Introduction
As one of the most common digestive malignancies in
humans worldwide, gastric cancer (GC) exhibits aggressive malignant
behavior. GC patients have a current 5-year survival rate of only
~24% (1). At present, most GC
patients are clinically diagnosed in the advanced stages and the
median survival time of GC patients with local invasion or
metastasis is less than 12 months (2). The challenges of discovering
associated tumor markers and understanding the mechanisms of GC
initiation, progression and metastasis, are the keys to diagnose,
prevent and treat GC appropriately in the early stages as well as
develop targeted treatment (3)
A disintegrin and metalloproteinases (ADAMs), are a
family of proteins which play a pivotal role in the proteolytic
process implicated in cell-cell and cell-matrix interactions
(4). Members of the ADAM family are
divided into two groups: membrane-anchored and the secreted-type
proteins (ADAMST) (5). Noteworthy,
membrane-anchored ADAMs belong to type I trans-membrane
proteins, which consist of a disintegrin-containing extracellular
domain and a metalloproteinase domain (6). The functions of ADAMs are multiple and
they are mainly involved in the proteolytic processing of
trans-membrane proteins, contributing to various
pathologies, including cell adhesion, cell signaling pathways and
human tumors (7). Ectodomain
shedding is a critical process conducted through proteolytic
cleavage of membrane-anchored molecules into the extracellular
microenvironment, and is related with tumorigenesis (8). ADAMs also participate in the
ectodomain shedding process by undergoing cleavage close to the
trans-membrane domains (9).
A disintegrin and metalloproteinase domain 9
(ADAM9), one of the ADAM family members, has been found and
described in a variety of solid tumors with overexpression and
dysregulation, in glioma, prostate, colon and breast cancer, which
suggest ADAM9 as an important molecule involved in tumorigenesis
(10–13). However, in GC, the role of ADAM9 is
still elusive and deserves to be elucidated.
MicroRNAs (miRs) are a class of non-coding RNAs
consisting of 22 nucleotides, which recognize a specific sequence
of messenger RNAs (mRNAs) on the 3′ untranslated region (3′UTR) as
targets, and consequentially induce either inhibition of mRNA
translation or degeneration of the targeted mRNAs (14). miRs functionally regulate and
control various pivotal pathophysiological processes
post-transcriptionally, including tumor initiation and progression
(15,16). For instance, miR-146 suppresses
gallbladder cancer cell proliferation by targeting epidermal growth
factor (EGF) (17); miR-99a
inhibits cell growth in osteosarcoma by negatively regulating
TNFAIP8 (18). In GC, numerous miRs
exhibit complex and marked effects either by suppressing or
promoting tumor progression. For example, miR-30a reportedly
targets RPA1 in GC cells and consequently suppresses the growth of
GC cells with cell cycle arrest (19); on the contrary, by decreasing the
expression of FBW7 through direct post-transcriptional regulation,
miR-363 significantly promotes the cell proliferation and
chemotherapy resistance in GC (20). Certainly, miRs provide us with
enormous possibilities to discover new targets in GC prevention,
diagnosis and therapeutic treatment.
Based on our previous research and studies from
other authors, microRNA-126 (miR-126) appears to be an extremely
important microRNA that functions as a suppressor in GC progression
and which is frequently downregulated in both GC tissues and cell
lines (21–24). As interactions between miRs and
their targets show complex networking, an individual miR could
target various mRNAs in multiple pathophysiological processes and
an individual mRNA could possibly be targeted by different miRs
simultaneously. Intensive detection of miR-126-targeted molecules
in GC is valuable to provide us with a clear sense of how this
suppressor functions in GC and also to provide us with sufficient
evidence in order to find new antitumor targets.
In the present study, by evaluating 76 pairs of GC
tissues compared with the adjacent non-cancerous tissues and 4 GC
cell lines (SGC-7901, MKN-45, MKN-28 and SUN-16), we ascertained
that ADAM9 was aberrantly overexpressed in GC. High levels of ADAM9
were significantly correlated with GC clinicopathological features,
such as tumor size, local invasion, lymph node metastasis and tumor
stage, which suggest a poorer prognosis for patients with a high
ADAM9 level. Knockdown of ADAM9 expression in SGC-7901 cells
significantly suppressed cell proliferation and arrested the cell
cycle at the G0/G1 phase. Moreover, by applying a dual-luciferase
reporter assay, we discovered that miR-126 could directly bind to
the 3′UTR of ADAM9 mRNA and markedly downregulate ADAM9 expression.
The promotional effect of ADAM9 on GC cell proliferation was
revealed through overexpression of miR-126. All the aforementioned
findings illustrate that ADAM9 functions as a tumor promoter in GC
and exerts a tumor-suppressive function.
Materials and methods
Cell culture and surgical
specimens
The immortalized gastric epithelium cell line and 4
GC cell lines (SGC-7901, MKN-45, MKN-28 and SUN-16) were purchased
from the Shanghai Institutes for Biological Sciences, Chinese
Academy of Science (Shanghai, China). SGC-7901 cells overexpressing
miR-126 (SGC-7901/miR-126) and the negative control (NigmiR) were
constructed in our previous study (25). All cells were cultured in RPMI-1640
medium supplemented with 10% heat-inactivated fetal bovine serum
(FBS), 100 µg/ml streptomycin and 100 U/ml penicillin in a
humidified cell incubator at 37°C with an atmosphere of 5%
CO2.
Seventy-six pairs of GC specimens and adjacent
non-cancerous tissues were collected from GC patients who had
undergone a radical gastrectomy without preoperative therapy at the
Department of Surgery, Ruijin Hospital, Shanghai Jiao Tong
University School of Medicine during 2012–2014. Ethical approval
was obtained from the Research Medical Ethics Committee of Rujin
Hospital, Shanghai Jiao Tong University School of Medicine.
Immunohistochemistry and western blot
analyses
Antibodies against ADAM9 and GAPDH (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), and horseradish
peroxidase-conjugated secondary antibody (Abcam, Cambridge, MA,
USA) were prepared. Immunohistochemical analysis was carried out
using antibody against ADAM9 following the manufacturers
instructions (1:50), and the tissues were individually examined by
two professional pathologists. GAPDH was used as a loading
control.
RIPA buffer containing a protease inhibitor cocktail
was used to lyse the cells, and the protein concentration was
measured by BCA Protein Assay kit (both from Pierce, Rockford, IL,
USA). Proteins were electrophoresed and electrotransferred.
Antibodies against ADAM9 (1:1,000) and GAPDH (1:5,000) were probed,
and a horseradish peroxidase-conjugated secondary antibody was used
for further probing. The protein quantity was detected using GAPDH
as a loading control.
RNA isolation and real-time qPCR
assay
Total RNA was extracted from the cell lines using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturers instructions. The first-strand cDNA was synthesized
using a HighCapacity cDNA Reverse Transcription kit (ABI, Foster
City CA, USA). RT-primers of ADAM9 mRNAs were synthesized as
follows: forward, 5′-GGAAGAGTGTGACTGTGGTAC-3′ and reverse,
5′-CCTCGGCATAAAGTACCTCC-3′ by Sangon Biotech Co. (Shanghai, China).
Real-time quantitative polymerase chain reaction (qRT-PCR) was
performed according to TaqMan Gene Expression Assays protocol
(ABI).
Cell transfection
SGC-7901 cells were transfected with pGU6/Neo
vectors (GenePharma, Shanghai, China) containing shRNA suppressing
ADAM9 translation or non-containing ones. Cells were cultured and
selected in medium containing 400 µg/ml G418 (Santa Cruz
Biotechnology, Inc.). The stable transfected cells aforementioned
were assessed by qRT-PCR and western blot analysis compared with
the negative control cells. All cells were cultured and maintained
in medium containing 200 µg/ml G418.
Recombinant adenovirus Ad5/F35 (Ad5/F35-ADAM9) was
constructed for overexpressing ADAM9 and Ad5/F35-Null was used as a
negative control (GenePharma). SGC-7901 cells overexpressing
miR-126 (SGC-7901/miR-126) and the negative control (NigmiR) were
further transfected with Ad5/F35-ADAM9 or Ad5/F35-Null, and were
assessed.
Cell proliferation assay and cell
cycle analysis
SGC-7901 cells (1×106) stably transfected
or the negative control cells were cultured in 96-well microtiter
plates in triplicate and incubated for 5 days at 37°C with an
atmosphere of 5% CO2. The OD was measured using
microplate computer software (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) according to the protocol from the Cell Counting Kit-8
(CCK-8) assay kit (Dojindo, Tokyo, Japan). The curves for cell
proliferation were plotted.
The aforementioned cells were treated with ethanol
fixation, RNase A treatment and propidium iodide staining, and were
then detected under flow cytometry by FACSCalibur
(Becton-Dickinson, Franklin Lakes, NJ, USA). Cell populations at
the G0/G1, S and G2/M phases were quantified by ModFit software
(Becton-Dickinson) excluding a calculation of cell debris and
fixation artifacts.
Dual-luciferase reporter assay
ADAM9 was predicted as a potential target of miR-126
by bioinformatics analysis (microcosm, http://mirecords.biolead.org). A 206 bp sequence from
the 3′UTR of ADAM9 mRNA including the putative miR-126 binding site
was selected as follows: 5′-uagagaaauuaauuuaaau
uagaauuucuauuaugaaucaugugaaagcaugacauucguucacaauagca
cuauuuuaaauaaauuauaagcuuuaagguacgaaguauuuaaugaucuaau
caaauauguugauucauggcuauaauaaagcaggagcaauuauaaaaucuuc
aaucaauugaacuuuuacaaaaccacuug-3′. The corresponding mutant sequence
was constructed by Sangon Biotech Co., as follows:
5′-aacacauaauuaauaauuuuaacaauuacaaauuucauugaag
aguauggaagucuuaccuacucuaaaccucaaauauuauuuauuaaaauggu
auuacgaagguacuuuauuaaacaacaauugauaaaagaucaaugaagccaaa
auuuauggacguggauuaaaauauuguacuaacuaaucaucauauucuauag cucauc-3′.
These sequences were cloned into pMIR-REPORT luciferase vectors
(Promega, Madison, WI, USA), containing Firefly luciferase, and
pRL-TK vectors containg Renilla luciferase which were used
as a control. SGC-7901 cells overexpressing miR-126 or the negative
control were co-transfected with the aforementioned vectors and the
luciferase activity was measured using a Dual-Glo Luciferase assay
system (Promega) 48 h post-transfection.
Statistical analysis
Statistical analysis was carried out using SPSS
18.0. P-values were calculated using a paired t-test and Fisher's
exact test. P-values <0.05 were considered to indicate a
statistically significant result.
Results
ADAM9 is overexpressed in GC tumor
specimens
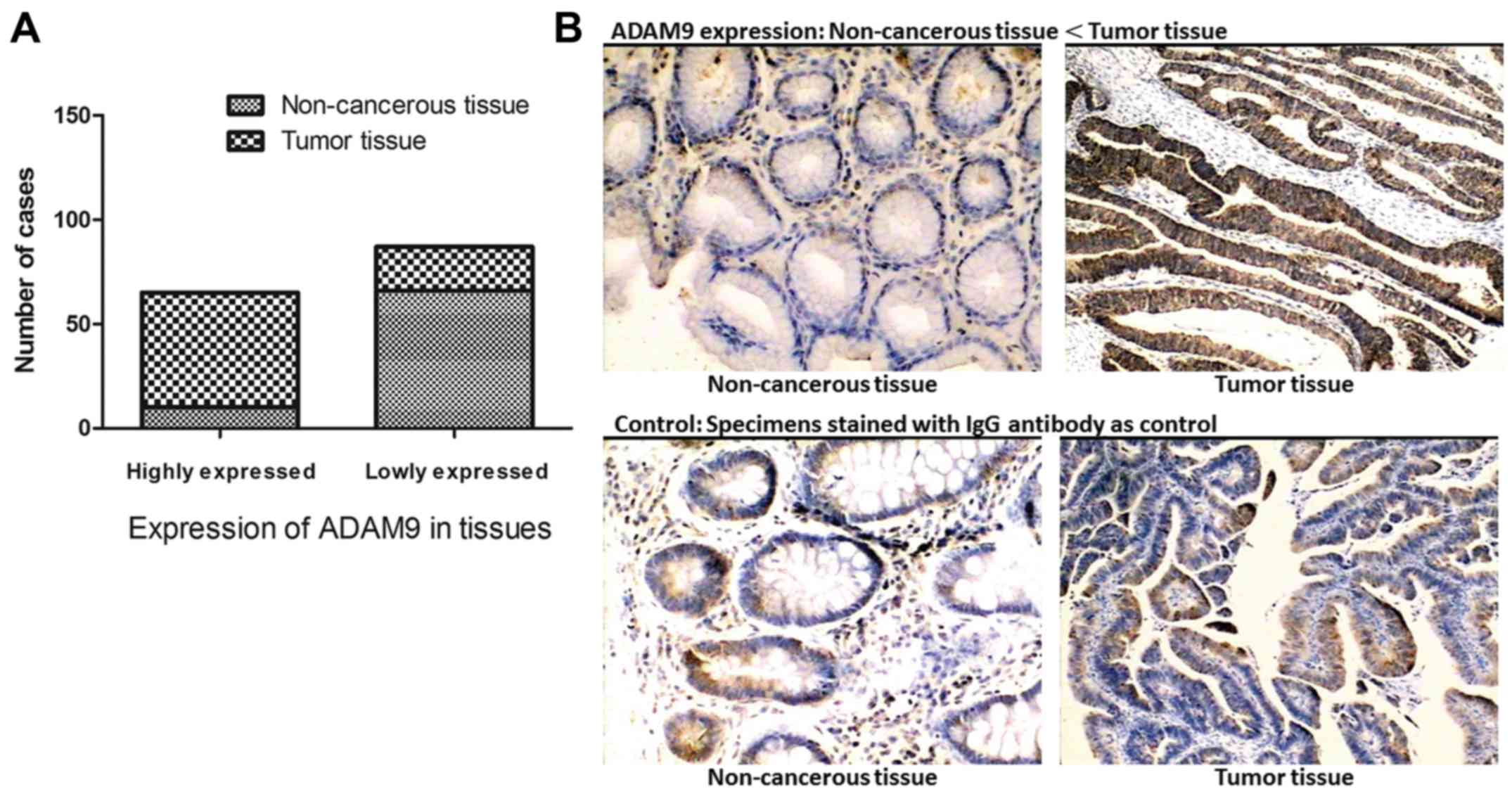
Seventy-six paired specimens from GC tumors and
adjacent non-cancerous tissues were examined using IHC. According
to the expression intensity of ADAM9, these cases were separated
into two groups: an ADAM9 low expression and an ADAM9 high
expression group. Among the tumor specimens, 72.3% (55/76) of the
cases presented high levels of ADAM9 expression, and only 27.6%
(21/76) of the cases expressed a relatively lower level of ADAM9.
On the contrary, in non-cancerous tissues, high expression of ADAM9
was detected in a small portion of the cases (13.2%, 10/76) and
86.8% (66/76) of the specimens presented a low level of ADAM9
expression. This proves that ADAM9 is expressed frequently higher
in GC tissues when compared with adjacent non-cancerous tissues
(Fig. 1).
ADAM9 expression is upregulated and
miR-126 is downregulated in GC cell lines
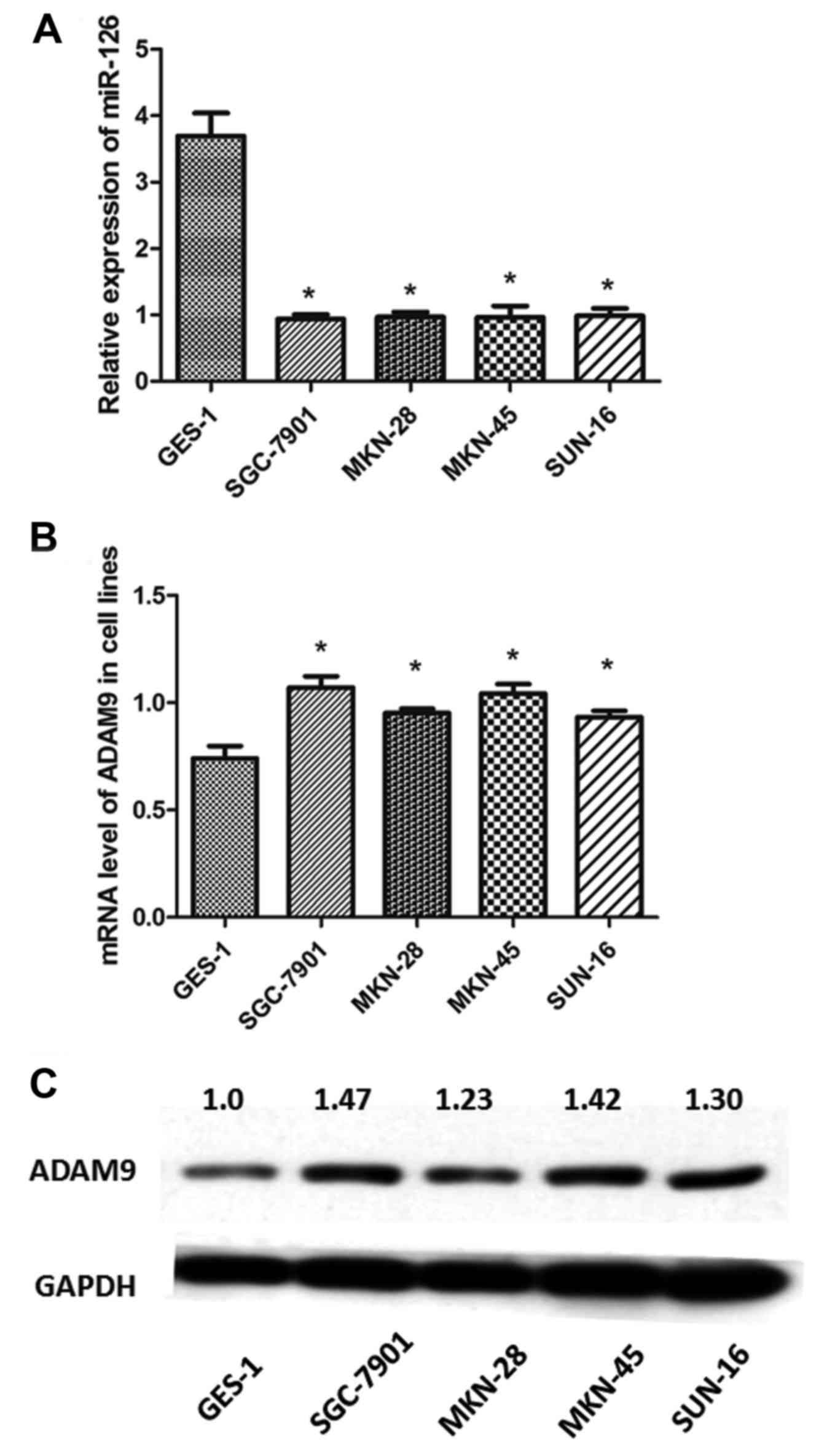
Analysis of the transcription levels of miR-126 in
the 4 GC cell lines (SGC-7901, MKN-28, MKN-45 and SUN-16) using
qRT-PCR revealed that the expression levels of miR-126 were
significantly downregulated in the GC cell lines compared with the
GES-1 cells (P<0.05) (Fig. 2A).
In contrast, the mRNA levels of ADAM9 were upregulated in the 4
different GC cell lines compared with the GES-1 cells (P<0.05)
(Fig. 2B). Similarly, western blot
analysis demonstrated that the protein levels of ADAM9 were
significantly higher in the GC cell lines than that in the GES-1
cells (Fig. 2C). These results were
consistent with the observations obtained in the IHC analysis of
the tumor tissues.
High expression of ADAM9 is correlated
with GC clinicopathological features
The correlation between the expression of ADAM9 and
the clinicopathological features of the 76 GC cases was analyzed.
According to Table I, there was no
significant correlation between ADAM9 expression and patient age,
gender or tumor location. However, a significant trend towards a
larger tumor size (P<0.05), deeper local invasion (P<0.05),
more frequent lymph node metastasis (P<0.05) and more advanced
tumor-node-metastasis (TNM) stage (P<0.05) in cases with higher
expression levels indicates a correlation between ADAM9
overexpression and certain GC clinicopathological features.
 | Table I.Correlation between ADAM9 expression
and clinicopathological features of the 76 GC cases. |
Table I.
Correlation between ADAM9 expression
and clinicopathological features of the 76 GC cases.
|
| ADAM9 expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
parameters | Low (n=21) | High (n=55) |
P-valuea |
|---|
| Age (years) |
|
| 0.592 |
| ≤60 | 8 | 17 |
|
|
>60 | 13 | 38 |
|
| Gender |
|
| 0.196 |
|
Male | 12 | 21 |
|
|
Female | 9 | 34 |
|
| Tumor diameter
(cm) |
|
| 0.023 |
| ≤5 | 15 | 19 |
|
|
>5 | 8 | 36 |
|
| Location |
|
| 0.778 |
| Distal
third | 16 | 39 |
|
| Middle
or proximal third | 5 | 16 |
|
| Histological
classification |
|
| 0.792 |
|
Poorly-differentiated
adenocarcinoma | 7 | 22 |
|
|
Middle/well-differentiated
adenocarcinoma | 4 | 6 |
|
| Signet
ring cell carcinoma | 3 | 6 |
|
|
Mucinous adenocarcinoma | 1 | 3 |
|
| Local invasion |
|
| 0.016 |
|
T1,T2 | 13 | 16 |
|
|
T3,T4 | 8 | 39 |
|
| Lymph node
metastasis |
|
| 0.030 |
| No | 12 | 15 |
|
|
Yes | 9 | 40 |
|
| TNM stage |
|
| 0.046 |
|
I,II | 10 | 12 |
|
|
III,IV | 11 | 43 |
|
Knockdown of ADAM9 suppresses cell
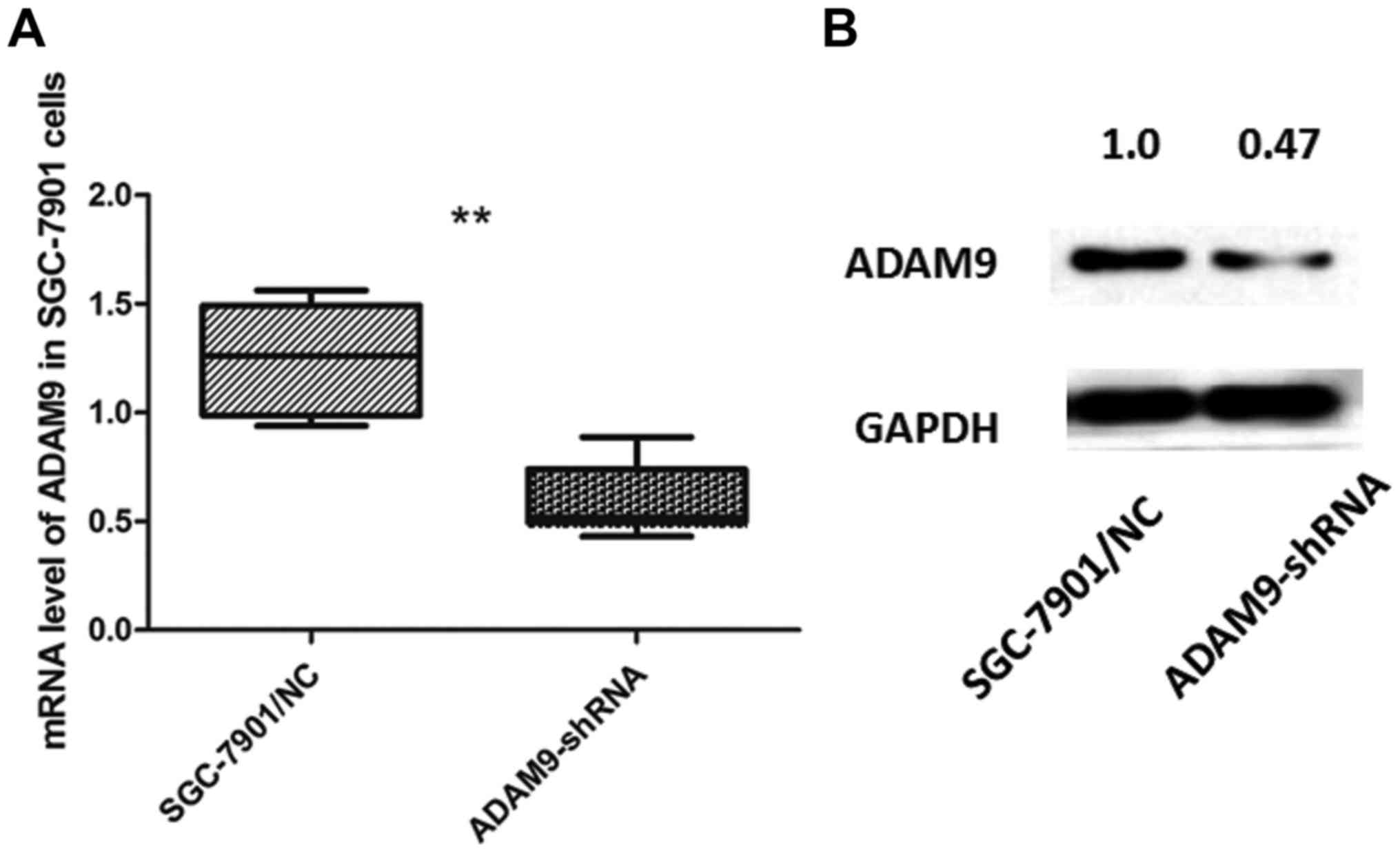
proliferation and arrests the cell cycle in SGC-7901 cells
SGC-7901 cells, which expressed the highest level of
ADAM9 among the 4 GC cell lines, were selected and transfected with
pGU6/Neo vectors to knock down the expression of ADAM9. We verified
the transfection effect through qRT-PCR and western blot analysis
(Fig. 3).
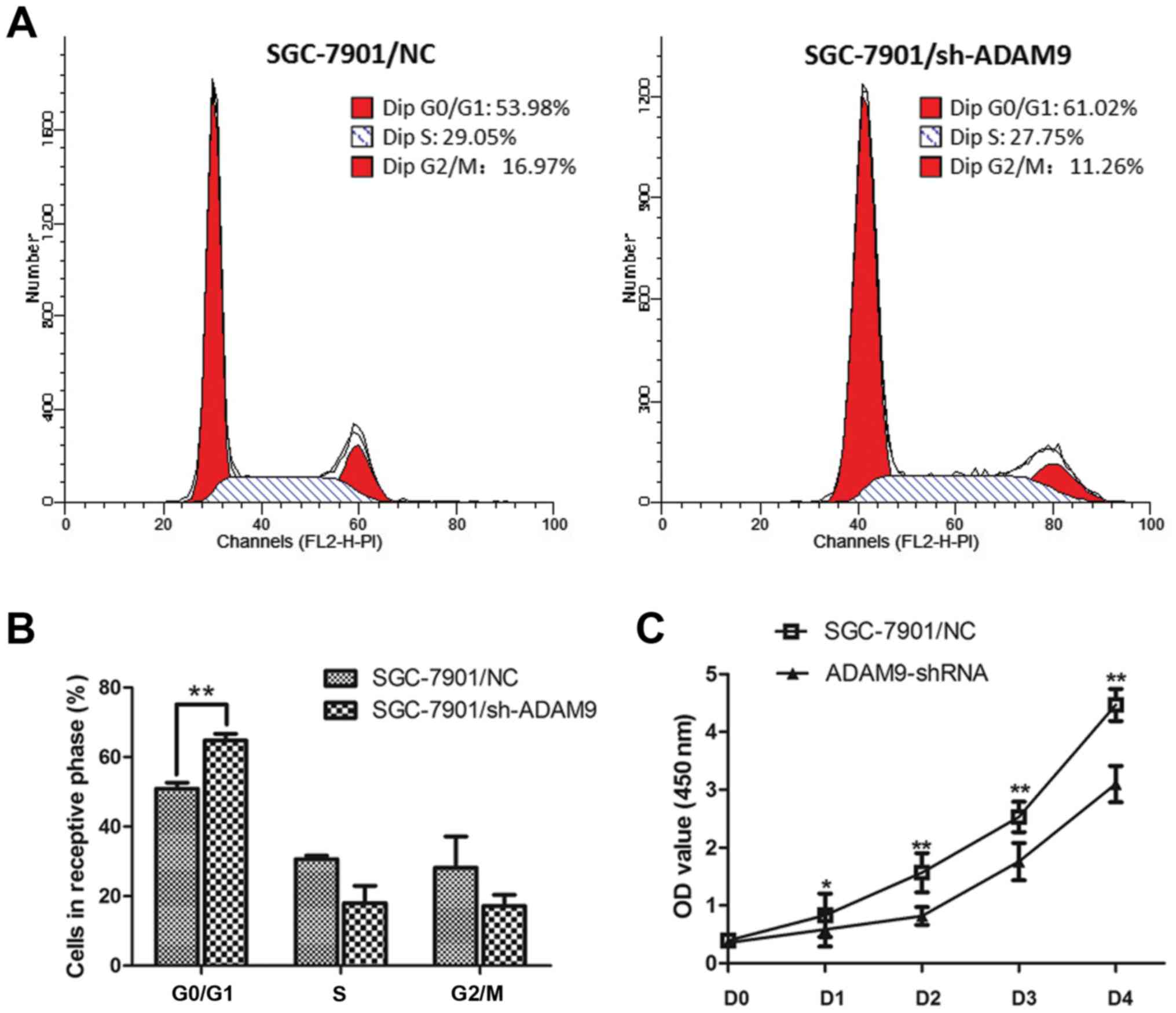
We conducted flow cytometric analysis and found
that, the cell cycle of SGC-7901 cells was significantly arrested
at the G0/G1 phase when ADAM9 was knocked down (Fig. 4A and B). The percentage of the
SGC-7901 cells in the G0/G1 phase was increased from 50.89 to
64.78% (P<0.01). The S phase was decreased from 30.63 to 18.05%,
and the G2/M phase was decreased from 28.16 to 17.16% (Fig. 4B). Meanwhile, as the CCK-8 assay
demonstrated, we observed a significant decrease in cell
proliferation in the ADAM9-knockdown SGC-7901 cells as the P-value
was <0.01 for day 1 and the P-value was <0.05 for days 2–4
(Fig. 4C). These results indicate
that knockdown of ADAM9 in SGC-7901 cells significantly impacts
tumor cell growth.
ADAM9 is a direct target
post-transcriptionally regulated by miR-126 in GC cells
We used microcosm, bioinformatics analysis employing
an online prediction software, to predict ADAM9 as a potential
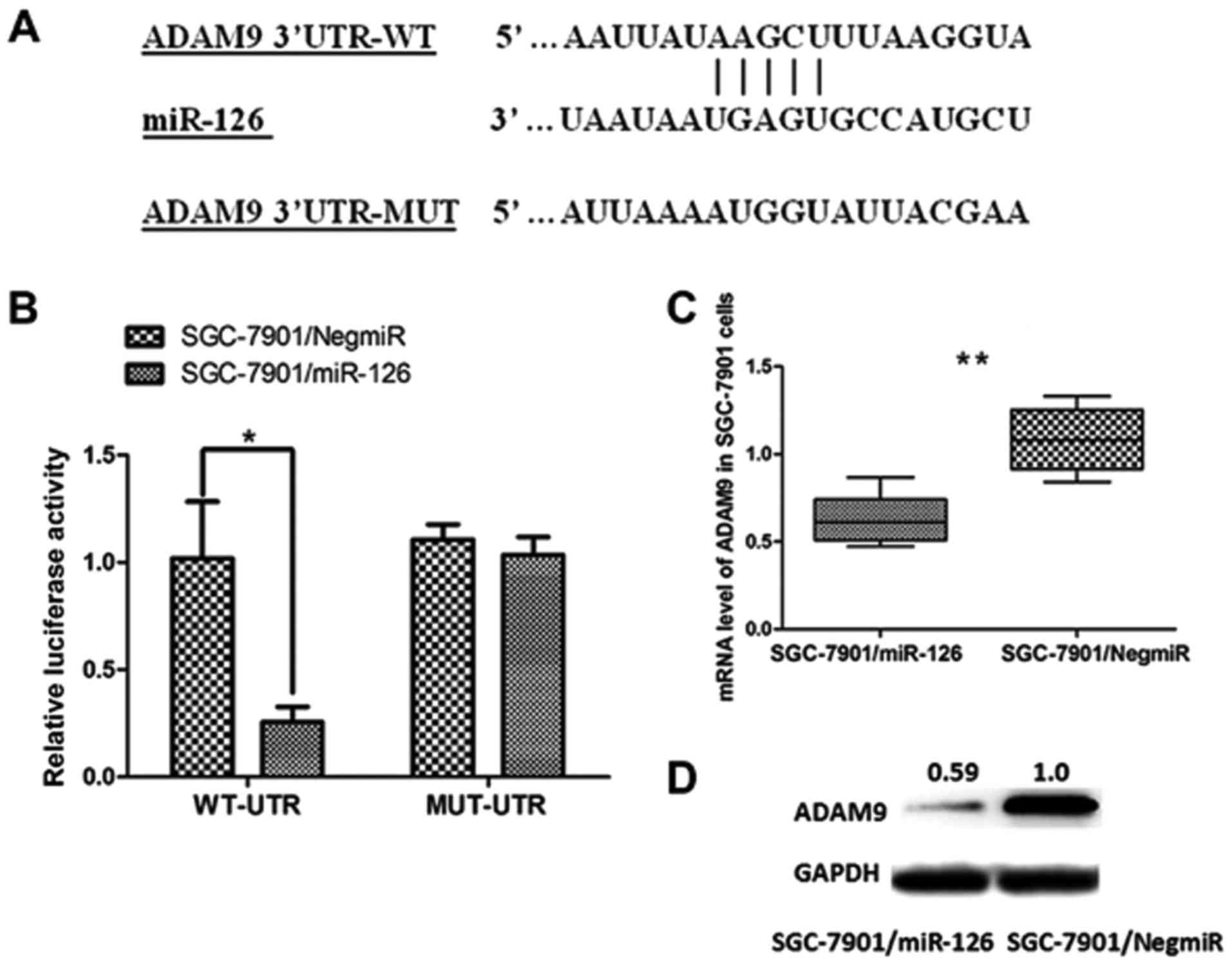
target of miR-126 (Fig. 5A).
Demonstration of the direct interaction between ADAM9 mRNA and
miR-126 was carried out using a dual-luciferase reporter assay.
Luciferase reporter vectors containing a 206-bp 3′UTR sequence of
ADAM9 (WT-UTR) and the corresponding control luciferase vectors
containing a mutated miR-126 target site (MUT-UTR) were
constructed. As shown in Fig. 5B,
overexpression of miR-126 in the SGC-7901 cells (SGC-7901/miR-126)
significantly decreased the luciferase signal of ADAM9/pMIR/WT,
compared with the negative control (SGC-7901/NigmiR). In addition,
this suppressive effect induced by miR-126 was significantly
abolished in the SGC-7901 cells with the putative binding site of
mutated miR-126 (Fig. 5B).
Moreover, both the mRNA level and the protein expression of ADAM9
were significantly decreased in SGC-7901/miR-126 cells (Fig. 5C and D). Collectively, these results
revealed that ADAM9 is a direct target of miR-126 in GC and was
post-transcriptionally downregulated by miR-126.
Introduction of ADAM9 in SGC-7901
cells reverses the phenotype of growth arrest induced by
overexpression of miR-126
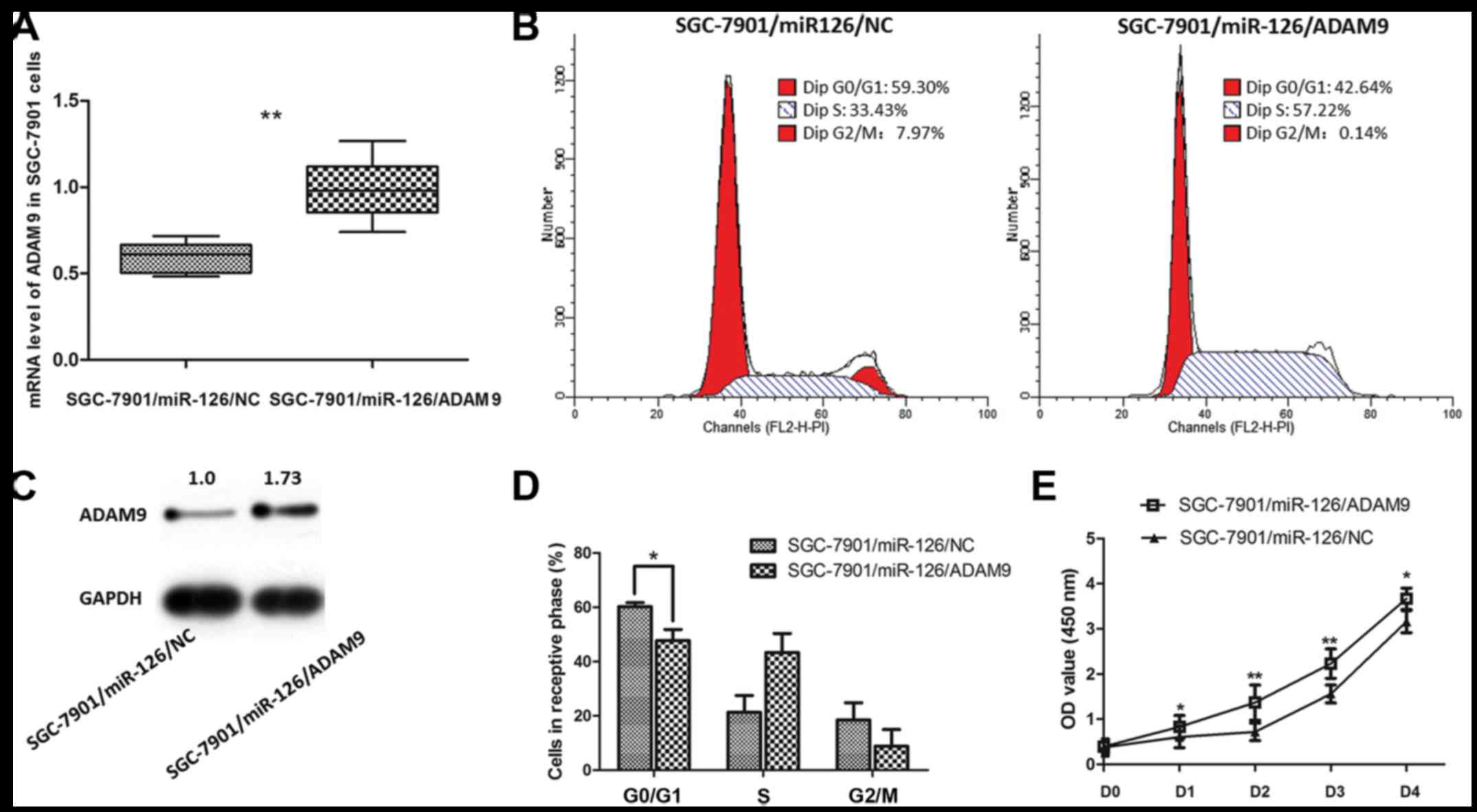
The aforementioned results indicate that ADAM9 is
one of the direct targets suppressed by miR-126 in GC. Based on
this, we assumed that introducing ADAM9 in the
miR-126-overexpressing SGC-7901 cells would at least relatively
reverse the phenotypes caused by overexpression of miR-126.
Recombinant adenovirus Ad5/F35 was applied in the present study to
upregulate the expression of ADAM9 in the SGC-7901 cells. As shown
in Fig. 6, the inhibitory effect on
SGC-7901 cell proliferation by miR-126 was significantly reversed
when ADAM9 expression was increased. Meanwhile, the cell cycle
arrest effect induced by miR-126 was also reverses after ADAM9
introduction. Thus, ADAM9 is a molecule that promotes GC cell
growth, which may be targeted by miR-126 as a part of its
post-transcriptional mechanism for suppressing GC.
Discussion
The initiation and development of human tumors are
under the control of a multitude of factors. With respect to
gastric cancer (GC), a large number of molecules and their relative
mechanisms, which are involved in GC tumorigenesis have been
discovered. A variety of molecules participate in the process of GC
with different types of mechanisms such as signaling pathways and
post-transcriptional regulation, which are accompanied by huge
networking between the molecules discovered or those which need to
be further studied. For example, nucleophosmin (NPM)/B23 was found
to be aberrantly overexpressed and regulated in GC, functioning as
an indicator of GC associated with advanced TNM stage, poor
prognosis and recurrence (26).
Plant homeodomain finger protein 10 (PHF10) was ascertained as a
promoter of GC enhancing the ability of cell proliferation
(27). In addition, concerning
non-coding RNAs, for example, miR-223 targets EPB41L3 in GC and
promotes tumor cell invasion and migration (28). miR-107 downregulates CDK6 mRNA, and
induces inhibition of GC cell invasion (29).
ADAM9 is a member of the ADAM family anchored to the
membrane, and is related to various human tumors as we previously
mentioned. In pancreatic ductal adenocarcinoma, ADAM9 is
upregulated at the mRNA level and over 70% of pancreatic carcinomas
present high protein levels (30).
In prostate cancer, ADAM9 appears to be regulated, and inhibition
of ADAM9 in vivo significantly suppressed tumor growth
(10). ADAM9 was also found to
modulate tumor-stromal cell interaction and sequentially promote
cell motility in human hepatocellular carcinoma and lung cancer
(31,32). Moreover, several reports have
demonstrated high expression of ADAM9 in GC (33,34).
However, the role of ADAM9 in GC and its relative upstream
regulatory mechanisms remain unclear.
In the present study, we validated the expression of
ADAM9 in 76 GC tumor tissues and cell lines. ADAM9 exhibited an
obvious high expression level in GC tissues. By analyzing the
clinicopathological features of the 76 patients, we found that high
levels of ADAM9 showed a significant correlation with larger tumor
size, deeper local invasion, more frequent lymph node metastasis
and more advanced tumor stages. Thus, ADAM9 is an independent
factor correlated with poor GC outcomes and prognosis.
Simultaneously, qRT-PCR and western blot analysis
showed that expression of ADAM9 was significantly higher at both
the mRNA and protein levels in GC cells than levels in GES-1 cells.
Among the 4 GC cell lines, SGC-7901 cells presented the highest
level of ADAM9. Considering what we observed from the specimens and
cells, we believe that ADAM9 is a potential functional molecule in
GC progression. To verify the function of ADAM9 in GC cells, we
selected SGC-7901 cells and knocked down the ADAM9 expression by
stable transfection. An in vitro cellular functional
experiment was carried out. As we had hypothesized, when ADAM9 was
knocked down in the SGC-7901 cells, the cell proliferation ability
was markedly suppressed. Additionally, flow cytometric analysis
demonstrated an obvious arrest of the cell cycle in GC cells at the
G0/G1 phase, indicating that inhibition of ADAM9 effectively
suppressed the growth of GC cells.
Furthermore, we speculated the mechanism by which
ADAM9 promotes the cell growth of GC. Through the use of online
bioinformatics tools, we found that a potential binding site for
miR-126 exists in the 3′UTR of ADAM9 mRNA. miR-126 is an important
non-coding RNA, which has been confirmed as a suppressor of GC
growth. In our recent study we found that miR-126 exerts its
tumor-suppressive function in various types of cancer by targeting
different mRNAs, and in GC, CRKL, LAT-1, VEGF-A and CADM-1, are all
targets of miR-126. We further speculated as to whether ADAM9 is a
functional target of miR-126 in GC. We then conducted a
dual-luciferase reporter assay to verify the direct interaction
between ADAM9 and miR-126. A combination of miR with a specific
3′UTR of the target mRNA could cause an impact on luciferase gene
expression. As the results showed, overexpression of miR-126
significantly decreased the luciferase signal intensity. On the
contrary, mutated 3′UTR of ADAM9 mRNA failed to bind with miR-126
and presented no significant change in luciferase signal intensity.
Thus, there is a direct correlation between miR-126 and the 3′UTR
of ADAM9 mRNA.
To further understand whether miR-126 suppresses GC
through ADAM9, we ectopically expressed ADAM9 in SGC-7901 cells
overexpressing miR-126. As expected, by introducing ADAM9, the cell
proliferation suppression induced by miR-126 was significantly
reversed. In addition, the percentage of the cells arrested in the
G0/G1 phase was notably decreased when ADAM9 was overexpressed. All
the aforementioned results suggest that ADAM9 is one of the direct
targets regulated by miR-126 in GC cells and by which miR-126
conducts its potential tumor suppressive function in GC.
In conclusion, according to the findings in the
present study, we conclude that ADAM9 is one of the direct targets
post-transcriptionally modulated by miR-126, which helps us to
understand the tumor-suppressive mechanism of miR-126. High levels
of ADAM9 in GC are correlated with a poor prognosis and aberrant
overexpression of ADAM9 leads to the promotion of GC cell growth.
ADAM9 should be considered as a potential target for GC prevention,
diagnosis and therapeutic treatment.
Acknowledgements
The authors thank Qucai, Minmin Shi, Hui Ye and Jun
Ji for providing valuable technical supports and assistance. The
present study was kindly supported by grants from the following:
the National Natural Science Foundation of China (nos. 81602544 and
81372187), and the Liu Haoqing Foundation for Medicine, Ruijin
Hospital Shanghai.
References
|
1
|
Russo F, Linsalata M and Orlando A:
Probiotics against neoplastic transformation of gastric mucosa:
Effects on cell proliferation and polyamine metabolism. World J
Gastroenterol. 20:13258–13272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang W, Sun XW, Li CF, Lv L, Li YF, Chen
YB, Xu DZ, Kesari R, Huang CY, Li W, et al: Comparison of the 6th
and 7th editions of the UICC TNM staging system for gastric cancer:
Results of a Chinese single-institution study of 1,503 patients.
Ann Surg Oncol. 18:1060–1067. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bringeland EA, Wasmuth HH, Fougner R,
Mjønes P and Grønbech JE: Impact of perioperative chemotherapy on
oncological outcomes after gastric cancer surgery. Br J Surg.
101:1712–1720. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bahudhanapati H, Bhattacharya S and Wei S:
Evolution of vertebrate Adam genes; Duplication of
testicular Adams from ancient Adam9/9-like loci. PLoS
One. 10:e01362812015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kelwick R, Desanlis I, Wheeler GN and
Edwards DR: The ADAMTS (A Disintegrin and Metalloproteinase with
Thrombospondin motifs) family. Genome Biol. 16:1132015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amendola RS, Martin AC, Selistre-de-Araújo
HS, Paula-Neto HA, Saldanha-Gama R and Barja-Fidalgo C: ADAM9
disintegrin domain activates human neutrophils through an autocrine
circuit involving integrins and CXCR2. J Leukoc Biol. Mar
12–2015.(Epub ahead of print). pii: jlb.3A0914-455R. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peduto L: ADAM9 as a potential target
molecule in cancer. Curr Pharm Des. 15:2282–2287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Quach HT, Hirano S, Fukuhara S, Watanabe
T, Kanoh N, Iwabuchi Y, Usui T and Kataoka T: Irciniastatin A
induces potent and sustained activation of extracellular
signal-regulated kinase and thereby promotes ectodomain shedding of
tumor necrosis factor receptor 1 in human lung carcinoma A549
cells. Biol Pharm Bull. 38:941–946. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones JC, Rustagi S and Dempsey PJ: ADAM
proteases and gastrointestinal function. Annu Rev Physiol.
78:243–276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu CM, Hsieh CL, He YC, Lo SJ, Liang JA,
Hsieh TF, Josson S, Chung LW, Hung MC and Sung SY: In vivo
targeting of ADAM9 gene expression using lentivirus-delivered shRNA
suppresses prostate cancer growth by regulating REG4 dependent cell
cycle progression. PLoS One. 8:e537952013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li J, Ji Z, Qiao C, Qi Y and Shi W:
Overexpression of ADAM9 promotes colon cancer cells invasion. J
Invest Surg. 26:127–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Micocci KC, Martin AC, Montenegro CF,
Durante AC, Pouliot N, Cominetti MR and Selistre-de-Araujo HS:
ADAM9 silencing inhibits breast tumor cell invasion in vitro.
Biochimie. 95:1371–1378. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen CM, Hsieh YH, Hwang JM, Jan HJ, Hsieh
SC, Lin SH and Lai CY: Fisetin suppresses ADAM9 expression and
inhibits invasion of glioma cancer cells through increased
phosphorylation of ERK1/2. Tumour Biol. 36:3407–3415. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kiselev FL: MicroRNA and cancer. Mol Biol.
48:232–242. 2014.(In Russian).
|
|
15
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Connerty P, Ahadi A and Hutvagner G: RNA
Binding proteins in the miRNA pathway. Int J Mol Sci. 17:E312015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cai J, Xu L, Cai Z, Wang J, Zhou B and Hu
H: MicroRNA-146b-5p inhibits the growth of gallbladder carcinoma by
targeting epidermal growth factor receptor. Mol Med Rep.
12:1549–1555. 2015.PubMed/NCBI
|
|
18
|
Xing B and Ren C: Tumor-suppressive
miR-99a inhibits cell proliferation via targeting of TNFAIP8 in
osteosarcoma cells. Am J Transl Res. 8:1082–1090. 2016.PubMed/NCBI
|
|
19
|
Zou Z, Ni M, Zhang J, Chen Y, Ma H, Qian
S, Tang L, Tang J, Yao H, Zhao C, et al: miR-30a can inhibit
DNA replication by targeting RPA1 thus slows down the proliferation
of cancer cells. Biochem J. 473:2131–2139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang PF, Sheng LL, Wang G, Tian M, Zhu
LY, Zhang R, Zhang J and Zhu JS: miR-363 promotes proliferation and
chemo-resistance of human gastric cancer via targeting of FBW7
ubiquitin ligase expression. Oncotarget. 7:35284–35292.
2016.PubMed/NCBI
|
|
21
|
Wang J, Chen X, Li P, Su L, Yu B, Cai Q,
Li J, Yu Y, Liu B and Zhu Z: CRKL promotes cell proliferation in
gastric cancer and is negatively regulated by miR-126. Chem Biol
Interact. 206:230–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Chen X, Su L, Li P, Cai Q, Liu B,
Wu W and Zhu Z: MicroRNA-126 inhibits cell proliferation in gastric
cancer by targeting LAT-1. Biomed Pharmacother. 72:66–73. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z, Wang R, Zhang T and Dong X:
MicroRNA-126 regulates migration and invasion of gastric cancer by
targeting CADM1. Int J Clin Exp Pathol. 8:8869–8880.
2015.PubMed/NCBI
|
|
24
|
Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N,
Zhou X and Chen C: Reduced miR-126 expression facilitates
angiogenesis of gastric cancer through its regulation on VEGF-A.
Oncotarget. 5:11873–11885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng R, Chen X, Yu Y, Su L, Yu B, Li J,
Cai Q, Yan M, Liu B and Zhu Z: miR-126 functions as a tumour
suppressor in human gastric cancer. Cancer Lett. 298:50–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong JC, Hasan MR, Rahman M, Yu AC, Chan
SK, Schaeffer DF, Kennecke HF, Lim HJ, Owen D and Tai IT:
Nucleophosmin 1, upregulated in adenomas and cancers of the colon,
inhibits p53-mediated cellular senescence. Int J Cancer.
133:1567–1577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wet M, Liu JY, Lv X, Nei H, Liu BY, Zhu
ZG, Yang ZY and Gu QL: Preparation of PHF10 antibody and analysis
of PHF10 expression gastric cancer tissues. Xi Bao Yu Fen Zi Mian
Yi Xue Za Zhi. 26:874–876. 2010.PubMed/NCBI
|
|
28
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng L, Xie Y, Zhang H and Wu Y: miR-107
targets cyclin-dependent kinase 6 expression, induces cell cycle G1
arrest and inhibits invasion in gastric cancer cells. Med Oncol.
29:856–863. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamada D, Ohuchida K, Mizumoto K, Ohhashi
S, Yu J, Egami T, Fujita H, Nagai E and Tanaka M: Increased
expression of ADAM 9 and ADAM 15 mRNA in pancreatic
cancer. Anticancer Res. 27:793–799. 2007.PubMed/NCBI
|
|
31
|
Zhang J, Chen N, Qi J, Zhou B and Qiu X:
HDGF and ADAM9 are novel molecular staging biomarkers, prognostic
biomarkers and predictive biomarkers for adjuvant chemotherapy in
surgically resected stage I non-small cell lung cancer. J Cancer
Res Clin Oncol. 140:1441–1449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tao K, Qian N, Tang Y, Ti Z, Song W, Cao D
and Dou K: Increased expression of a disintegrin and
metalloprotease-9 in hepatocellular carcinoma: Implications for
tumor progression and prognosis. Jpn J Clin Oncol. 40:645–651.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim JM, Jeung HC, Rha SY, Yu EJ, Kim TS,
Shin YK, Zhang X, Park KH, Park SW, Chung HC, et al: The effect of
disintegrin-metalloproteinase ADAM9 in gastric cancer progression.
Mol Cancer Ther. 13:3074–3085. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Carl-McGrath S, Lendeckel U, Ebert M,
Roessner A and Röcken C: The disintegrin-metalloproteinases ADAM9,
ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol.
26:17–24. 2005.PubMed/NCBI
|