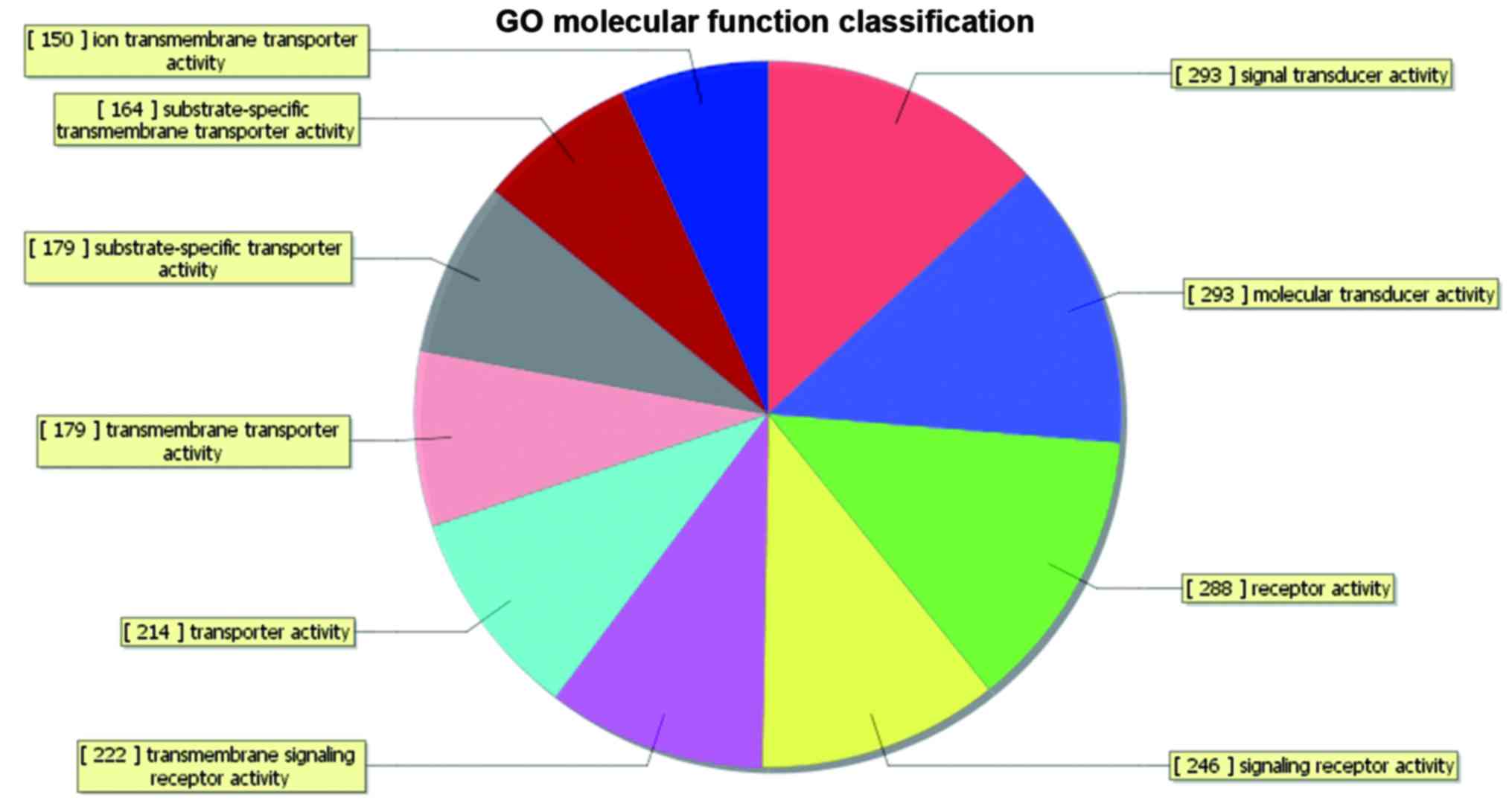
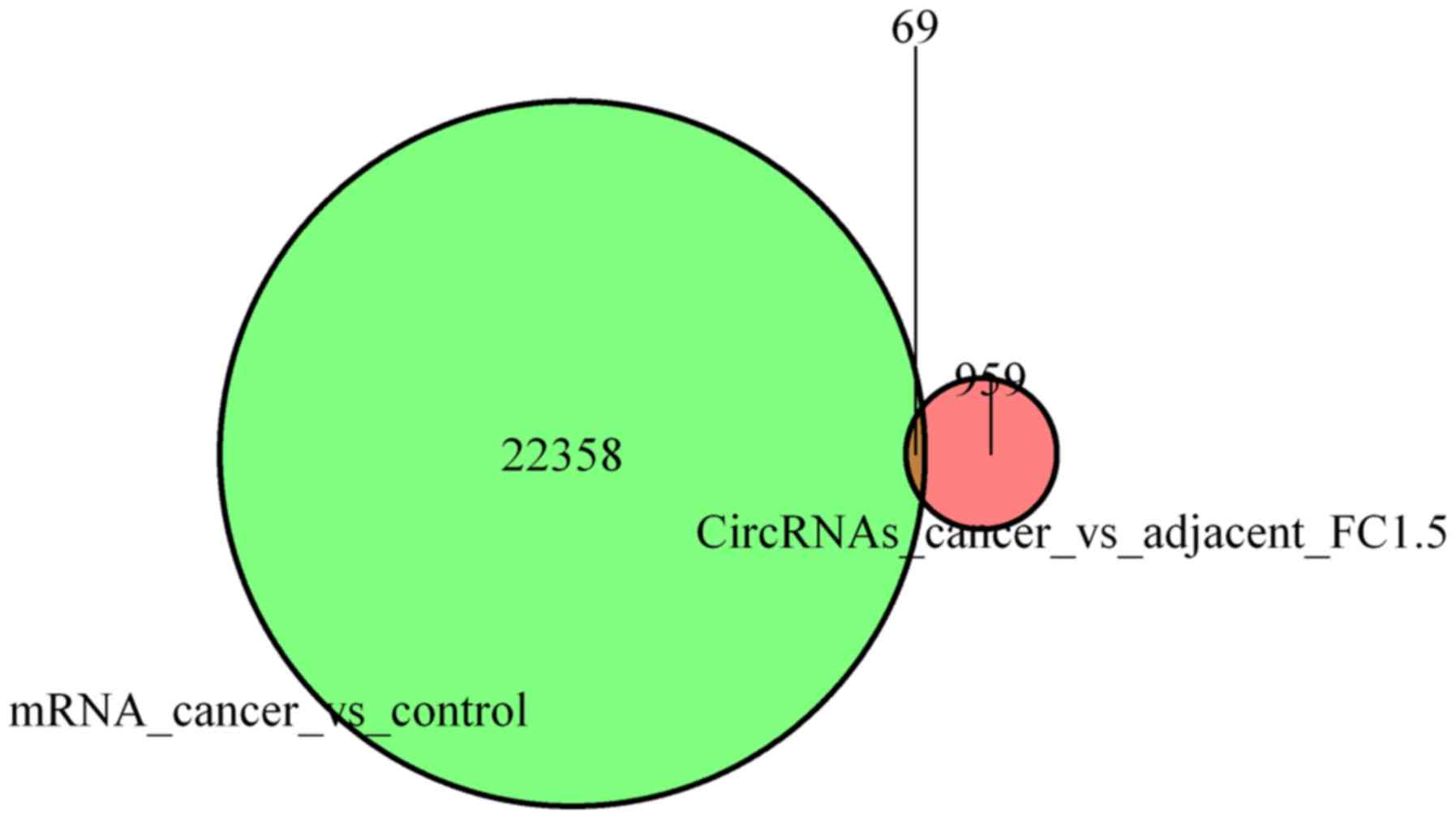
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park DJ, Thomas NJ, Yoon C and Yoon SS:
Vascular endothelial growth factor a inhibition in gastric cancer.
Gastric Cancer. 18:33–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Degiuli M and Calvo F: Survival of early
gastric cancer in a specialized European center. Which
lymphadenectomy is necessary? World J Surg. 30:2193–2203. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schwarz RE and Smith DD: Clinical impact
of lymphadenectomy extent in resectable gastric cancer of advanced
stage. Ann Surg Oncol. 14:317–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valdmanis PN and Kay MA: The expanding
repertoire of circular RNAs. Mol Ther. 21:1112–1114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai Y, Sui W, Lan H, Yan Q, Huang H and
Huang Y: Comprehensive analysis of microRNA expression patterns in
renal biopsies of lupus nephritis patients. Rheumatol Int.
29:749–754. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doberstein K, Bretz NP, Schirmer U, Fiegl
H, Blaheta R, Breunig C, Müller-Holzner E, Reimer D, Zeimet AG and
Altevogt P: miR-21-3p is a positive regulator of L1CAM in several
human carcinomas. Cancer Lett. 354:455–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fukuhara S, Sakurai A, Sano H, Yamagishi
A, Somekawa S, Takakura N, Saito Y, Kangawa K and Mochizuki N:
Cyclic AMP potentiates vascular endothelial cadherin-mediated
cell-cell contact to enhance endothelial barrier function through
an Epac-Rap1 signaling pathway. Mol Cell Biol. 25:136–146. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arai K, Sakamoto R, Kubota D and Kondo T:
Proteomic approach toward molecular backgrounds of drug resistance
of osteosarcoma cells in spheroid culture system. Proteomics.
13:2351–2360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cho HJ, Baek KE, Kim IK, Park SM, Choi YL,
Nam IK, Park SH, Im MJ, Yoo JM, Ryu KJ, et al: Proteomics-based
strategy to delineate the molecular mechanisms of RhoGDI2-induced
metastasis and drug resistance in gastric cancer. J Proteome Res.
11:2355–2364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
PLoS One Editors, . Retraction: GREB1
functions as a growth promoter and is modulated by IL6/STAT3 in
breast cancer. PLoS One. 9:e1022872014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akiyama H, Watanabe T, Wakabayashi K,
Nakade S, Yasui S, Sakata K, Chiba R, Spiegelhalter F, Hino A and
Maitani T: Quantitative detection system for maize sample
containing combined-trait genetically modified maize. Anal Chem.
77:7421–7428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feigin VL and Findlay M: Advances in
subarachnoid hemorrhage. Stroke. 37:305–308. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van der Voet M, Olson JM, Kuivaniemi H,
Dudek DM, Skunca M, Ronkainen A, Niemelä M, Jääskeläinen J,
Hernesniemi J, Helin K, et al: Intracranial aneurysms in Finnish
families: Confirmation of linkage and refinement of the interval to
chromosome 19q13.3. Am J Hum Genet. 74:564–571. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Juvela S: Natural history of unruptured
intracranial aneurysms: Risks for aneurysm formation, growth, and
rupture. Acta Neurochir Suppl. 82:27–30. 2002.PubMed/NCBI
|
|
27
|
Liang S, He L, Zhao X, Miao Y, Gu Y, Guo
C, Xue Z, Dou W, Hu F, Wu K, et al: MicroRNA let-7f inhibits tumor
invasion and metastasis by targeting MYH9 in human gastric cancer.
PLoS One. 6:e184092011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Naor D, Nedvetzki S, Golan I, Melnik L and
Faitelson Y: CD44 in cancer. Crit Rev Clin Lab Sci. 39:527–579.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1 - a paradigm for long noncoding RNA function in cancer. J
Mol Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang JH, Ren Y, Zhang R, Han Y, Sheng YH,
Hou WJ and Ao HF: Effects of CXXC finger protein 5 up-regulated
expression in epithelial ovarian cancer. China Oncol. 25:260–268.
2015.(In Chinese). http://www.china-oncology.com/EN/Y2015/V25/I4/260
|