Introduction
According to the statistics of WHO, breast cancer
has become the number one killer among female cancers. Though, the
cure percentage of early stage breast cancer has reached 80–90%,
the late stage of this disease is still indisputable (1). Accordingly, to search for more
sensitive diagnostic indicators and therapeutic methods is of great
importance. A recent molecular testing of breast cancer has shown,
during the carcinogenesis and progression of cancer, a number of
molecules and signaling pathways participate having respective
functions. Thus breast cancer can be divided into four subtypes
depending on the molecular features: Luminal A, Luminal B,
HER2+ and basal like (2). Cancer cells exist in a complicated
microenvironment, the surrounding cells cross-talk with malignant
cancer cells and confer to the progression of cancer (3). Various cytokines and regulators are
involved. Interleukin (IL)-24 transforms the tumor microenvironment
in colon cancer (4). β-catenin,
PPAR-γ, and FGFR3 pathways are activated and drives non-T
cell-inflamed tumor microenvironment in urothelial bladder cancer
(5). Cytokines produced by stromal
cells are connected with tumor grades and survival percentage. As
reported, IL-1β and IL-17 are positively associated with
histological grade; IFNβ expressed higher, and NF-κB lower in
HER-2-positive tumors; and IL-6 was shown with a higher global
expression in node-negative tumors (6). Recent data suggests that immunity
associated molecules of tumor microenvironment play an important
role in breast cancer malignancy (7), STATs family is one of them and has
been studied in recent years as vital molecules in inflammation
induced cancers.
Signal transducers and activators of transcription
(STAT) is a class of cytoplasmic and unclear signaling pathway
molecules. The activation of STAT3 is mostly conducted by JAK
family. STAT3 is the crucial member of the STAT family. It
medicates transcription of several kinds of cytokines and growth
factors (8). As is reported, STAT3
is continuously activated in diverse human cancers. It also
enhances transcription of oncogenes, and inhibits cell apoptosis in
cancers. STAT3 especially plays a core role in inflammation induced
cancers, IL6-JAK-STAT3 is the key signaling pathway of
carcinogenesis and epigenetic transformation (9). Besides, some extrinsic carcinogenic
factors including sunlight, pathogens, chemical cancerogens can
also active STAT3 (10). High
percentage of activated STAT3 has been found in certain cancers,
such as thyroid cancer, colorectal cancer, liver cancer, lung
cancer, breast cancer, and cancer associated microenvironments
(11). Thus, to find a new
mechanism that can specifically inhibit STAT3 would be of great
therapeutic value.
MicroRNAs are a class of 19–25 nt short non-coding
RNAs, which exist in many physiological and pathologic processes
(12,13). Mature miRNAs form RNA-induced
silencing complex (miRISC) to conversely complement with 3′UTR of
the target mRNA, which results in silence or merely degradation of
a specific mRNA (14). It has been
reported, nearly 30% of human genes are regulated by microRNAs.
Genes of microRNAs surrounded by aberrantly histidine modified CpG
island can be named as onco-microRNAs (15), which are always upregulated.
MicroRNA expresses abnormally in most of human diseases,
particularly in cancers. At present, microRNA expression profiles
have been applied for diagnosing and classifying human cancers
(16,17).
MicroRNAs have a vital role in the progress of
diseases. A microRNA can have completely opposite functions
depending on the tissue and the target gene (18,19).
MicroRNA-494 promotes cervical cancer proliferation by regulating
PTEN (20), but it represses the
expression of HOXA10 to inhibit cell proliferation in oral cancer
(21).
Some microRNAs are correlated with cancer drug
resistance, miR519a has been reported to induce tamoxifen
resistance and decrease the survival rate in breast cancer
(22). miRNA221/222, known as
oncomirs, can cause acquired fulvestrant resistance in MCF cell
lines (23). Some are tumor
suppressors, miR-520c suppresses NF-κB and TGF-β in estrogen
receptor negative breast cancer (24).
Herein, we confirmed miR520c was the corresponding
small non-coding RNA for binding with STAT3 3′UTR. Then we
discovered miR520c and STAT3 expressed abnormally in different
degrees of breast cancer cells. Additionally, miR520c negatively
regulated the protein synthesis of STAT3 by direct targeting of
STAT3 mRNA. Importantly, the degradation of STAT3 generated
prevention consequence on EMT, which abolished progression of
breast cancer. No research has been reported on the relevance
between miR-520c and STAT3 in cancers. Our study presented here,
explain the unknown mechanism between the miR-520c and STAT3, and
the inhibition effect on breast cancer in EMT.
Materials and methods
Cell lines and culture conditions
The human breast cancer cell lines MCF-7, SK-BR-3,
MDA-MB-231 and the HEK293T cells were obtained from ATCC and
maintained in Dulbecco's modified Eagle's medium or RPMI-1640
(Gibco) with 10% fetal bovine serum (BI) and antibiotics (100 mg/ml
streptomycin, 100 U/ml penicillin, Beyotime) cultured in incubator
at 37°C and in 5% CO2.
Plasmids construction
The pri-microRNAs and full-length of STAT3 3′UTR
were amplified from human genome (extracted from HEK293T cells with
Universal Genomic DNA kit, CWBIO CW2298) and then STAT3 3′UTR was
constructed into pCDNA3.1-luc (this vector and other empty plasmids
were aquired from Professor Qin Zhou, Laboratory of Molecular
Nephrology, Chongqing Medical University) namely STAT3 3′UTR
wild-type (WT). The STAT3 3′UTR mutation (Mut) was obtained through
site mutation PCR (PrimeStar; Takara). hsa-pri-miR196b,
hsa-pri-miR298, hsa-pri-miR494, hsa-pri-miR495, hsa-pri-miR519b,
hsa-pri-miR517a/519d, hsa-pri-miR520c microRNA overexpression
plasmids were constructed into pdsAAV-CB-EGFP. The primer sequences
are in Table I.
 | Table I.Primers for plasmid construction. |
Table I.
Primers for plasmid construction.
| Pimers | Sequences |
|---|
| hsa-mir196b | F:
atttcaggtcccggattactggggcctgtggcttccc |
| hsa-mir196b | R:
caccaccaccggatccaacctaaccctacctgctgtg |
| hsa-mir298 | F:
aatttcaggtcccggacagccctagctgggttcctaat |
| hsa-mir298 | R:
caccaccaccggatcgccttgccattcatcttctgaa |
| hsa-mir494 | F:
aatttcaggtcccggattcattgtgaaggcttgaagag |
| hsa-mir494 | R:
caccaccaccggatctccttcaaccacagaagcacag |
| hsa-mir495 | F:
aatttcaggtcccggagcctctgctcagtgtcagcc |
| hsa-mir495 | R:
caccaccaccggatcaggcctcgccaactgtgcct |
| hsa-mir519b | F:
aatttcaggtcccggaggatttccccttgatgaacaag |
| hsa-mir519b | R:
caccaccaccggatcaagaggaccgtttgagcctaaa |
| hsa-mir520c | F:
aatttcaggtcccggaggaggattgcccgttgatga |
| hsa-mir520c | R:
caccaccaccggatcctacatactagtgcttgggc |
| pCDNA3.1-luc-STAT3
3′UTR | F:
acgccgtgtaaaagctccatgtgaggagctgagaac |
| pCDNA3.1-luc-STAT3
3′UTR | R:
agccctctagactcgatgaatgcagtggccaggaca |
Luciferase reporter assay
Cells (0.1×105) were seeded into 24-well
plate per well. In 16–24 h, following with primary microRNAs (500
ng), pCDNA3.1-luc STAT3 3′UTR WT/Mut (500 ng), pRL-SV40 (10 ng)
were co-transfected into each well by Lipofectamine 2000
(Invitrogen). After 24 h, cells were washed with PBS (pH 7.4) and
lysed with diluted 5X lysis buffer on ice for 30 min the proteins
were collected, luciferase and Renilla activity was measured
by Dual-Luciferase Reporter assay system (Promega, USA).
Western blotting
Cells were washed with ice-cold PBS (pH 7.4) and
cleaved by RIPA (consisting of 50 mM Tris (pH 7.4), 150 mM NaCl, 1%
Triton X-100, 1% sodium deoxycholate, 0.1% SDS, sodium
orthovanadate, sodium fluoride, EDTA, and leupeptin, Beyotime)
pre-added 100 nM PMSF (phenylmethanesulfonyl fluoride, Sigma).
Protein concentration was determined with Bradford protein dye
reagent (Beyotime). The loading volumes were evaluated for the
equal amount of proteins. Lysate was separated by 8% SDS-PAGE and
blotted onto polyvinylidene fluoride (PVDF) membrane. The membrane
was blocked with 5% fat-free milk then incubated with primary
antibodies at 4°C overnight. Primary antibodies were diluted at
ratios of 1:1,000 (β-actin, ZSBio TA-09), 1:500 (STAT3, Boister
1621), 1:1,000 (p-STAT3 (Tyr705), CST #4113), 1:1,000 (E-cadherin,
Bioworld BS1098), 1:1,000 (Vimentin, Bioworld BS1491).
Antigen-antibody complex was visualized with immobilon Western
chemiluminescent HRP substrate (Millipore, WBKLS0500).
Quantitive RT-PCR
Total RNA was extracted with RNA isoPlus (Takara).
Reverse transcription PCR was conducted with random/oligodT and
microRNA specific primers (PrimScript RT reagent kit with gDNA
Eraser, Takara): miR520c reverse oligos
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGAAAGC-3′; hU6
reverse oligos
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3′.
Real-time PCR was performed with SYBR Premix Ex Taq II (Takara) and
the following primers: miR520c real-time forward,
5′-TGCGGCTCTAGAGGGAAGCGTT-3′; hU6 real-time forward,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′; microRNA universal real-time reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′. The microRNA expession level was
normalized by hU6 with 2−∆∆t method.
Cell scratch assay
MCF-7 cells were seeded at the density of
0.25×105 cells/well into a 6-well plate. Forty-eight
hours after cells were transfected, a wound was created in each
well with a 10-µl tip and then washed with PBS three times,
followed by imaging at 0 h, and 24 h under a microscope to record
the migration distance at ×40.
Boyden chamber assay
The wells (Millipore, 24-well Millicell) for the
24-well plate were coated with 50 µl Matrix gel (Matrigel Matrix,
356234; Corning) at 37°C for 30 min. Pre-transfected cells
(2×104) were seeded into the top of the insert with 1%
fetal bovine serum media, while 10% fetal bovine serum media was
placed in the well below. Then cultured cultured for 24 h, cells
still in the top were wiped off with cotton swab, and the migratory
cells were stained with crystal violet and captured under the
microscope. The invasive cell number in each field was recorded at
×200.
Statistical analysis
All statistical analyses were performed by GraphPad
Prim 5 software and subjected to Student's t-test (two-tailed, with
P<0.05) to compare significant differences between the sample
means obtained from three independent experiments. Error bars
depict SD. Asterisks were used to represent statistical signifiance
of P-values in the figures.
Results
The 3′UTR of STAT3 was targeted by
miR520c
STAT3 is consistently activated in several kinds of
human cancers and cancer microenvironments. To find which microRNA
could directly down regulate STAT3 is of great value in cancer
targeted therapy. We predicted microRNAs according to the three
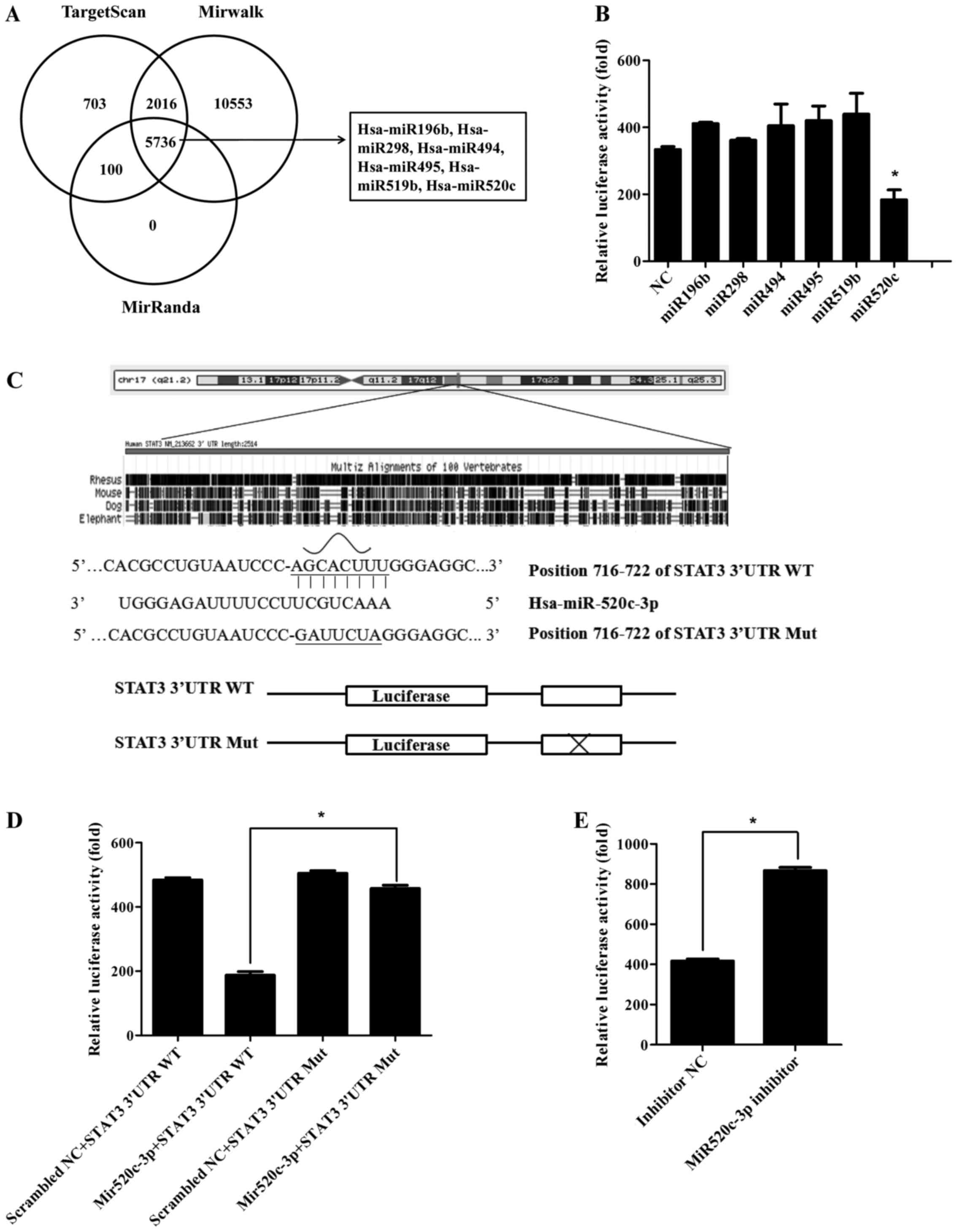
authoritative microRNA databases: targetscan, mirwalk and miRanda.
Then selected the six top ranking and cross-database microRNAs
(p<0.01) (Fig. 1A). Afterwards,
we screened these putative miRNAs by Dual-luciferase reporter
assay. Results indicated miR520c downregulated the relative
luciferase activity approximately by half compared with the
negative control. The other miRNAs were not significantly
downregulated (Fig. 1B). So we
chose miR520c for further mechanism study. According to database
analysis, we found miR520c had only one conservative seed sequence
on STAT3 3′UTR (WT) among other species (Fig. 1C). So we constructed the following
seed sequence mutated plasmid to verify whether miR520c regulated
STAT3 3′UTR through this vital site. Constant with prediction,
miR520c had no effect on STAT3 Mut (Fig. 1D). Concordantly, relative luciferase
activity could be increased (2-fold) when cells were transfected
with miR520c inhibitor (Fig. 1E).
Hence, as demonstrated, STAT3 3′UTR was directly regulated by
miR520c.
The expression of STAT3 and miR520c in
breast cancer cell lines
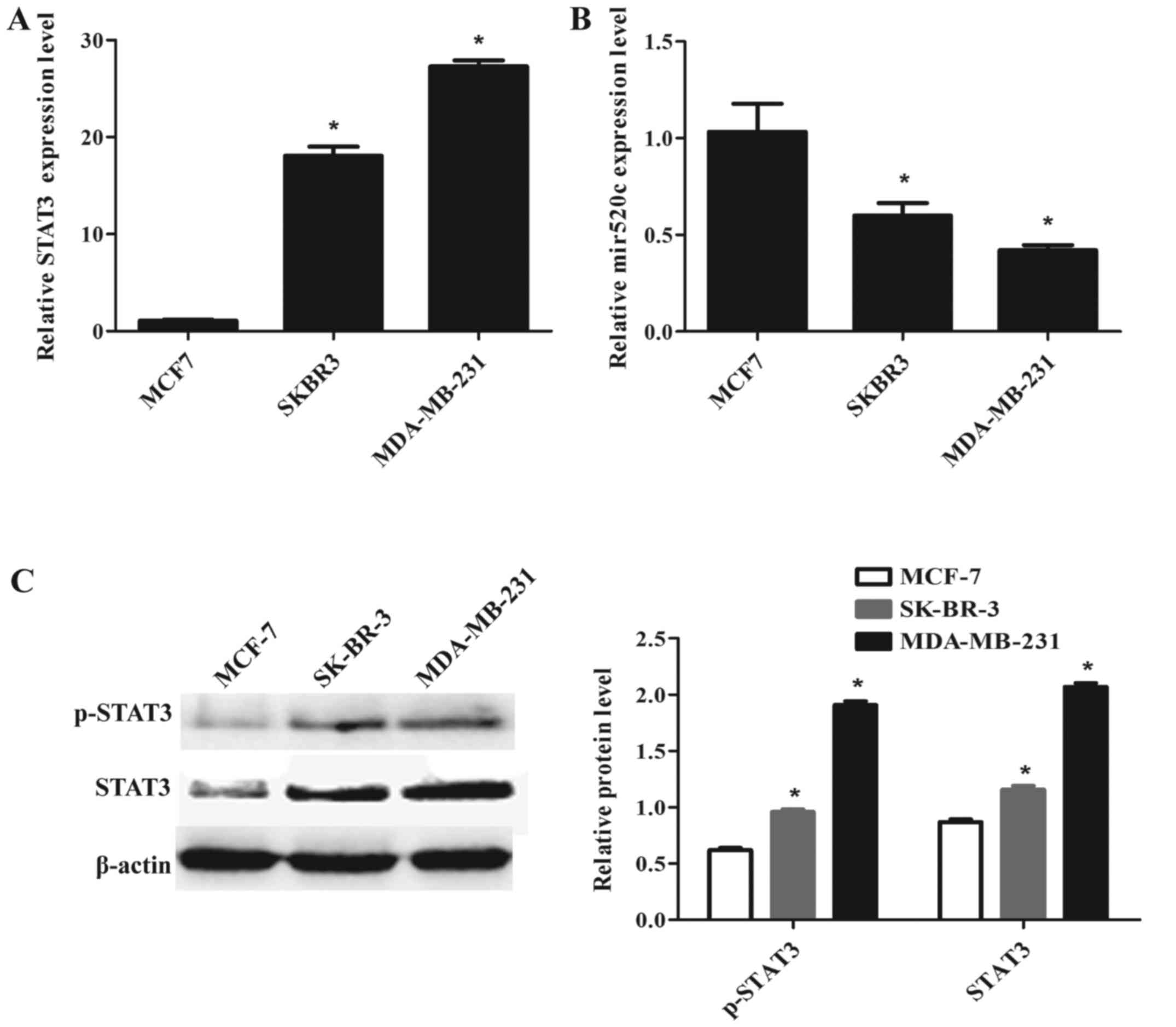
In 2014, Liu et al (25) reported that STAT3 was elevated in
high grade breast cancers. We detected the mRNA and protein levels
of STAT3 in three different grades of breast cancer cell lines:
MCF-7, SK-BR-3 and MDA-MB-231. STAT3 was obviously upregulated
(18-fold) in SK-BR-3 and (27-fold) in MDA-MB-231 compared to the
low malignant MCF-7 (Fig. 2A). The
higher metastatic cell line MDA-MB-231 expressed the highest mRNA
and protein levels among the three cell lines. Activated form of
STAT3-phosphorylated STAT3 (p-STAT3) of MCF-7 was significantly
lower than the other two cell lines (Fig. 2C). Then we detected the expression
level of miR520c in three different progressions of breast cancer
cells. Q-PCR results showed that the mRNA level of miR520c was
decreased in MDA-MD-231, and MCF-7 expressed the highest level
among the three breast cancer cell lines. Additionally, miR520c
showed negative correlation with STAT3 expression level.
miR520c downregulates STAT3 in breast
cancer cells
For further understanding of mechanism between
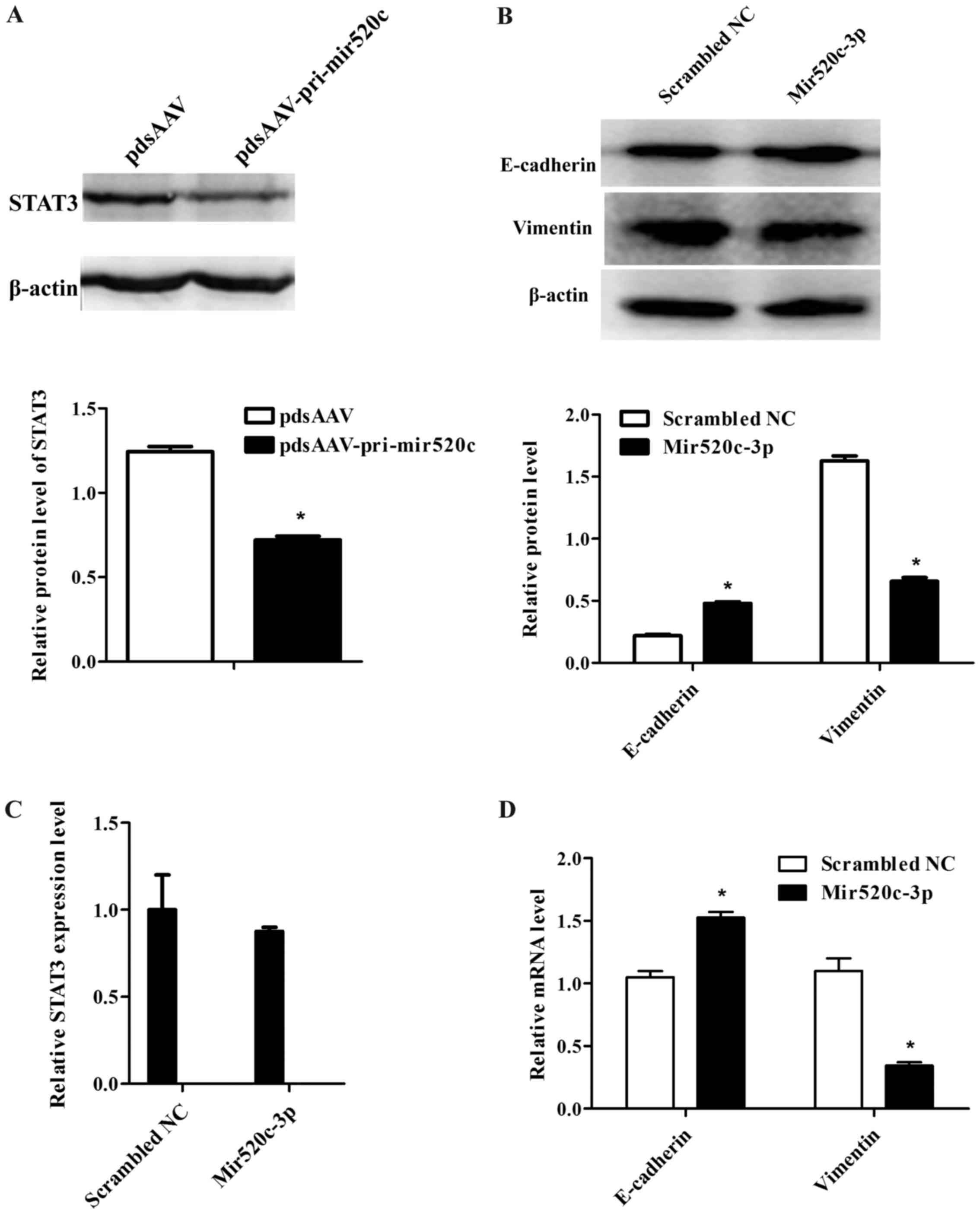
miR520c and STAT3, we overexpressed miR520c mimics in MCF-7 cells.
Western blotting result indicated that STAT3 protein level was
significantly downregulated in miR520c overexressed group compared
with the negative control group, it meant miR520c could inhibit
STAT3 (Fig. 3A). Furthermore, we
detected the mRNA level of STAT3, it was shown in the Q-PCR results
that STAT3 was scarcely downregulated in miR520c overexpressed
group. which indicated STAT3 was regulated by miR520c
post-transcriptionally (Fig.
3C).
The breast cancer cells EMT
progression is inhibited by miR520c targeting STAT3
EMT is one of the malignant cancer properties. There
is remarkable change of tumor metastasis in epithelial to
mesenchymal transition. When STAT3 was inhibited by miR520c, it
demonstrated the EMT phenotype. Epthelial marker E-cadherin was
significantly upregulated in miR520c overexpression group, and
mesenchymal marker vimentin was downregulated at both protein and
mRNA levels (Fig. 3B and D). These
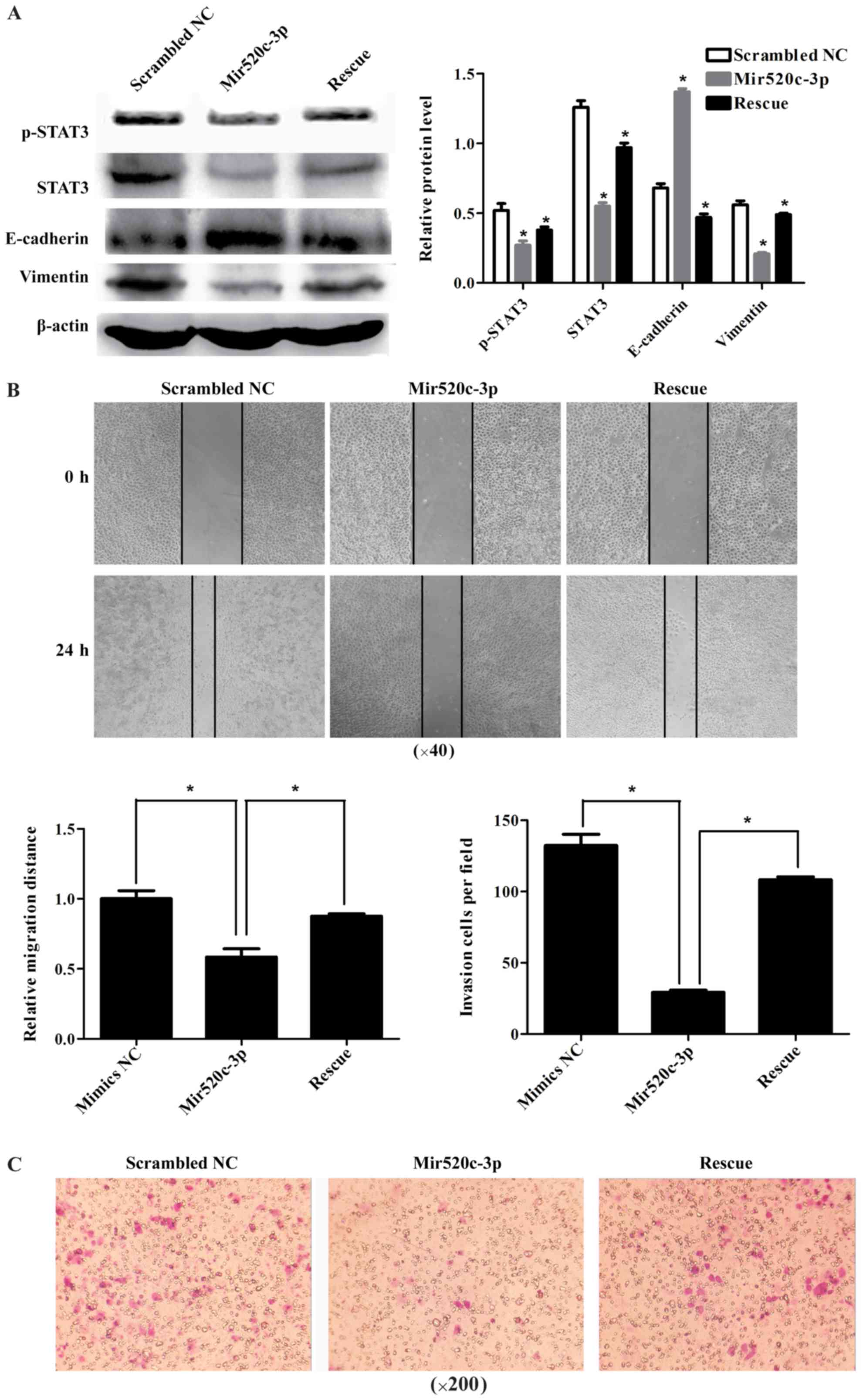
changes could be reversed by overexpression of STAT3 (Fig. 4A). Later on, we observed cell
migration and invasion. It was shown in cell scratch assay, that
miR520c inhibited cell motility and when we overexpressed STAT3 to
rescue the transformation, we found the inhibition was partially
abrogated (Fig. 4B). Similarly,
miR520c suppressed cells invasion and migration, which could be
rescued by STAT3 (Fig. 4C). These
results demonstrated, miR520c suppressed STAT3, which led to
inhibition of EMT.
Discussion
STAT3 is hyper-activated in cancers, approaches to
inhibit STAT3 can be a key therapeutic task. In glioblastoma,
BIS-mediated STAT3 stabilization can regulate stem cell-like
phenotypes (26). In gastric
cancer, suppressing the activation of STAT3 resensitized cells to
chemotherapy (27). Inhibition of
STAT3 has influence on cell cytotoxic and cytostatic effect, it
also reinforces anticancer immunosurveillance to increase
therapeutic efficacy and stem cell-like phenotype (28). In human Wilms tumor SK-NEP-1 cells,
inhibiting STAT3 and CDDP reduced cell growth in vivo
(29).
Besides, STAT3 can be downregulated by series of
microRNAs. miR-23a, miR-27a and miR-24 coordinately exert
JAK1/Stat3 cascade in human acute erythroid leukemia (30). miR-124 repressed proliferation in
glioblastoma by targeting STAT3 (31). miR-125a promoted paclitaxel
sensitivity via downregulation of STAT3 in cervical cancer
(32).
It has been reported that mir520c also have
functions in cancers. In isolated melanoma cancer stem cells,
mir520c was identified to have relations with
epithelial-to-mesenchymal transition and stem cell potential
(33). miR-520c-3p suppresses
diffuse large B cell lymphoma development by decreasing eIF4GII
(34). Mir520c has dual functions
in different types of cancers. It can act as a tumor suppressor,
but it is also an oncomiR. The role of miR-520c in regulating
activities of MMP2 and MMP9 was contrary (35). The mechanisms of miR520c inhibition
in breast cancer carcinogenesis have not been fully illuminated. In
our study, we selected miR520c as a potential regulator of STAT3.
To make clear whether it promoted cancer progression, we screened
the expression level of miR520c as well as STAT3, consistent with
our expectations, with the rising of malignant grades, the mRNA
level became lower and lower. Nevertheless, the
post-transcriptional regulation of STAT3 in breast cancer may be
multifactorial, including microRNAs. Afterwards, we validated
miR520c bound with STAT3 3′UTR but not with the mutated one.
Conversely, when we inhibited miR520c, the binding effect was
attenuated. Furthermore, we verified overexpression of miR520c
strongly decreased protein level of STAT3. It has been reported
that STAT3 facilitates inflammation, proliferation, stem cell
phenotype, metastasis. In our previous study, we detected apoptosis
of breast cancer cells after transfected with miR520c, but no
significant differences was observed. However, it has suppressing
influence on EMT phenotypes. The epithelial marker E-cadherin was
increased, on the contrary, mesenchymal marker vimentin was
decreased. Cell motility and invasion ability were abrogated, but
the rescue experiments partially recovered the repression effects,
which means other mechanisms are needed to be regulated by
miR520c.
Taken together, our data indicate STAT3 was a direct
target of miR520c in breast cancer cell, and through this
mechanism, the EMT process was repressed. It may lay a basis for
breast cancer therapy and prognostic evaluation. Further work will
be devoted to uncover the relationships between miR520c and STAT3
in cancer microenvironments and clinical cases.
References
|
1
|
De Martel C, Ferlay J, Franceschi S,
Vignat J, Bray F, Forman D and Plummer M: Global burden of cancers
attributable to infections in 2008: A review and synthetic
analysis. Lancet Oncol. 13:607–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parker JS, Mullins M, Cheang MC, Leung S,
Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al:
Supervised risk predictor of breast cancer based on intrinsic
subtypes. J Clin Oncol. 27:1160–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yaacoub K, Pedeux R, Tarte K and
Guillaudeux T: Role of the tumor microenvironment in regulating
apoptosis and cancer progression. Cancer Lett. 378:150–159. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma YF, Ren Y, Wu CJ, Zhao XH, Xu H, Wu DZ,
Xu J, Zhang XL and Ji Y: Interleukin (IL)-24 transforms the tumor
microenvironment and induces anticancer immunity in a murine model
of colon cancer. Mol Immunol. 75:11–20. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sweis RF, Spranger S, Bao R, Paner GP,
Stadler WM, Steinberg GD and Gajewski TF: Molecular drivers of the
non-T cell-inflamed tumor microenvironment in urothelial bladder
cancer. Cancer Immunol Res. 4:563–568. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fernandez-Garcia B, Eiró N, Miranda MA,
Cid S, González LO, Domínguez F and Vizoso FJ: Prognostic
significance of inflammatory factors expression by stroma from
breast carcinomas. Carcinogenesis. 37:768–776. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dieci MV, Griguolo G, Miglietta F and
Guarneri V: The immune system and hormone-receptor positive breast
cancer: Is it really a dead end? Cancer Treat Rev. 46:9–19. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Griesinger AM, Josephson RJ, Donson AM,
Levy JM Mulcahy, Amani V, Birks DK, Hoffman LM, Furtek SL, Reigan
P, Handler MH, et al: Interleukin-6/STAT3 pathway signaling drives
an inflammatory phenotype in group A ependymoma. Cancer Immunol
Res. 3:1165–1174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bromberg JF, Wrzeszczynska MH, Devgan G,
Zhao Y, Pestell RG, Albanese C and Darnell JE Jr.: Stat3 as an
oncogene. Cell. 98:295–303. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garcia R and Jove R: Activation of STAT
transcription factors in oncogenic tyrosine kinase signaling. J
Biomed Sci. 5:79–85. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meltzer PS: Cancer genomics: Small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Benhamed M, Herbig U, Ye T, Dejean A and
Bischof O: Senescence is an endogenous trigger for
microRNA-directed transcriptional gene silencing in human cells.
Nat Cell Biol. 14:266–275. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kunej T, Godnic I, Ferdin J, Horvat S,
Dovc P and Calin GA: Epigenetic regulation of microRNAs in cancer:
An integrated review of literature. Mutat Res. 717:77–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ferdin J, Kunej T and Calin GA: Non-coding
RNAs: Identification of cancer-associated microRNAs by gene
profiling. Technol Cancer Res Treat. 9:123–138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fabbri M, Ivan M, Cimmino A, Negrini M and
Calin GA: Regulatory mechanisms of microRNAs involvement in cancer.
Expert Opin Biol Ther. 7:1009–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang YK, Xi WY, Xi RX, Li JY, Li Q and Gao
YE: MicroRNA-494 promotes cervical cancer proliferation through the
regulation of PTEN. Oncol Rep. 33:2393–2401. 2015.PubMed/NCBI
|
|
21
|
Libório-Kimura TN, Jung HM and Chan EK:
miR-494 represses HOXA10 expression and inhibits cell proliferation
in oral cancer. Oral Oncol. 51:151–157. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ward A, Shukla K, Balwierz A, Soons Z,
König R, Sahin O and Wiemann S: MicroRNA-519a is a novel oncomir
conferring tamoxifen resistance by targeting a network of
tumour-suppressor genes in ER+ breast cancer. J Pathol.
233:368–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keklikoglou I, Koerner C, Schmidt C, Zhang
JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T,
Schneeweiss A, et al: MicroRNA-520/373 family functions as a tumor
suppressor in estrogen receptor negative breast cancer by targeting
NF-κB and TGF-β signaling pathways. Oncogene. 31:4150–4163. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu LY, Chang LY, Kuo WH, Hwa HL, Lin YS,
Jeng MH, Roth DA, Chang KJ and Hsieh FJ: Correction to: Prognostic
features of signal transducer and activator of transcription 3 in
an ER(+) breast cancer model system. Cancer Inform. 13:125–129.
2014.PubMed/NCBI
|
|
26
|
Im CN, Yun HH, Song B, Youn DY, Cui MN,
Kim HS, Park GS and Lee JH: BIS-mediated STAT3 stabilization
regulates glioblastoma stem cell-like phenotypes. Oncotarget.
7:35056–35070. 2016.PubMed/NCBI
|
|
27
|
Yang H, Yamazaki T, Pietrocola F, Zhou H,
Zitvogel L, Ma Y and Kroemer G: Improvement of immunogenic
chemotherapy by STAT3 inhibition. OncoImmunology. 5:e10780612015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang JL, Liu XZ, Wang PY, Chen GW, Jiang
Y, Qiao SK, Zhu J, Wang X, Pan YS and Liu YC: Targeting HCCR
expression resensitizes gastric cancer cells to chemotherapy via
down-regulating the activation of STAT3. Sci Rep. 6:241962016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Zhang N, Qu H, You G, Yuan J, Chen
C, Li W and Pan F: Inhibitory effect of STAT3 gene combined with
CDDP on growth of human Wilms tumour SK-NEP-1 cells. Biosci Rep.
36:e003422016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su R, Dong L, Zou D, Zhao H, Ren Y, Li F,
Yi P, Li L, Zhu Y, Ma Y, et al: microRNA-23a, −27a and −24
synergistically regulate JAK1/Stat3 cascade and serve as novel
therapeutic targets in human acute erythroid leukemia. Oncogene.
35:6001–6014. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li W, Huang H, Su J, Ji X, Zhang X, Zhang
Z and Wang H: miR-124 acts as a tumor suppressor in glioblastoma
via the inhibition of signal transducer and activator of
transcription 3. Mol Neurobiol. Mar 18–2016.(Epub ahead of
print).
|
|
32
|
Fan Z, Cui H, Yu H, Ji Q, Kang L, Han B,
Wang J, Dong Q, Li Y, Yan Z, et al: MiR-125a promotes paclitaxel
sensitivity in cervical cancer through altering STAT3 expression.
Oncogenesis. 5:e1972016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fomeshi MR, Ebrahimi M, Mowla SJ,
Khosravani P, Firouzi J and Khayatzadeh H: Evaluation of the
expressions pattern of miR-10b, 21, 200c, 373 and 520c to find the
correlation between epithelial-to-mesenchymal transition and
melanoma stem cell potential in isolated cancer stem cells. Cell
Mol Biol Lett. 20:448–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mazan-Mamczarz K, Zhao XF, Dai B,
Steinhardt JJ, Peroutka RJ, Berk KL, Landon AL, Sadowska M, Zhang
Y, Lehrmann E, et al: Down-regulation of eIF4GII by miR-520c-3p
represses diffuse large B cell lymphoma development. PLoS Genet.
10:e10041052014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu S, Zhu Q, Zhang Y, Song W, Wilson MJ
and Liu P: Dual-Functions of miR-373 and miR-520c by differently
regulating the activities of MMP2 and MMP9. J Cell Physiol.
230:1862–1870. 2015. View Article : Google Scholar : PubMed/NCBI
|