Introduction
Gastric cancer (GC) is the fourth most frequently
occurring malignant tumor, and exhibits considerable geographic
variation. GC ranks as the second leading cause of cancer-related
deaths worldwide. Environmental factors and the accumulation of
genetic alterations are currently thought to be the leading causes
of GC. Benefiting from advances in the early diagnosis and adjuvant
treatment of GC, the 5-year disease-free survival rate of patients
has increased in the last few decades. However, many patients who
are treated with such therapies still experience disease
progression (1). Thus, new
treatment choices are critically required.
Tanshinone IIA (TSN), a commonly used Chinese
traditional drug for the treatment of cardiovascular and
cerebrovascular diseases such as atherosclerosis, angina pectoris
and acute ischemic stroke (2,3), has
also exhibited a variety of anticancer effects since Wang et
al first reported that TSN suppressed breast cancer progression
by inhibiting the proliferation and by inducing apoptosis of cancer
cells (4). Subsequent research
further confirmed the anticancer effect of TNS on esophageal,
prostate, colorectal, lung and GC (5–8).
However, the mechanism by which TSN suppresses GC progression
remains unclear.
The forkhead box M1 (FOXM1) gene is a member of the
FOX family, and has been shown to play important roles in cell fate
decisions. In tumor genesis, numerous studies have shown that FOXM1
is significantly increased in multiple human cancers such as
esophageal and breast cancer, hepatocellular carcinoma, colorectal
cancer and GC (9–12). In addition, its overexpression is
closely correlated with tumor progression and metastasis (11) and downregulation of FOXM1 inhibits
tumor progression. However, the function of FOXM1 in TSN-induced
inhibition of gastric tumor metastasis has not been reported.
In the present study, we demonstrated that TSN
inhibits the proliferation and migration of GC cells, and also
demonstrated that downregulation of FOXM1 is the key underlying
mechanism.
Materials and methods
Cell culture and transfection
The human GC cell line (SGC-7901) was obtained from
the American Type Culture Collection (ATCC; Rockville, MD, USA) and
kept in RPMI-1640 medium with 10% fetal bovine serum (FBS) (Gibco,
Carlsbad, CA, USA), 1% of 100 U/ml penicillin and 1% of 100 mg/ml
streptomycin sulfates. The cells were incubated in humidified
incubators with 5% CO2 at 37̊C.
Human FOXM1 gene or FOXM1-siRNA was constructed into
the pcDNA3.1+HA vector by Life Technologies (GeneChem, Shanghai,
China), and the empty vector served as the negative control. For
transfection, after the cells were cultured to 70–80% confluency,
pcDNA3.1+HA-FOXM1, pcDNA3.1+HA empty vector, pcDNA3.1+FOXM1-siRNA
or pcDNA3.1+NC-siNRA was transfected using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
instructions.
Quantitative real-time PCR
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen), and then miRNAs were
reverse-transcribed to cDNA using a reverse transcription kit
(Takara, Tokyo, Japan). Quantitative real-time PCR (qRT-PCR) was
performed using SYBR-Green PCR kit on an ABI 7500 Fast Real-Time
PCR system according to the manufacturer's instructions. The
expression of FOXM1 mRNA was normalized to U6. All experiments were
carried out in triplicate. The 2−ΔΔCt calculation method
was used to calculate the relative expression of the genes.
Cell proliferation assay
The proliferation ability of SGC-7901 cells was
assessed by the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich, St. Louis, MO, USA) assay. Approximately
104 cells were seeded into each well of 96-well plates.
Then, SGC-7901 cells were transfected with pcDNA3.1+HA-FOXM1,
pcDNA3.1+HA empty vector, pcDNA3.1+FOXM1-siRNA or pcDNA3.1+NC-siNRA
following the manufacturer's instructions. After 6 h of
transfection, the cells were treated with TSN or a placebo. After
12, 24 or 48 h of incubation, 25 µl of MTT (5 mg/ml) was added to
each well and the plates were incubated for 4 h at 37°C. Then, the
precipitates in each well were solubilized with 150 µl of dimethyl
sulfoxide (DMSO; Sigma-Aldrich), and the plates were read on a
microplate reader (Anthos Labtec Instruments, Salzburg, Austria) at
490 nm. Values were normalized using the control value.
Cell migration assay
The migration ability of the SGC-7901 cells was
measured by a wound healing assay. Approximately 106
cells were seeded into each well of 6-well plates. Then, SGC-7901
cells were transfected with pcDNA3.1+HA-FOXM1, pcDNA3.1+HA empty
vector, pcDNA3.1+FOXM1-siRNA or pcDNA3.1+NC-siNRA following the
manufacturer's instructions. After 6 h of transfection, a cell
scratch spatula was used to scratch the cell layer when cells
reached ~90% confluency. After being washed with warm
phosphate-buffered saline (PBS) 3 times, the cells were treated
with TSN or a placebo. Then, the cells continued to be incubated at
37°C for 18 h. A digital camera system (Olympus Corp., Tokyo,
Japan) was used to acquire images of the scratches of the cells
after incubation at 0 and 18 h.
Western blotting
Cells after treatment, were extracted with RIPA
lysis buffer (Biyuntian, Hangzhou, China). Protein lysates were
then separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinylidene
fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After
blocking with 5% non-fat milk for 2 h at room temperature, the
membranes were incubated with the primary antibodies: P21
(1:1,000), Ki-67 (1:1,000), PCNA (1:1,000), MMP-2 (1:1,000), MMP-9
(1:2,000), FOXM1 (1:1,000) and β-actin (1:1,000) (all from Abcam,
Cambridge, MA, USA) overnight at 4°C. Then, the membranes were
incubated in HRP-linked secondary antibodies (Santa Cruz
Biotechnology, Santa Cruz, CA, USA) for 2 h. Western blotting
signals were detected using the ECL Plus kit (Biyuntian). Each
experiment was repeated 3 times, independently.
Statistical analysis
All statistical analyses were performed using SPSS
20.0. Data are presented as the mean ± SD. Differences between
groups were analyzed using Student's t-test or one-way ANOVA
analysis. All experiments were repeated at least 3 times. The value
of p<0.05 was considered to indicate a statistically significant
result.
Results
TSN inhibits SGC-7901 cell
proliferation
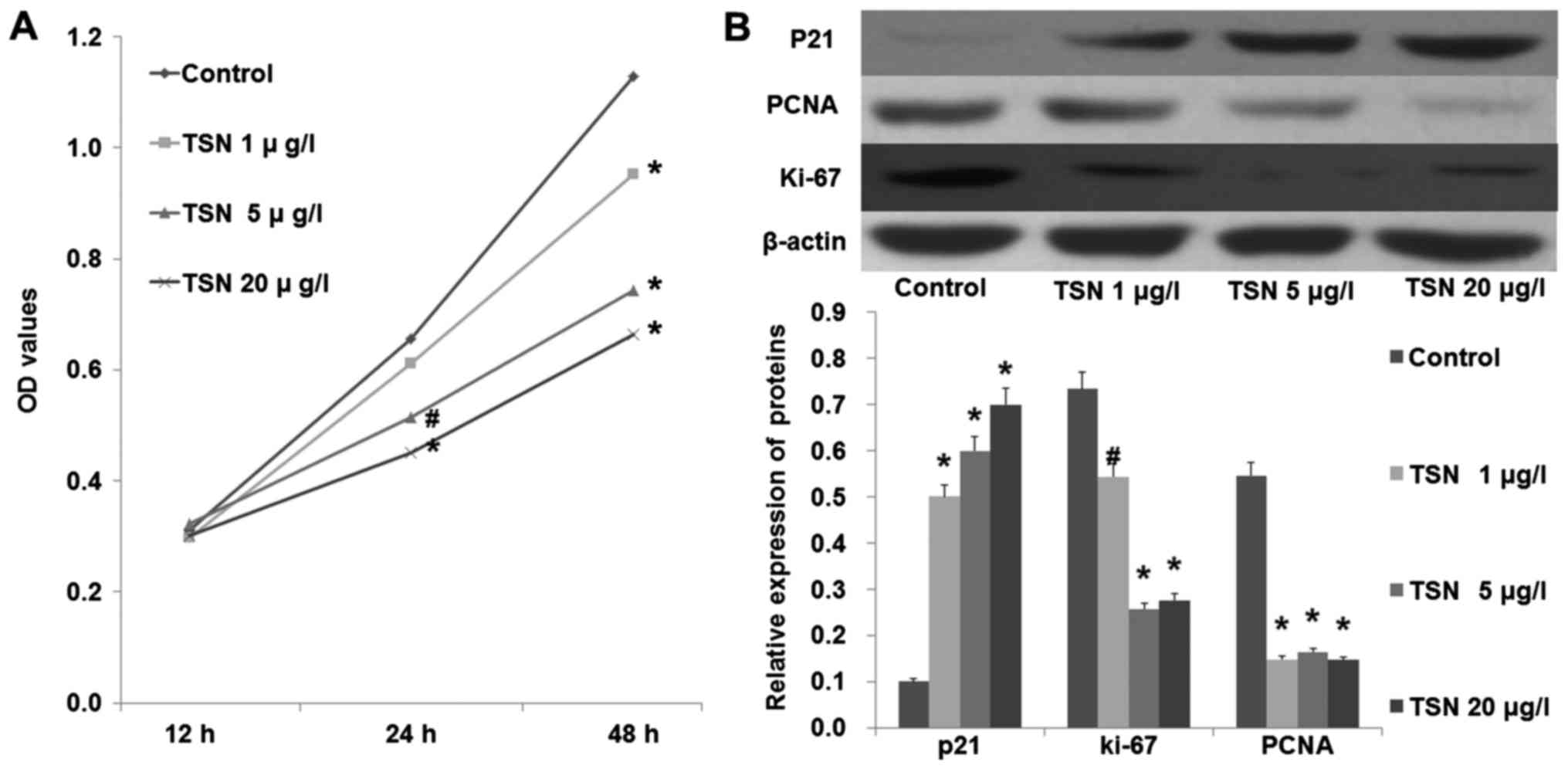
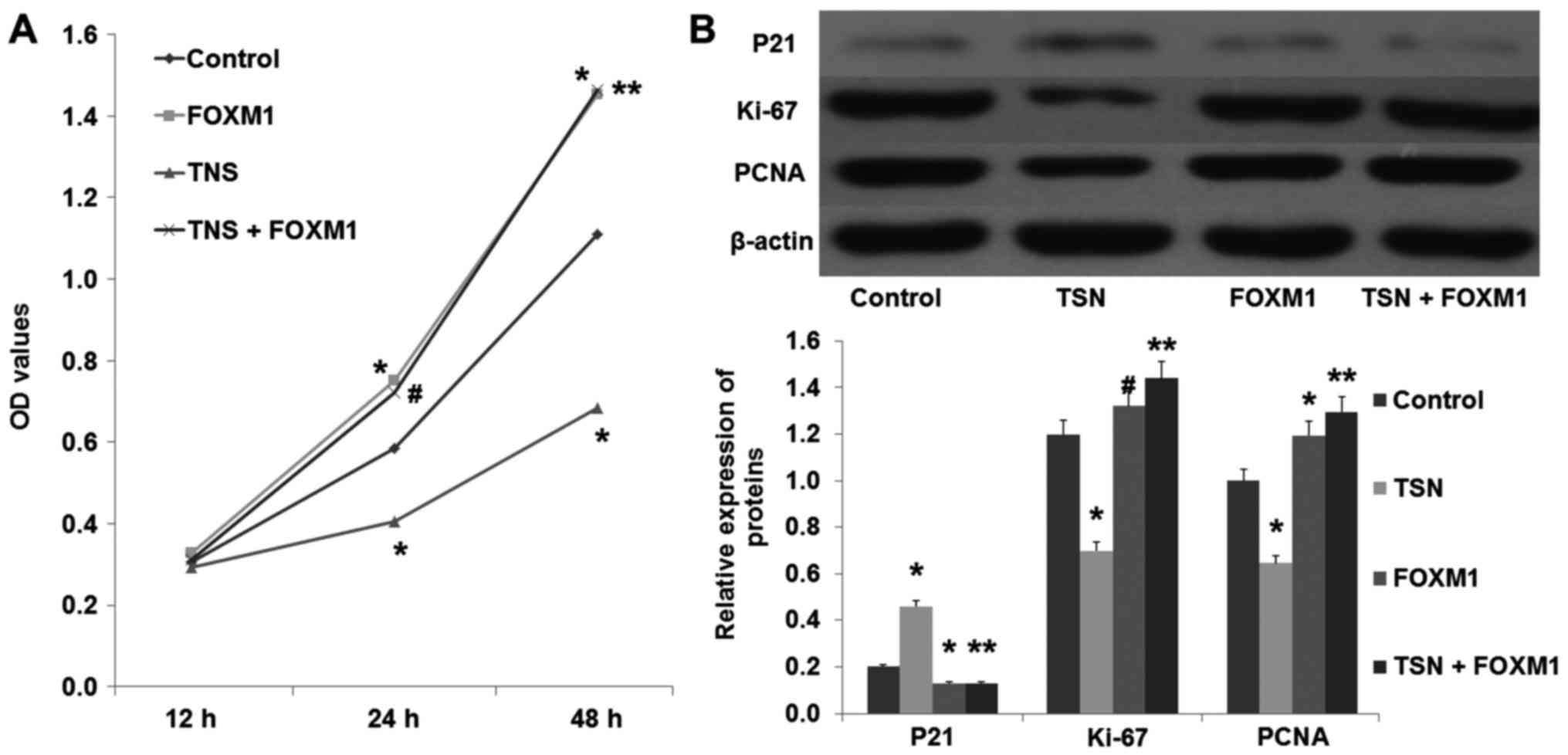
An MTT assay was performed to test the effect of TSN
on SGC-7901 cell proliferation. As shown in Fig. 1A, TSN markedly inhibited the
proliferation ability of SGC-7901 cells in a dose-dependent manner
(p<0.05). Moreover, we also found that TSN increased the
expression of P21 and decreased the expression levels of PCNA and
Ki-67 in the SGC-7901 cells in a dose-dependent manner (Fig. 1B; p<0.05). These results
indicated that TSN inhibited SGC-7901 cell proliferation.
TSN suppresses SGC-7901 cell
migration
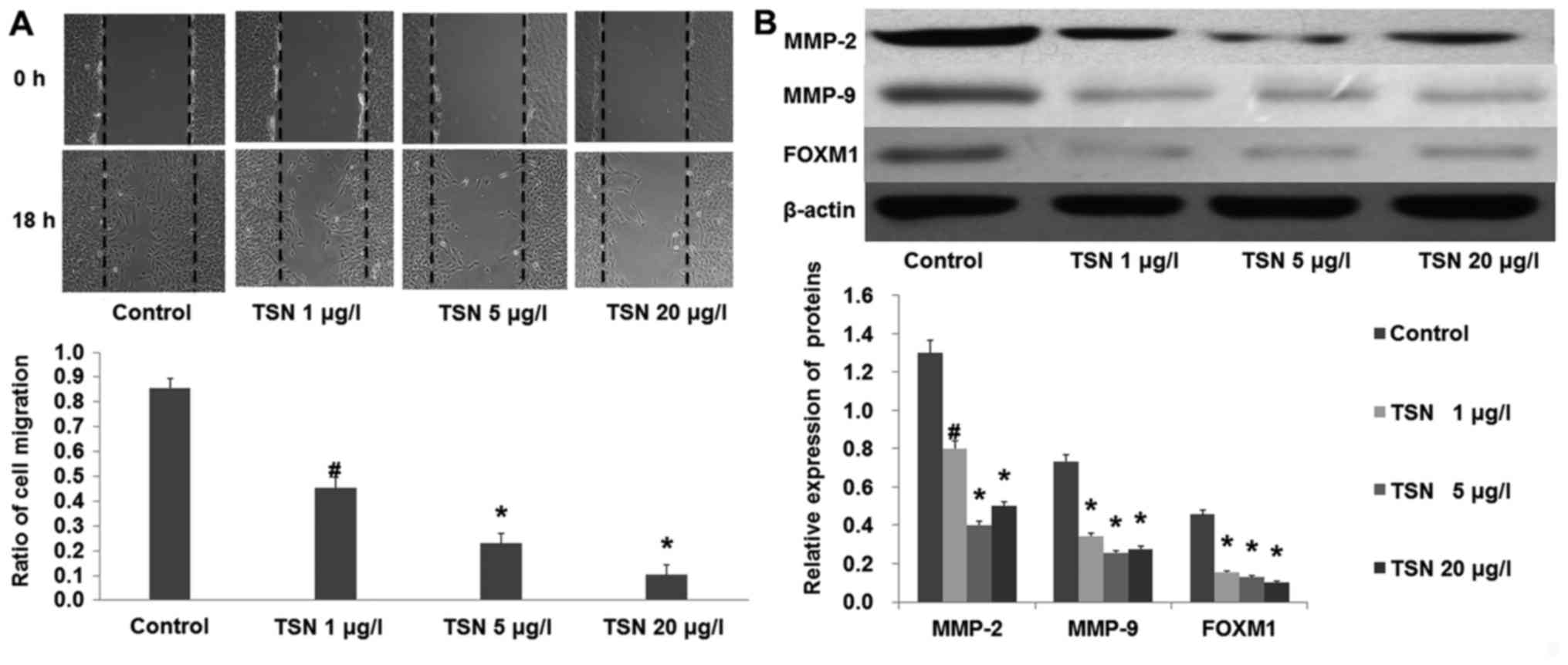
Cells were incubated with increasing concentrations
of TSN. Images of the scratches were captured at 0 and 18 h after
TSN was added. We found that TSN markedly inhibited SGC-7901 cell
migration after 18 h (Fig. 2A;
p<0.01). In agreement, the results from western blot analysis
showed that TSN also suppressed the expression of MMP-2 and MMP-9
(Fig. 2B). These results indicated
that TSN inhibited SGC-7901 cell migration.
TSN decreases the expression of FOXM1
in SGC-7901 cells
After SGC-7901 cells were incubated with increasing
concentrations of TSN for 48 h, western blot analysis showed that
the expression of FOXM1 was significantly decreased by TSN in a
dose-dependent manner (Fig. 2B;
p<0.01). Considering the critical role of FOXM1 in the
development of cancer, these results prompted us to focus on FOXM1
in the following experiment.
Downregulation of FOXM1 inhibits
SGC-7901 cell proliferation and migration similarly to TSN
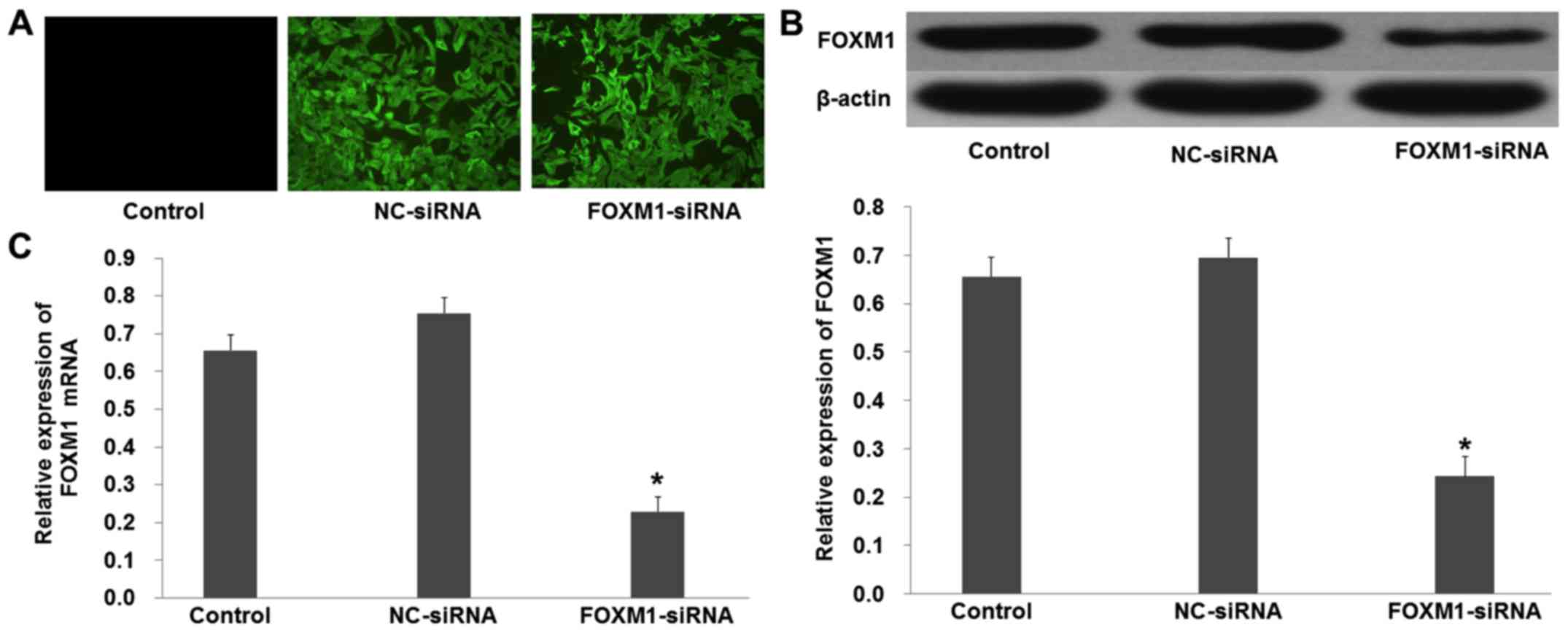
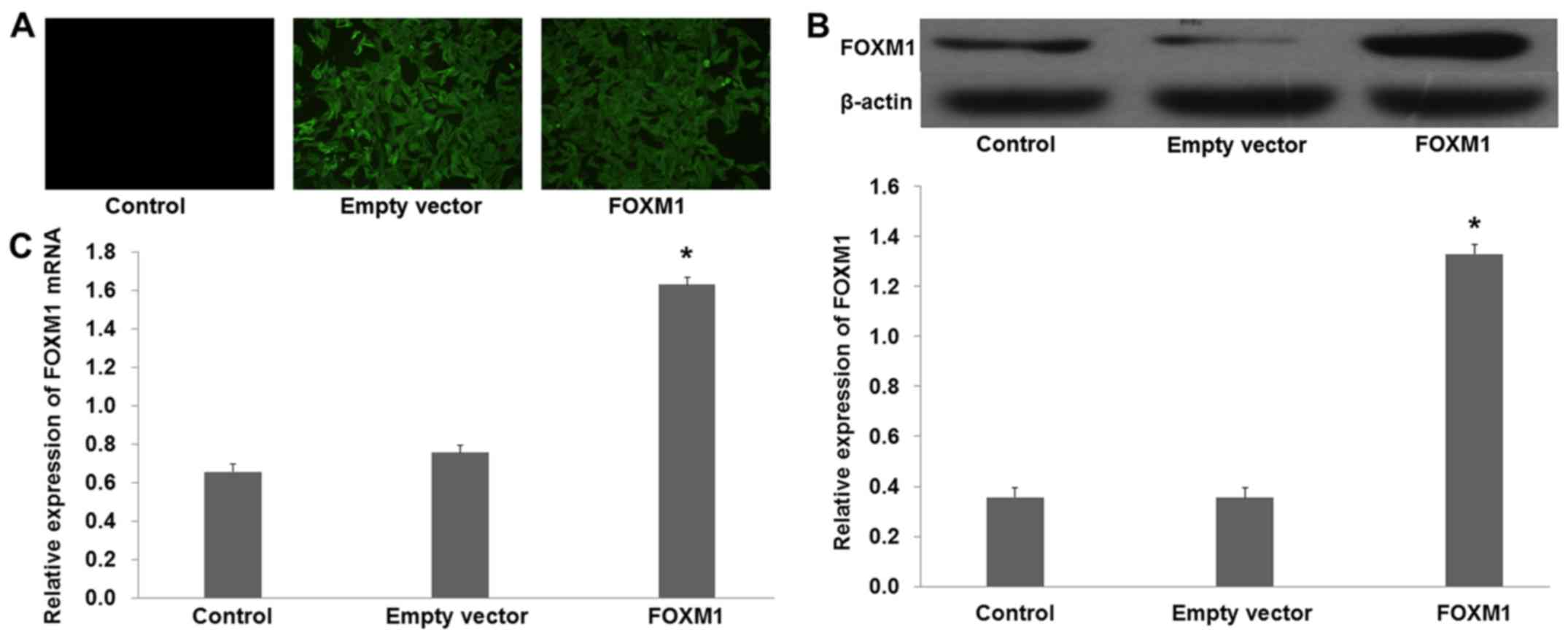
FOXM1-siRNA was used to knock out the expression of
FOXM1 in the SGC-7901 cells. Fluorescence showed that the plasmid
was successfully transfected into the SGC-7901 cells (Fig. 3A). Western blot analysis showed that
the protein levels of FOXM1 were decreased in the cells transfected
with pcDNA3.1+FOXM1-siRNA compared with the NC-siRNA (Fig. 3B; p<0.01). Moreover, quantitative
real-time PCR demonstrated similar results that were obtained from
the western blot analysis (Fig. 3C;
p<0.01).
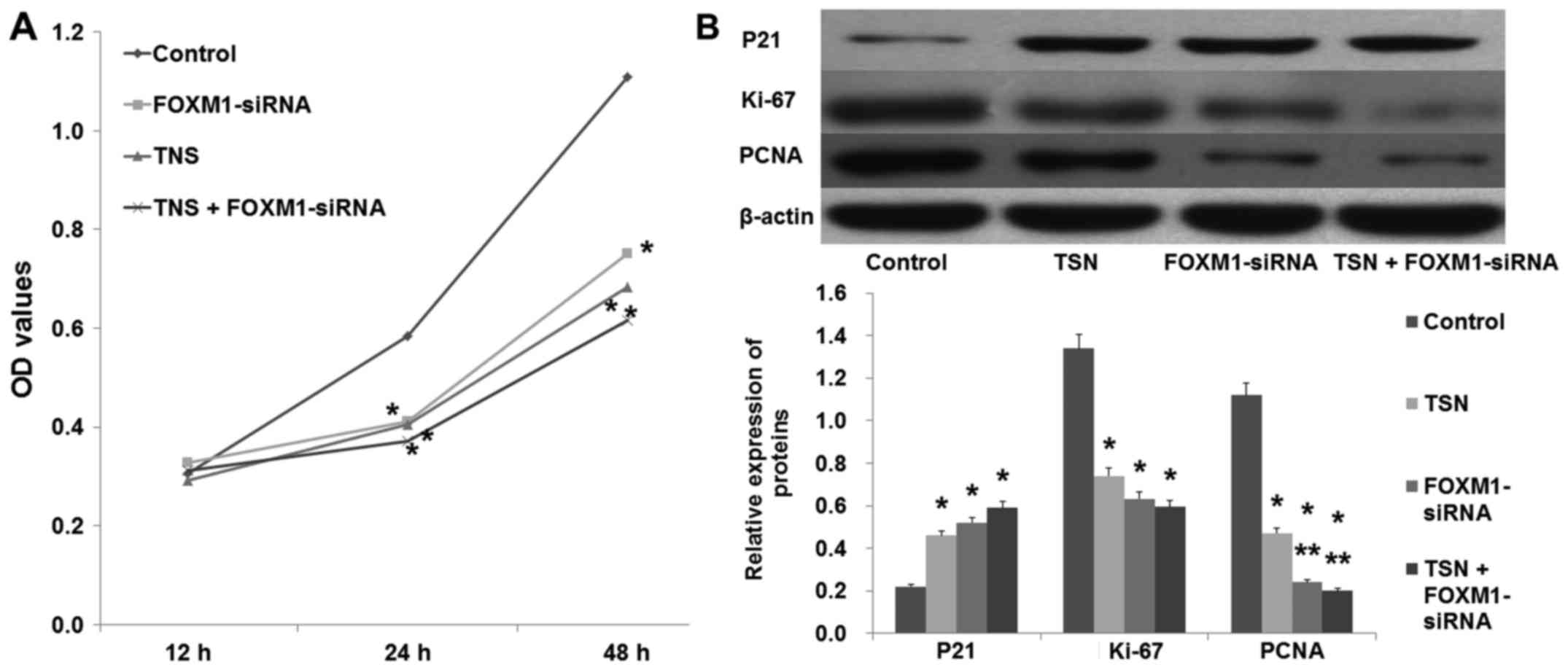
In a similar manner to TSN (5 µg/l), downregulation
of FOXM1 by FOXM1-siRNA also suppressed SGC-7901 proliferation
(Fig. 4A; p<0.05). In agreement,
western blot analysis revealed that similarly to TSN,
downregulation of FOXM1 increased the expression of P21 and
decreased the expression of Ki-67 and PCNA (Fig. 4B; p<0.05). In addition, the
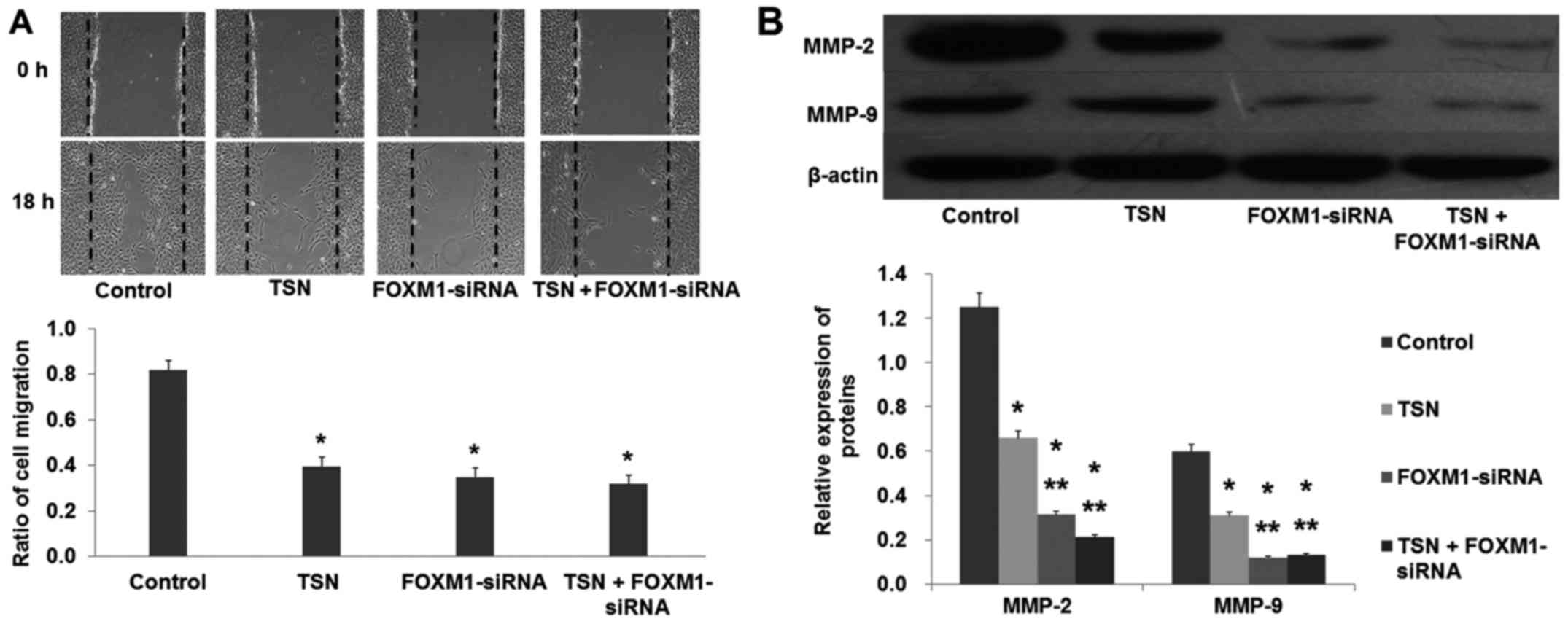
migration ability of SGC-7901 cells was also suppressed after being
transfected with FOXM1-siRNA (Fig.
5; p<0.05). These data showed that downregulation of FOXM1
had the same effect as TSN on SGC-7901 cell proliferation and
migration.
Overexpression of FOXM1 reverses the
effect of TSN on SGC-7901 cells
To further elucidate the role of FOXM1 in
TSN-induced inhibition of SGC-7901 cell proliferation and
migration, pcDNA3.1+HA-FOXM1 was transfected into SGC-7901 cells to
increase the expression of FOXM1. Fluorescence revealed that the
plasmid was successfully transfected into the SGC-7901 cells
(Fig. 6A). Western blot analysis
demonstrated that the protein level of FOXM1 was increased in the
cells transfected with pcDNA3.1+HA-FOXM1 compared with the empty
vector (Fig. 6B; p<0.01).
Furthermore, quantitative real-time PCR revealed similar results to
those obtained from the western blot analysis (Fig. 6C; p<0.01).
After transfection with pcDNA3.1+HA-FOXM1 or the
empty plasmid, SGC-7901 cells were incubated with TSN (5 µg/l) or
the vehicle. Compared with the control group, overexpression of
FOXM1 increased the proliferation ability of the SGC-7901 cells
(Fig. 7A; p<0.01). In addition,
we found that overexpression of FOXM1 also decreased the expression
of P21 and increased the expression of PCNA and Ki-67 (Fig. 7B; p<0.01). Moreover, compared
with the TSN group, overexpression of FOXM1 not only suppressed the
TSN-induced inhibition of SGC-7901 proliferation, but also reversed
the TSN-induced decreased expression of PCNA and Ki-67 (Fig. 4; p<0.01). These results suggested
that FOXM1 was involved in TSN-induced inhibition of SGC-7901 cell
proliferation.
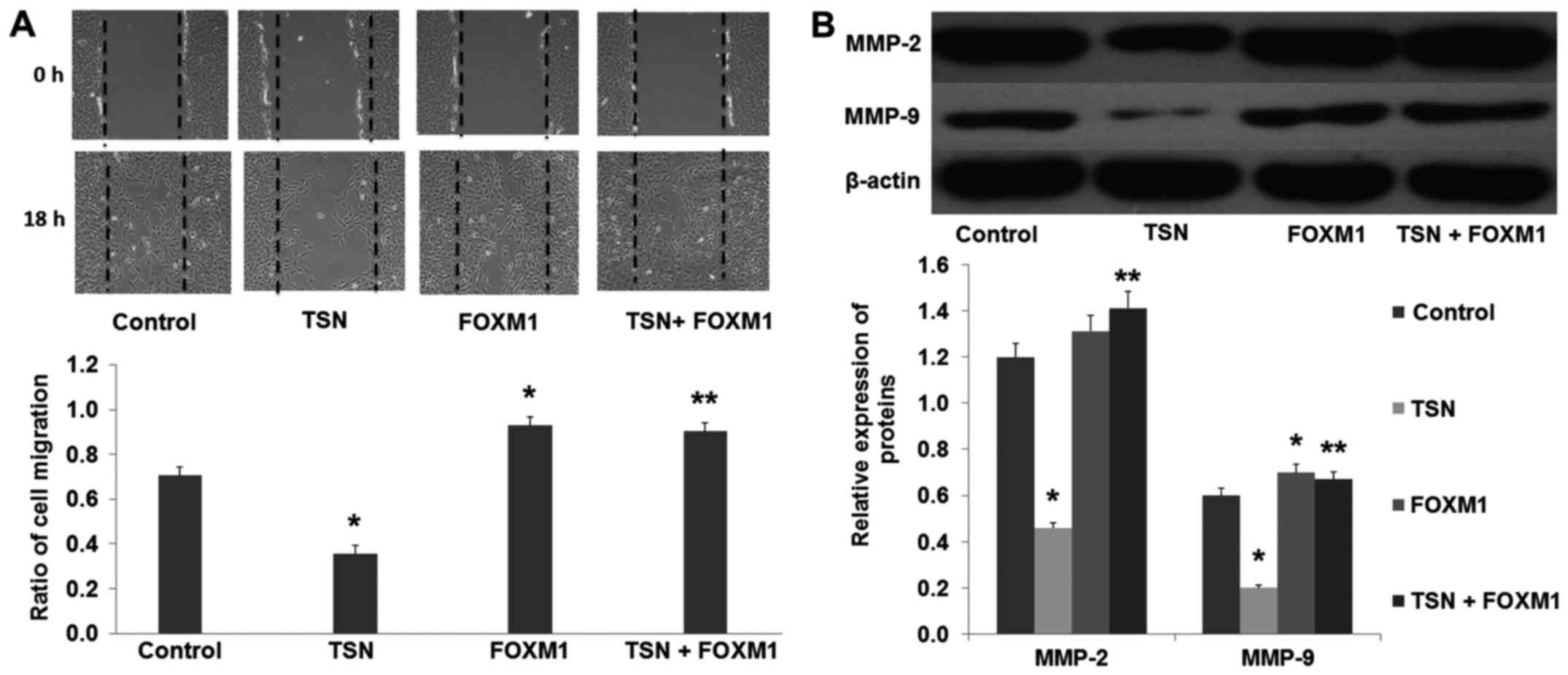
The results from the wound healing assay showed that
overexpression of FOXM1 increased the migration ability of the
SGC-7901 cells compared with the control group (Fig. 8A; p<0.01). Moreover, we found
that overexpression of FOXM1 also increased the expression of MMP-2
and MMP-9 (Fig. 8B; p<0.01). In
addition, our results revealed that compared with the TSN group,
overexpression of FOXM1 suppressed the TSN-induced inhibition of
SGC-7901 cell migration, and reversed the TSN-induced decreased
expression of MMP-2 and MMP-9 (Fig.
8; p<0.01). These results strongly suggested that FOXM1 was
involved in the TSN-induced inhibition of SGC-7901 cell
migration.
Discussion
Abnormal proliferation and migration of tumor cells
are crucial pathological processes involved in malignant tumors
(13–16). Tumor metastasis is a complex process
which includes migratory tumor cells leaving the primary tumor by
invasion, disseminating throughout the body via the circulatory
system, and colonizing eventually at distant organs (17). It has already been proven that tumor
cells acquiring the excessive ability of proliferation and
migration and departing from the original locality is a
prerequisite for metastasis. Therefore, it is important to find an
effective anti-gastric cancer (GC) cell proliferation and migration
approach in order to improve the prognosis of patients with GC.
Tanshinone is a herbal medicine derived from the
dried root of Salvia miltiorrhiza, which has been widely
used in China for hundreds of years. Recently, the pharmacological
properties of tanshinone have attracted great interest. Tanshinone
IIA (TNS) is the main component of tanshinone, and a variety of
clinical trials and experimental studies have demonstrated the
protective effect of TSN on cardiovascular diseases, diabetes and
tumors (2,18–21).
In GC, Jing et al first reported that TSN induced apoptosis
and growth inhibition in vitro and in vivo.
Subsequent research showed that TSN could reverse the malignant
phenotype of GC cells and induce pro-survival autophagy in GC cells
(22,23). In accordance with these studies, our
research found that TSN inhibited the proliferation and migration
ability of SGC-7901 cells in a dose-dependent manner. As a
multi-target drug, the molecular targets of TSN include
apoptotic-regulating proteins, transcription and growth factors,
ion channels and inflammatory mediators (6,7,24,25).
In the present study, we found that TSN decreases the expression of
FOXM1 in SGC-7901 cells in a dose-dependent manner.
FOXM1 is an important transcription factor required
for tissue development and differentiation in vertebrates (26). FOXM1 binds to sequence-specific
motifs on DNA (C/TAAACA) through its DNA-binding domain (DBD) and
activates proliferation-, migration- and EMT-associated genes.
Aberrant overexpression of FOXM1 is a key feature in oncogenesis
and progression of many types of human cancer (27). Recently, overexpression of FOXM1 was
correlated with the poor prognosis of patients with malignant
tumors and has been reported in many types of cancers including GC
(28–32). Zhang et al reported that
downregulation of FOXM1 suppressed PLK1-regulated cell cycle
progression in renal cancer cells (33). Additionally, Inoguchi et al
found that microRNA-24-1 inhibited bladder cancer cell
proliferation by targeting FOXM1 (34). Therefore, we inferred that TSN
inhibited SGC-7901 cell proliferation and migration via the
downregulation of FOXM1. Consistent with these studies, our results
showed that knockdown of FOXM1 by siRNA had the same effect as TSN
on SGC-7901 cells including suppression of cell proliferation and
migration, inhibition of the expression of Ki-67, PCNA and MMP-2/−9
and an increase in the expression of P21, which indicated that
FOXM1 plays an important role in the regulation of SGC-7901 cell
proliferation and migration. Additionally, we also found that
overexpression of FOXM1 increases the expression of Ki-67, PCNA,
MMP-2/−9 and promotes the proliferation and migration abilities of
the SGC-7901 cells. Moreover, our results demonstrated that
overexpression of FOXM1 reverses TSN-induced inhibition of SGC-7901
cell proliferation and migration. These results demonstrated that
TSN inhibits SGC-7901 cell proliferation and migration via the
downregulation of FOXM1.
In summary, the present study provides new insights
into the effect of TSN on SGC-7901 cells and the related mechanism.
The present study suggests that TSN inhibits proliferation and
migration of SGC-7901 cells through, at least in part, the
downregulation of FOXM1.
References
|
1
|
Wang T, Chen T, Niu H, Li C, Xu C, Li Y,
Huang R, Zhao J and Wu S: MicroRNA-218 inhibits the proliferation
and metastasis of esophageal squamous cell carcinoma cells by
targeting BMI1. Int J Mol Med. 36:93–102. 2015.PubMed/NCBI
|
|
2
|
Xu S, Little PJ, Lan T, Huang Y, Le K, Wu
X, Shen X, Huang H, Cai Y, Tang F, et al: Tanshinone II-A
attenuates and stabilizes atherosclerotic plaques in
apolipoprotein-E knockout mice fed a high cholesterol diet. Arch
Biochem Biophys. 515:72–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Jiang Q, Wan L, Yang K, Zhang Y,
Chen Y, Wang E, Lai N, Zhao L, Jiang H, et al: Sodium tanshinone
IIA sulfonate inhibits canonical transient receptor potential
expression in pulmonary arterial smooth muscle from pulmonary
hypertensive rats. Am J Respir Cell Mol Biol. 48:125–134. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Wei Y, Yuan S, Liu G, Lu Y, Zhang
J and Wang W: Potential anticancer activity of tanshinone IIA
against human breast cancer. Int J Cancer. 116:799–807. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang HS, Zhang FJ, Li H, Liu Y, Du GY and
Huang YH: Tanshinone IIA inhibits human esophageal cancer cell
growth through miR-122-mediated PKM2 down-regulation. Arch Biochem
Biophys. 598:50–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu S, Granet R, Mbakidi JP, Brégier F,
Pouget C, Micallef L, Sothea-Ouk T, Leger DY, Liagre B, Chaleix V,
et al: Delivery of tanshinone IIA and α-mangostin from
gold/PEI/cyclodextrin nanoparticle platform designed for prostate
cancer chemotherapy. Bioorg Med Chem Lett. 26:2503–2506. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu M, Wang C and Wang J: Tanshinone I
induces human colorectal cancer cell apoptosis: The potential roles
of Aurora A-p53 and survivin-mediated signaling pathways. Int J
Oncol. 49:603–610. 2016.PubMed/NCBI
|
|
8
|
Kim EO, Kang SE, Im CR, Lee JH, Ahn KS,
Yang WM, Um JY, Lee SG and Yun M: Tanshinone IIA induces TRAIL
sensitization of human lung cancer cells through selective ER
stress induction. Int J Oncol. 48:2205–2212. 2016.PubMed/NCBI
|
|
9
|
Sanders DA, Ross-Innes CS, Beraldi D,
Carroll JS and Balasubramanian S: Genome-wide mapping of FOXM1
binding reveals co-binding with estrogen receptor alpha in breast
cancer cells. Genome Biol. 14:R62013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ahmed M, Hussain AR, Siraj AK, Uddin S,
Al-Sanea N, Al-Dayel F, Al-Assiri M, Beg S and Al-Kuraya KS:
Co-targeting of Cyclooxygenase-2 and FoxM1 is a viable strategy in
inducing anticancer effects in colorectal cancer cells. Mol Cancer.
14:1312015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang N, Xie Y, Li B, Ning Z, Wang A and
Cui X: FoxM1 influences mouse hepatocellular carcinoma metastasis
in vitro. Int J Clin Exp Pathol. 8:2771–2778. 2015.PubMed/NCBI
|
|
12
|
Zhang J, Zhang J, Cui X, Yang Y, Li M, Qu
J, Li J and Wang J: FoxM1: A novel tumor biomarker of lung cancer.
Int J Clin Exp Med. 8:3136–3140. 2015.PubMed/NCBI
|
|
13
|
Zhang Y, Li CF, Ma LJ, Ding M and Zhang B:
MicroRNA-224 aggrevates tumor growth and progression by targeting
mTOR in gastric cancer. Int J Oncol. 49:1068–1080. 2016.PubMed/NCBI
|
|
14
|
Kim HY, Cho Y, Kang H, Yim YS, Kim SJ,
Song J and Chun KH: Targeting the WEE1 kinase as a molecular
targeted therapy for gastric cancer. Oncotarget. Jun 23–2016.(Epub
ahead of print). doi: 10.18632/oncotarget.10231.
|
|
15
|
Kanda M, Shimizu D, Fujii T, Tanaka H,
Tanaka Y, Ezaka K, Shibata M, Takami H, Hashimoto R, Sueoka S, et
al: Neurotrophin receptor-interacting melanoma antigen-encoding
gene homolog is associated with malignant phenotype of gastric
cancer. Ann Surg Oncol. 23 Suppl 4:S532–S539. 2016. View Article : Google Scholar
|
|
16
|
Kanda M, Shimizu D, Fujii T, Tanaka H,
Shibata M, Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, et
al: Protein arginine methyltransferase 5 is associated with
malignant phenotype and peritoneal metastasis in gastric cancer.
Int J Oncol. 49:1195–1202. 2016.PubMed/NCBI
|
|
17
|
Han TS, Hur K, Xu G, Choi B, Okugawa Y,
Toiyama Y, Oshima H, Oshima M, Lee HJ, Kim VN, et al: MicroRNA-29c
mediates initiation of gastric carcinogenesis by directly targeting
ITGB1. Gut. 64:203–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu ZL, Wang JN, Wu XH, Xie HJ, Han Y, Guan
YT, Qin Y and Jiang JM: Tanshinone IIA prevents rat basilar artery
smooth muscle cells proliferation by inactivation of PDK1 during
the development of hypertension. J Cardiovasc Pharmacol Ther.
20:563–571. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang C, Du X, Yang R, Liu J, Xu D, Shi J,
Chen L, Shao R, Fan G, Gao X, et al: The prevention and treatment
effects of tanshinone IIA on oestrogen/androgen-induced benign
prostatic hyperplasia in rats. J Steroid Biochem Mol Biol.
145:28–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan X, Yang Y, Cheng J, Li P, Inoue I and
Zeng X: Unique action of sodium tanshinone II-A sulfonate (DS-201)
on the Ca2+ dependent BKCa activation in mouse cerebral arterial
smooth muscle cells. Eur J Pharmacol. 656:27–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu Q, Sheng L, Yang M, Zhu M, Huang X and
Li Q: Tanshinon IIA injection accelerates tissue expansion by
reducing the formation of the fibrous capsule. PLoS One.
9:e1057562014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jing X, Xu Y, Cheng W, Guo S, Zou Y and He
L: Tanshinone I induces apoptosis and pro-survival autophagy in
gastric cancers. Cancer Chemother Pharmacol. 77:1171–1181. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu M, Cao FL, Li NY, Liu YQ, Li YP and Lv
CL: Tanshinone IIA reverses the malignant phenotype of SGC7901
gastric cancer cells. Asian Pac J Cancer Prev. 14:173–177. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Li JX, Wang YQ and Miao ZH:
Tanshinone I inhibits tumor angiogenesis by reducing Stat3
phosphorylation at Tyr705 and hypoxia-induced HIF-1α accumulation
in both endothelial and tumor cells. Oncotarget. 6:16031–16042.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin JY, Ke YM, Lai JS and Ho TF:
Tanshinone IIA enhances the effects of TRAIL by downregulating
survivin in human ovarian carcinoma cells. Phytomedicine.
22:929–938. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang CY, Hua L, Sun J, Yao KH, Chen JT,
Zhang JJ and Hu JH: MiR-211 inhibits cell proliferation and
invasion of gastric cancer by down-regulating SOX4. Int J Clin Exp
Pathol. 8:14013–14020. 2015.PubMed/NCBI
|
|
27
|
Gormally MV, Dexheimer TS, Marsico G,
Sanders DA, Lowe C, Matak-Vinković D, Michael S, Jadhav A, Rai G,
Maloney DJ, et al: Suppression of the FOXM1 transcriptional
programme via novel small molecule inhibition. Nat Commun.
5:51652014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song IS, Jeong YJ, Jeong SH, Heo HJ, Kim
HK, Bae KB, Park YH, Kim SU, Kim JM, Kim N, et al: FOXM1-induced
PRX3 regulates stemness and survival of colon cancer cells via
maintenance of mitochondrial function. Gastroenterology.
149:1006–1016.e9. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Buchner M, Park E, Geng H, Klemm L, Flach
J, Passegué E, Schjerven H, Melnick A, Paietta E, Kopanja D, et al:
Identification of FOXM1 as a therapeutic target in B-cell lineage
acute lymphoblastic leukaemia. Nat Commun. 6:64712015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wiseman EF, Chen X, Han N, Webber A, Ji Z,
Sharrocks AD and Ang YS: Deregulation of the FOXM1 target gene
network and its coregulatory partners in oesophageal
adenocarcinoma. Mol Cancer. 14:692015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
32
|
Hui MK, Chan KW, Luk JM, Lee NP, Chung Y,
Cheung LC, Srivastava G, Tsao SW, Tang JC and Law S: Cytoplasmic
Forkhead box M1 (FoxM1) in esophageal squamous cell carcinoma
significantly correlates with pathological disease stage. World J
Surg. 36:90–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Zhang G and Kong C: FOXM1
participates in PLK1-regulated cell cycle progression in renal cell
cancer cells. Oncol Lett. 11:2685–2691. 2016.PubMed/NCBI
|
|
34
|
Inoguchi S, Seki N, Chiyomaru T, Ishihara
T, Matsushita R, Mataki H, Itesako T, Tatarano S, Yoshino H, Goto
Y, et al: Tumour-suppressive microRNA-24-1 inhibits cancer cell
proliferation through targeting FOXM1 in bladder cancer. FEBS Lett.
588:3170–3179. 2014. View Article : Google Scholar : PubMed/NCBI
|