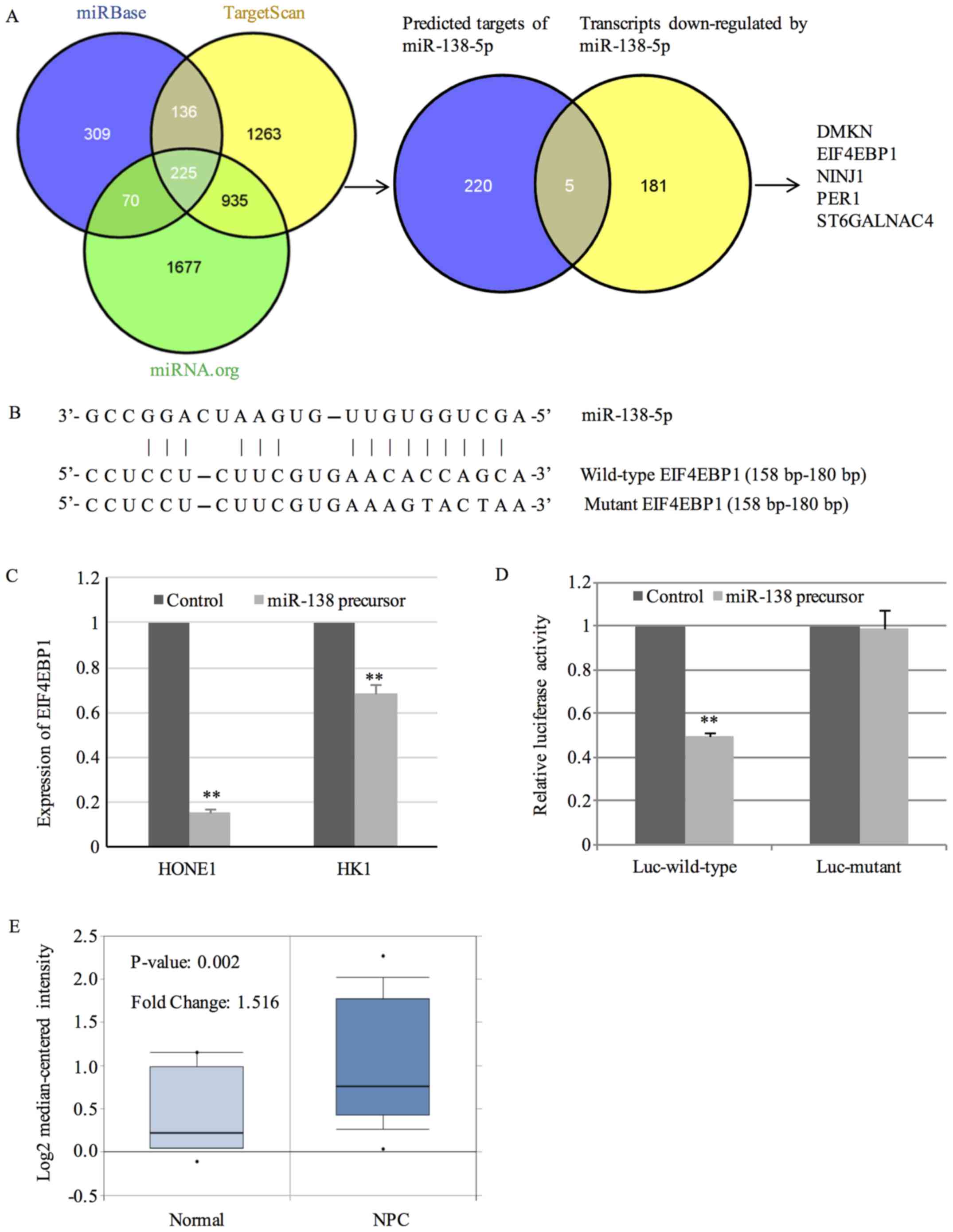
First, we used prediction databases to predict the
targets of miR-138-5p. Considering that individual prediction
database probably generated false positive targets, we employed
Bioinformatics resource manager v2.3 (BRM) software (http://www.sysbio.org/dataresources/brm.stm) for
prediction (17). BRM utilizes
three databases including microCosm/miRBase, TargetScan and
miRNA.org to predict targets. Those targets that are
conserved across all three databases were considered as
high-confidence ones. By the prediction function of BRM, 225 genes
were predicted to be targets of miR-138-5p by all three databases.
Given that miRNA promoted the degradation of target mRNA, we
carried out microarray analysis to identify transcripts
downregulated by miR-138-5p, which were its putative targets. The
global gene expression profiling of HONE1 cells transfected with
miR-138 precursor was examined. With a fold-change cut-off of 3 and
p<0.05, 186 genes were found to be downregulated by miR-138-5p.
Then, the 225 high-confidence targets were integrated with the 186
transcripts downregulated by miR-138-5p. By this novel strategy, we
identified 5 potential targets including DMKN, EIF4EBP1, NINJ1,
PER1 and ST6GALNAC4 (Fig. 3A).
The binding sites between miR-138-5p and 3′-UTR of
EIF4EBP1 are shown in Fig. 3B.
Results from QPCR further confirmed the downregulation of EIF4EBP1
by miR-138-5p (Fig. 3C). To explore
whether miR-138-5p modulated the expression of EIF4EBP1 by binding
to the 3′-UTR, luciferase reporter assay was performed in HONE1
cells transfected with luciferase vector containing 3′-UTR of
EIF4EBP1 together with miR-138 precursor or negative control.
Overexpression of miR-138-5p significantly reduced the luciferase
activity of luciferase vector harboring wild-type 3′-UTR of
EIF4EBP1 (Fig. 3D). In contrast,
mutation of the binding sites within 3′-UTR of EIF4EBP1 abrogated
the suppressing effect (Fig.
3D).
To gain insights into the clinical implications of
EIF4EBP1 in NPC, the expression of EIF4EBP1 was analyzed in
publicly available microarray dataset GSE12452 in the Oncomine
database (www.oncomine.org) (18). GSE12452 contained expression data of
31 NPC tissues and 10 normal controls. A 1.516-fold increase in
EIF4EBP1 expression was observed in NPC tissues in comparison with
normal controls (Fig. 3E).
Silence of EIF4EBP1 increased the
sensitivity of NPC cells to radiation
To investigate the role of EIF4EBP1 in
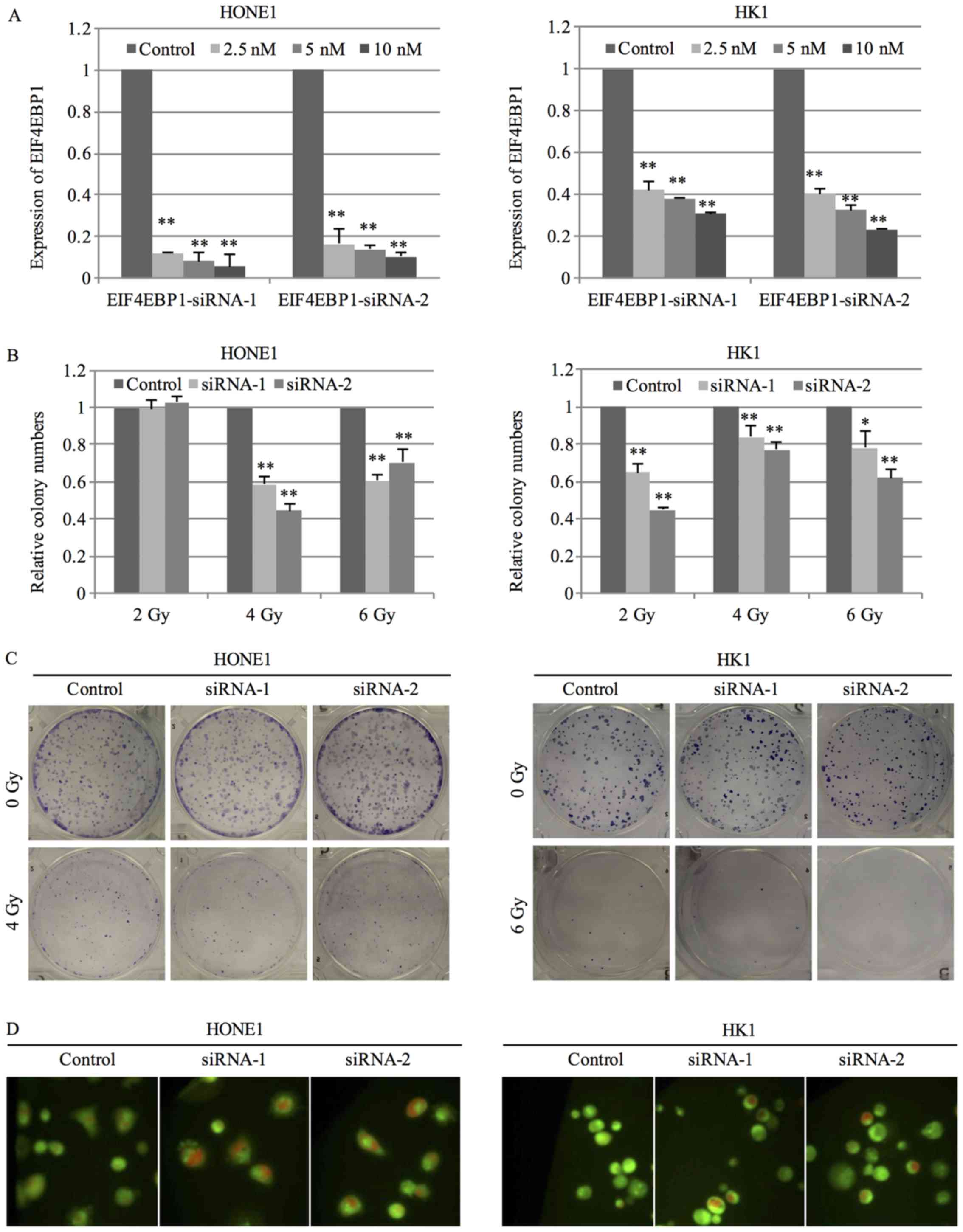
radiosensitivity of NPC cells, the expression of EIF4EBP1 was
silenced by 2 specific siRNA. Transfection of siRNA at the
concentration of 2.5 nM successfully inhibited the expression of
EIF4EBP1 in HONE1 cells, while siRNA at the concentration of 10 nM
was required to suppress the expression in HK1 cells (Fig. 4A). NPC cells transfected with
EIF4EBP1 siRNA were exposed to radiation treatment at the dose of
2, 4 or 6 Gy and the ability to form colony was measured after 14
days. NPC cells with silenced expression of EIF4EBP1 displayed a
significant decrease in the number of colonies after radiation
treatment compared to negative control siRNA (Fig. 4B and C). Results from AO staining
showed that silence of EIF4EBP1 enhanced radiation-induced
autophagy as evidenced by an increase in red signal under radiation
treatment (Fig. 4D).
Discussion
We found that expression of miR-138-5p is one of the
determining factors involved in modulating the sensitivity of NPC
cells to radiation treatment. miR-138-5p controls the cell cycle
related genes in hepatocellular carcinoma (19). In NPC, the oncogenic cell cycle
regulator CCND1 is a reported target of miR-138-5p (9). In small cell lung cancer, inhibition
of cell cycle progression is also observed in miR-138-5p
overexpressing cells (20). It is
suggested that miR-138-5p might be involved in DNA damage response
by targeting the histone H2AX (20). Recently, the role of miR-138-5p in
modulating the sensitivity to cancer therapeutics is reported. In
prostate cancer, miR-138-5p regulates Kindlin-2, a regulator of
integrin adhesion receptor which is frequently overexpressed in
castration-resistant prostate cancer (21). Suppressing Kindlin-2 could improve
the sensitivity of prostate cancer cells to docetaxel (21). Upregulating miR-138-5p could
increase the responsiveness of lung cancer cells to radiation
treatment (22).
eIF4E is an oncogenic driver which could promote
malignant transformation (23,24).
Increased eIF4F expression is rarely found in the benign lesions
(25). High expression has been
observed in cancer with aggressive cancer and poorly differentiated
phenotype (26). Overexpression of
eIF4F is also found in cancers of the head and neck regions
including oral cavity, oropharynx, larynx and nasopharynx (27–29).
In NPC, high eIF4F expression promotes proliferation and cell cycle
progression with high correlation to the oncogenic protein c-Myc
and MMP9 (29). As the cancer cell
responses to radiation damage by activating the cell cycle
checkpoints, it is suggested that eIF4E might function in radiation
response by initiating expression of genes involved in cell cycle
regulation (30,31). Apart from the protein translation
function, eIF4E can promote oncogenic transformation by
preferential export of capped mRNAs containing 50-nucleotide
structural element in the 3′ UTR (32).
Inactivation of the eIF4E inhibitor EIF4EBP1 could
be achieved by phosphorylation. The serine/threonine protein kinase
mammalian target of rapamycin (mTOR) is a critical player involved
in EIF4EBP1 phosphorylation and dissociating from eIF4E (33). Another serine/threonine protein
kinase Akt, upstream of mTOR, is an indicator of poor prognosis of
NPC patients. Activated Akt showed high correlation with the
inactivated EIF4EBP1, which is a strong indication of the
involvement of Akt signaling in EIF4EBP1 regulation (34). Akt/mTOR signaling can be activated
by radiation. Thus, the use of Akt/mTOR inhibitors could increase
the sensitivity of cancer cells to radiation (35).
In general, hypoxic cells are less sensitive to
radiation treatment due to the specific expression of particular
proteins in the hypoxic environment (36). It has been shown that EIF4EBP1
expression will affect the cancer proteomic leading to the
differential expression of different proteins under normoxic and
hypoxic conditions (37). The
quantity of EIF4EBP1 protein and activated EIF4EBP1 was induced in
response to radiation treatment (38). In glioblastoma, inhibiting EIF4EBP1
could sensitize the xenograft to radiation (39). Thus, it is important to explore the
functional impact and the role of EIF4EBP1 in the radiation
responses.
In conclusion, our results indicate that miR-138-5p
mediates the responsiveness of NPC to ionizing radiation by
targeting EIF4EBP1. Further study on the impact of miR-138-5p and
EIF4EBP1 on the radiosensitivity of tumor cells will be useful for
the development of therapeutic regimens to enhance the therapeutic
radiosensitivity of NPC.
Acknowledgements
This study was supported by S.K. Yee Medical
Foundation Grant and Seed Funding of Basic Research, The University
of Hong Kong.
References
|
1
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng XP, Yi H, Li MY, Li XH, Yi B, Zhang
PF, Li C, Peng F, Tang CE, Li JL, et al: Identification of
biomarkers for predicting nasopharyngeal carcinoma response to
radiotherapy by proteomics. Cancer Res. 70:3450–3462. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leung SF, Teo PM, Shiu WW, Tsao SY and
Leung TW: Clinical features and management of distant metastases of
nasopharyngeal carcinoma. J Otolaryngol. 20:27–29. 1991.PubMed/NCBI
|
|
4
|
Lee AW, Poon YF, Foo W, Law SC, Cheung FK,
Chan DK, Tung SY, Thaw M and Ho JH: Retrospective analysis of 5037
patients with nasopharyngeal carcinoma treated during 1976–1985:
Overall survival and patterns of failure. Int J Radiat Oncol Biol
Phys. 23:261–270. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan JY, Chow VL, Tsang R and Wei WI:
Nasopharyngectomy for locally advanced recurrent nasopharyngeal
carcinoma: Exploring the limits. Head Neck. 34:923–928. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee AW, Ng WT, Chan YH, Sze H, Chan C and
Lam TH: The battle against nasopharyngeal cancer. Radiother Oncol.
104:272–278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pause A, Belsham GJ, Gingras AC, Donzé O,
Lin TA, Lawrence JC Jr and Sonenberg N: Insulin-dependent
stimulation of protein synthesis by phosphorylation of a regulator
of 5′-cap function. Nature. 371:762–767. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meric F and Hunt KK: Translation
initiation in cancer: A novel target for therapy. Mol Cancer Ther.
1:971–979. 2002.PubMed/NCBI
|
|
12
|
Yu L, Shang ZF, Wang J, Wang H, Huang F,
Zhang Z, Wang Y, Zhou J and Li S: PC-1/PrLZ confers resistance to
rapamycin in prostate cancer cells through increased 4E-BP1
stability. Oncotarget. 6:20356–20369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Glaser R, Zhang HY, Yao KT, Zhu HC, Wang
FX, Li GY, Wen DS and Li YP: Two epithelial tumor cell lines (HNE-1
and HONE-1) latently infected with Epstein-Barr virus that were
derived from nasopharyngeal carcinomas. Proc Natl Acad Sci USA.
86:9524–9528. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang DP, Ho JH, Poon YF, Chew EC, Saw D,
Lui M, Li CL, Mak LS, Lai SH and Lau WH: Establishment of a cell
line (NPC/HK1) from a differentiated squamous carcinoma of the
nasopharynx. Int J Cancer. 26:127–132. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuo LJ and Yang LX: Gamma-H2AX - a novel
biomarker for DNA double-strand breaks. In Vivo. 22:305–309.
2008.PubMed/NCBI
|
|
16
|
Paglin S, Hollister T, Delohery T, Hackett
N, McMahill M, Sphicas E, Domingo D and Yahalom J: A novel response
of cancer cells to radiation involves autophagy and formation of
acidic vesicles. Cancer Res. 61:439–444. 2001.PubMed/NCBI
|
|
17
|
Tilton SC, Tal TL, Scroggins SM, Franzosa
JA, Peterson ES, Tanguay RL and Waters KM: Bioinformatics Resource
Manager v2.3: An integrated software environment for systems
biology with microRNA and cross-species analysis tools. BMC
Bioinformatics. 13:3112012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
MiR-138 induces cell cycle arrest by targeting cyclin D3 in
hepatocellular carcinoma. Carcinogenesis. 33:1113–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang H, Luo J, Liu Z, Zhou R and Luo H:
MicroRNA-138 regulates DNA damage response in small cell lung
cancer cells by directly targeting H2AX. Cancer Invest. 33:126–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sossey-Alaoui K and Plow EF:
miR-138-mediated regulation of KINDLIN-2 expression modulates
sensitivity to chemotherapeutics. Mol Cancer Res. 14:228–238. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang H, Tang Y, Guo W, Du Y, Wang Y, Li P,
Zang W, Yin X, Wang H, Chu H, et al: Up-regulation of microRNA-138
induce radiosensitization in lung cancer cells. Tumour Biol.
35:6557–6565. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Larsson O, Li S, Issaenko OA, Avdulov S,
Peterson M, Smith K, Bitterman PB and Polunovsky VA: Eukaryotic
translation initiation factor 4E induced progression of primary
human mammary epithelial cells along the cancer pathway is
associated with targeted translational deregulation of oncogenic
drivers and inhibitors. Cancer Res. 67:6814–6824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Avdulov S, Li S, Michalek V, Burrichter D,
Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman
PB, et al: Activation of translation complex eIF4F is essential for
the genesis and maintenance of the malignant phenotype in human
mammary epithelial cells. Cancer Cell. 5:553–563. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng J, Li J, Xu L, Xie G, Wen Q, Luo J,
Li D, Huang D and Fan S: Phosphorylated Mnk1 and eIF4E are
associated with lymph node metastasis and poor prognosis of
nasopharyngeal carcinoma. PLoS One. 9:e892202014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
De Benedetti A and Harris AL: eIF4E
expression in tumors: Its possible role in progression of
malignancies. Int J Biochem Cell Biol. 31:59–72. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nathan CO, Franklin S, Abreo FW, Nassar R,
de Benedetti A, Williams J and Stucker FJ: Expression of eIF4E
during head and neck tumorigenesis: Possible role in angiogenesis.
Laryngoscope. 109:1253–1258. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sorrells DL Jr, Ghali GE, De Benedetti A,
Nathan CO and Li BD: Progressive amplification and overexpression
of the eukaryotic initiation factor 4E gene in different zones of
head and neck cancers. J Oral Maxillofac Surg. 57:294–299. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu M, Liu Y, Di X, Kang H, Zeng H, Zhao Y,
Cai K, Pang T, Wang S, Yao Y, et al: EIF4E over-expresses and
enhances cell proliferation and cell cycle progression in
nasopharyngeal carcinoma. Med Oncol. 30:4002013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:928–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Culjkovic B, Topisirovic I, Skrabanek L,
Ruiz-Gutierrez M and Borden KL: eIF4E is a central node of an RNA
regulon that governs cellular proliferation. J Cell Biol.
175:415–426. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Culjkovic-Kraljacic B, Baguet A, Volpon L,
Amri A and Borden KL: The oncogene eIF4E reprograms the nuclear
pore complex to promote mRNA export and oncogenic transformation.
Cell Rep. 2:207–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goberdhan DCI and Boyd CAR: mTOR:
Dissecting regulation and mechanism of action to understand human
disease. Biochem Soc Trans. 37:213–216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang W, Wen Q, Xu L, Xie G, Li J, Luo J,
Chu S, Shi L, Huang D, Li J, et al: Activation of Akt/mTOR pathway
is associated with poor prognosis of nasopharyngeal carcinoma. PLoS
One. 9:e1060982014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Albert JM, Kim KW, Cao C and Lu B:
Targeting the Akt/mammalian target of rapamycin pathway for
radiosensitization of breast cancer. Mol Cancer Ther. 5:1183–1189.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brown JM: The hypoxic cell: A target for
selective cancer therapy - eighteenth Bruce F. Cain Memorial Award
lecture. Cancer Res. 59:5863–5870. 1999.PubMed/NCBI
|
|
37
|
Magagnin MG, van den Beucken T, Sergeant
K, Lambin P, Koritzinsky M, Devreese B and Wouters BG: The mTOR
target 4E-BP1 contributes to differential protein expression during
normoxia and hypoxia through changes in mRNA translation
efficiency. Proteomics. 8:1019–1028. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Braunstein S, Badura ML, Xi Q, Formenti SC
and Schneider RJ: Regulation of protein synthesis by ionizing
radiation. Mol Cell Biol. 29:5645–5656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dubois L, Magagnin MG, Cleven AH, Weppler
SA, Grenacher B, Landuyt W, Lieuwes N, Lambin P, Gorr TA,
Koritzinsky M, et al: Inhibition of 4E-BP1 sensitizes U87
glioblastoma xenograft tumors to irradiation by decreasing hypoxia
tolerance. Int J Radiat Oncol Biol Phys. 73:1219–1227. 2009.
View Article : Google Scholar : PubMed/NCBI
|