Introduction
Cervical cancer is the second most common type of
malignancy in women worldwide, and is the leading cause of
cancer-related deaths among women in developing countries (1). Despite improvements in surgery as well
as in radiotherapy and chemotherapy, the 5-year survival rate of
patients with advanced cervical cancer is still unsatisfactory
(2). Therefore, understanding the
pathogenesis of cervical cancer is beneficial to identifying
optimal therapy and determining patient prognosis.
In recent years, accumulating evidence has shown
that epigenetic silencing and activation of genes are essential to
tumorigenesis and metastasis. For example, epigenetic silencing of
ADAMTS19 is commonly involved in colorectal and mucinous ovarian
cancer (3). GAS7C, a metastasis
suppressor, was found to be hypermethylated in its promoter region
in lung cancer (4). Hypomethylation
of the ELMO3 promoter induced upregulation of its mRNA and
ultimately resulted in the metastasis of lung cancer (5). Aberrant hypomethylation of TKTL1
functions as an oncogene that induces a malignant phenotype of head
and neck cancer (6). Therefore,
identifying candidate epigenetic targets would be instructive for
better understanding of carcinogenesis.
Wolf-Hirschhorn syndrome candidate 1 (WHSC1) is
located in the Wolf-Hirschhorn syndrome (WHS) critical region on
chromosome 4p16.3 and is closely related with multiple myeloma (MM)
(7). The oncogenic role of WHSC1 in
MM was first reported when knockdown of WHSC1 led to cell cycle
arrest (8). In addition to
regulating the expression of many genes related to the mediation of
cell proliferation and adhesion, WHSC1 also regulates the Wnt and
NF-κB signaling pathways in carcinogenesis (9–11).
Clinically, two studies have shown that overexpression of WHSC1
occurs in a large variety of tumors and is associated with tumor
aggressiveness by analysis of a publicly available gene expression
database and conducting experimental analyses, respectively
(12,13). All these data suggest that WHSC1 is
activated in many types of cancer. However, the biological function
and potential mechanism responsible for dysregulated WHSC1
signaling in solid tumors remain unclear.
In the present study, we first identified the
epigenetic regulation of WHSC1 in cervical cancer cells and tissues
as hypomethylation of its promoter CpG island. Both methylation and
mRNA expression of WHSC1 were significantly correlated with lymph
node metastasis and the overall survival of patients. Knockdown of
WHSC1 inhibited cell proliferation, migration and invasion in
vitro and tumorigenicity in vivo. In addition, the
AKT/metalloproteinase-2 (MMP-2) signaling pathway was activated by
overexpression of WHSC1.
Materials and methods
Clinical samples, cell culture and
transfection
All protocols in the present study were approved by
the Ethics Committee of Linyi People's Hospital, and written
informed consent was obtained from patients before surgery. Primary
cervical carcinoma samples (n=65) and their matched non-tumor
tissues were collected from patients undergoing surgery at the
Department of Obstetrics and Gynecology between February 2009 and
July 2011. Non-cancerous tissues were excised at 1.5 cm from the
tumor-free margin. Pathological features were independently
evaluated by two pathologists and classified according to the World
Health Organization (WHO) classification standard. The clinical
data of the patients were extracted from their medical records.
Tissue specimens were stored in liquid nitrogen at −80̊C until
further use.
Human cervical cancer cell lines HeLa, CaSki and
C33A were obtained from the Chinese Center for Type Culture
Collection (Wuhan, China), and cultured under recommended
conditions. HaCaT cells [an immortalized human papillomavirus
(HPV)-negative skin keratinocyte line] were used as the normal
control. All cells were cultured in a humidified incubator at 37̊C
and 5% CO2.
The expression of WHSC1 was silenced by siRNA
interference with the following sequence:
5′-ACTCGTACACAGCGTGGAGTT-3′. Human WHSC1 cDNA (NM_133330.2) was
PCR-amplified, and then subcloned into a lentiviral vector. The
siRNA and cDNA were transfected into 293T cells with the packaging
plasmids, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. The stable
WHSC1-silencing and negative control cells were named as siWHSC1
and siRNA, respectively. Stable WHSC1-overexpressing cells and the
negative control were named as WHSC1 and vector, respectively.
RNA isolation and quantitative
real-time PCR (qRT-PCR)
Total RNA from cell lines and tissue specimens were
isolated using TRIzol reagent (Invitrogen). The quantity of RNA was
measured using NanoDrop 2000 (Thermo, Waltham, MA, USA). The
expression of WHSC1 was determined using qRT-PCR with GAPDH as an
internal reference. Isolated mRNA from each sample was transcribed
to complementary DNA (cDNA) using a First-Strand cDNA Synthesis kit
(Roche, Basel, Switzerland). Primers used for WHSC1 were as
follows: 5′-TAGTCATCACAGACACTTCAC-3′ (forward) and
5′-TGACTCTCGCTCACAACTCT-3′ (reverse); for GAPDH, primers were:
5′-GGAAAGCTGTGGCGTGAT-3′ (forward) and 5′-AAGGTGGAAGAATGGGAGTT-3′
(reverse). Each sample was analyzed in triplicate and comparative
quantification of the target gene was performed based on the cycle
threshold (Ct) normalized to GAPDH using the ΔΔCt method.
DNA isolation, bisulfite modification
and methylation specific-PCR (MSP)
Genomic DNA was isolated from cells and frozen
tissues using DNeasy Blood and Tissue kit (Qiagen, Duesseldorf,
Germany). Bisulfite treatment of genomic DNA (2 µg) was conducted
using EpiTect Bisulfite kit (Qiagen), and the resulting DNA was
used as a template in the MSP assay. The primers of MSP were
designed by Methyl Primer Express v1.0 [Applied Biosystems (ABI)
Foster City, CA, USA]. The primers for the methylated sequence of
WHSC1 were as follows: 5′-CGAGAGTTTCGGTTTGGTC-3′ (forward) and
5′-CTACGATACGTCGCATCCTC-3′ (reverse). For the unmethylated
sequence, primers were: 5′-GTGAGAGTTTTGGTTTGGTT-3′ (forward) and
5′-ACTACAATACATCACATCCTC-3′ (reverse). PCR conditions were 1 cycle
at 95̊C for 15 min; 48 cycles at 94̊C for 30 sec, 56̊C for 20 sec
and 72̊C for 20 sec; and final extension at 72̊C for 10 min.
Bisulphite genomic sequencing
The primers were designed to amplify the promoter
and exon 1 (from −272 to +47) for bisulfite sequencing-PCR (BSP)
analysis. The following primers used were as follows:
5′-GGATTTGAAAAGTTTGGTT-3′ (forward) and 5′-ATCCAACCCAAATACTTCC-3′
(reverse). PCR products were purified and then subjected to
TA-cloning using pUC18-T vector (Shenggong Biological Engineering
Co., Ltd., Shanghai, China). Five clones for each sample were
selected for sequencing and analyzed on DNA sequence analyzer
(ABI). The methylation data obtained from the sequencing were
analyzed using BiQ Analyzer software (Max-Planck Institute,
Saarbrücken, Germany) to generate a lollipop diagram.
Quantitative methylation specific-PCR
assay
A total of 20 ng of converted DNA was used as a
template in the quantitative methylation-specific PCR (QMSP) assay,
and the reaction was conducted on a LightCycler 480 (Roche).
Leukocyte DNA was methylated with excess SssI methyltransferase
(NEB, Beverly, MA, USA) to generate completely methylated DNA, and
serial dilutions (50–0.005 ng) of this DNA were used to construct a
calibration curve. Each plate contained tissue samples, water
blanks and positive controls. Amplifications were performed in
384-well plates, and the reaction (20 µl) included 1.2 mmol/l of
primers, 1 U of platinum Taq DNA polymerase, and 200 mmol/l
of each dNTP. The cycling conditions were as follows: 95̊C for 10
min; 50 cycles of amplification (95̊C for 30 sec, 57̊C for 10 sec,
and 72̊C for 20 sec); and a final step at 72̊C for 10 min. The
relative methylation levels for WHSC1 in each sample were
determined as the ratio of QMSP-amplified WHSC1 to ACTB and then
multiplied by 100 for easier tabulation.
Demethylation analysis
When cell cultures reached 40% confluence, HaCaT
cells were treated with 0 or 5 µM of 5-Aza-2′-deoxycytidine (5-Aza;
Sigma-Aldrich, St. Louis, MO, USA) for 72 h. The fresh media
containing 5-Aza was changed every 24 h for 3 days. After 72 h,
cells were harvested and subjected to qRT-PCR analysis.
Cell proliferation assay
Cell proliferation was evaluated by absorbance using
MTT assay. Briefly, cervical cancer cells (3,000 cells/well) were
plated in 96-well plates. At the indicated time-points (24, 48, 72
and 96 h), 5 mg/ml MTT (Sigma-Aldrich) was added to the cells and
incubated at 37̊C for another 4 h. The absorbance at 490 nm was
assessed using an automatic immunosorbent assay reader (BioTek,
Winooski, VT, USA).
Wound-healing and Transwell invasion
assays
A wound-healing assay was conducted to assess the
migratory ability of the cells. Cells were plated onto 6-well
plates and cultured in complete culture medium until near
confluence (80–90%). The medium was discarded and an artificial
wound was made with a 100-µl sterile pipette tip. The cells were
then washed twice and incubated in serum-free medium for 24 h,
after which images were captured.
The BioCoat Matrigel Invasion Chamber (BD
Biosciences, Franklin Lakes, NJ, USA) was used to assess cell
invasiveness. Cells were resuspended in serum-free medium at a
density of 1×106 cells/ml. Cells (1×105) were
plated into the upper chambers coated with Matrigel. Dulbeccos
modified Eagles medium (DMEM) containing 10% fetal bovine serum
(FBS) was added to the lower chambers. After 24 h, non-migratory
cells on the upper surface of the membrane were carefully scraped
away. Cells on the lower side of the membrane were fixed, stained
with 1% crystal violet for 2 min, visualized at a magnification of
×200 and counted in 6 randomly selected fields. The assays were
performed in triplicate.
Tumor xenografts in vivo
A total of 5×106 cells transfected with
either WHSC1 (siWHSC1) or control siRNA (siRNA) were subcutaneously
injected into the right and left flanks, respectively, of male
nu/nu mice (6-weeks old). After the tumor growth had been monitored
for 5 weeks, the mice were sacrificed, and the tumors were
harvested for immunohistochemistry (IHC) analysis. The tumor volume
was calculated by the following formula: Width × length2
× 0.5.
Western blot analysis
Proteins were extracted by RIPA lysis buffer, and
concentrations were quantified by the BCA protein assay kit (both
from Thermo Fisher Scientific). Equal amounts of protein were
separated by SDS-polyacrylamide gel electrophoresis (PAGE), and
then transferred to polyvinylidine difluoride (PVDF) membranes
(Bio-Rad, Hercules, CA, USA). The membranes were blocked and then
incubated with the primary antibodies overnight at 4̊C, and then
with the horseradish peroxidase-conjugated secondary antibodies.
Bands were visualized using the ECL western blotting detection
system (GE Healthcare, Fairfield, CT, USA).
IHC analysis
Xenograft tumors were sliced into 4-µm sections and
subjected to standard procedures to assess the expression of WHSC1
protein and Ki-67, a proliferation marker. The staining intensity
for WHSC1 was scored as 0 (no staining), 1 (weak), 2 (moderate) and
3 (strong), and the staining area was scored as 0 (0%), 1 (1–25%),
2 (26–50%), 3 (51–75%), or 4 (76–100%) on the basis of the
percentage of positively stained cells. The sum of the area and the
intensity score was calculated to obtain a total score that
indicated the expression of the WHSC1 protein. We also used this
procedure to assess the protein levels of p-AKT, MMP-2 and MMP-9 in
cervical cancer tissues.
Statistical analysis
Data are presented as the mean ± SD from at least 3
independent experiments. Statistical significance was analyzed
using either one-way ANOVA or a two-tail Student's t-test with SPSS
16.0 (IBM, Chicago, IL, USA) and Prism 5 (GraphPad, San Diego, CA,
USA). Associations among categorical data were analyzed using a
Chi-square or Fisher's exact tests. Overall survival curves were
generated by the Kaplan-Meier method and compared using the
log-rank test. Cox's proportional hazard regression analysis was
used to evaluate independent prognostic factors, the hazard ratio
(HR) and the confidence interval (CI). All tests were two-sided,
and a p-value of <0.05 was considered to indicate a
statistically significant result.
Results
WHSC1 is hypomethylated in cervical
cancer cells
First, we evaluated the endogenous mRNA levels of
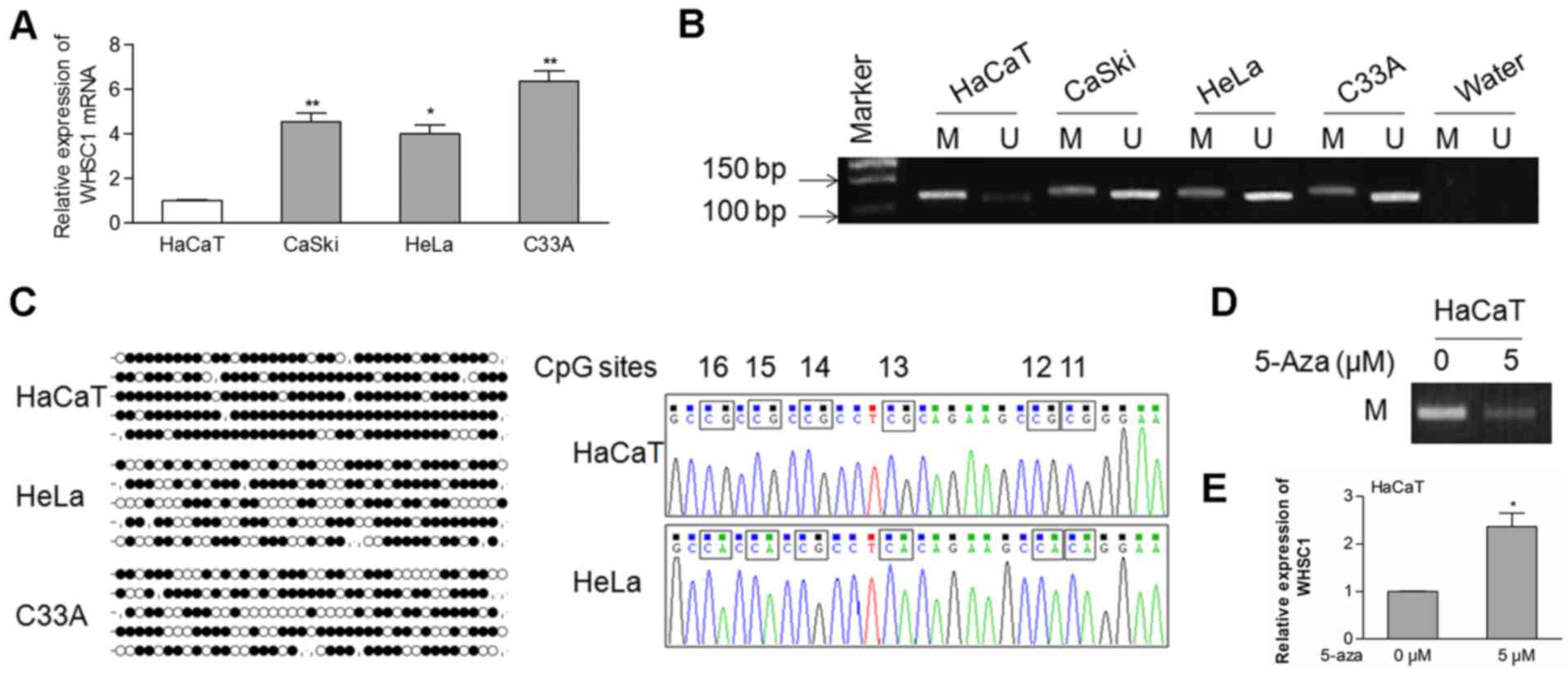
WHSC1 in cervical cancer cells. As shown in Fig. 1A, the mRNA expression of WHSC1 was
significantly upregulated in cervical cancer cells compared to that
in the HaCaT cells. Then, a CpG island located in the promoter
region of WHSC1 was identified using the UCSC Genome Browser
(GRCH38/hg38), suggesting the possibility of epigenetic
modifications of the WHSC1 gene. Using MSP analysis, we observed
that the WHSC1 promoter was hypomethylated in the CaSki, HeLa and
C33A cells, but hypermethylated in the HaCaT cells (Fig. 1B).
To identify the methylation status of specific CpG
sites, we performed the BSP assay. Our data showed that most CpG
sites were methylated in the HaCaT cells, whereas fewer sites were
methylated in the HeLa and C33A cells (Fig. 1C). To further analyze whether
hypomethylation of the WHSC1 gene is correlated with its mRNA
expression, HaCaT cells were treated with the DNA methyltransferase
inhibitor 5-Aza. The methylation of the allele was decreased in the
HaCaT cells treated with 5 µM of 5-Aza in comparison with the
control cells (Fig. 1D). In
contrast, the expression of WHSC1 was increased in cells that
received the 5-Aza treatment (Fig.
1E). These data indicated that hypomethylation of the WHSC1
promoter at least partially regulates its transcriptional
activation in cervical cancer.
Upregulation of WHSC1 is associated
with its hypomethylation in clinical cases
We also ascertained the correlation between the
expression of WHSC1 and its promoter methylation status in clinical
samples. Genomic DNA extracted from 65 pairs of cervical cancer
specimens, and matched normal tissues was amplified by QMSP, and
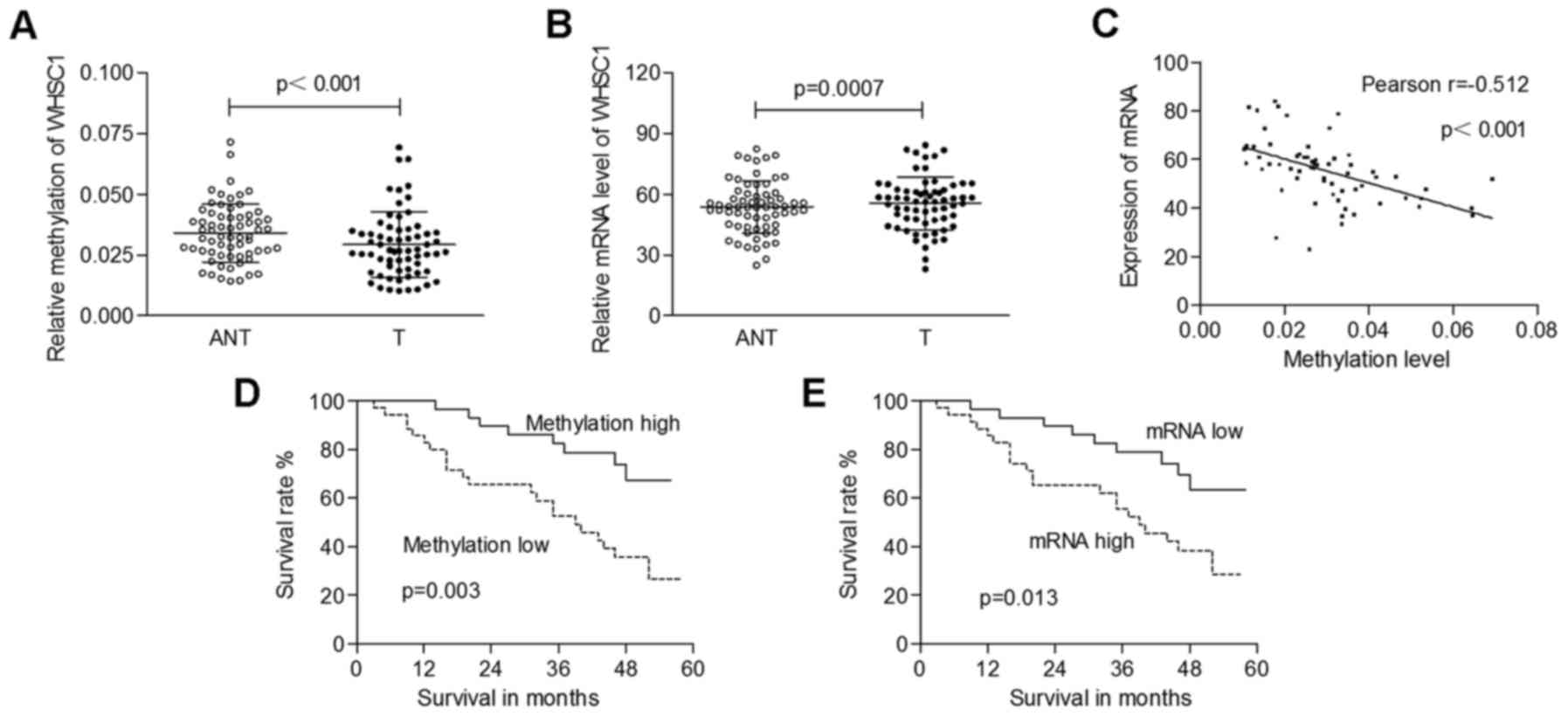
total mRNA was isolated for qRT-PCR analysis. As shown in Fig. 2A, the methylation levels of WHSC1
were significantly decreased in tumors (0.029±0.013) compared to
those in adjacent normal tissues (0.034±0.012; p<0.001). The
mRNA levels of WHSC1 in cancerous tissues (55.64±12.97) were
significantly higher than those in normal tissues (52.76±12.88;
p=0.0007; Fig. 2B). Furthermore, we
identified a significantly negative correlation between WHSC1
methylation and its mRNA expression (Pearson's correlation,
r=−0.512; p<0.001; Fig. 2C).
Dysregulation of WHSC1 is correlated
with tumor progression and clinical outcomes
The mean value of the methylation levels of WHSC1 in
tumors was used as a cut-off to classify cases into 2 groups (low
methylation, n=35; high methylation, n=30). The hypomethylation of
WHSC1 was correlated with higher tumor stage and lymph node
metastasis (p=0.015 and 0.004, respectively; Table I). The mean value of the mRNA levels
of WHSC1 in the tumors was used as the threshold to classify
patients into 2 groups (low expression, n=30; high expression,
n=35). Upregulation of WHSC1 was significant in cases with lymph
node metastasis (p=0.019; Table
I).
 | Table I.Correlations between
clinicopathological characteristics of the cervical cancer patients
and WHSC1 methylation or mRNA levels. |
Table I.
Correlations between
clinicopathological characteristics of the cervical cancer patients
and WHSC1 methylation or mRNA levels.
|
|
| Methylation
level |
| Expression of
mRNA |
|
|---|
|
|
|
|
|
|
|
|---|
| Characteristics | No. of patients
n=65 | Low, n=35 | High, n=30 | P-value | Low, n=30 | High, n=35 | P-value |
| Age (years) |
|
|
| 0.940 |
|
| 0.679 |
|
<55 | 30 | 16 | 14 |
| 13 | 17 |
|
| ≥55 | 35 | 19 | 16 |
| 17 | 18 |
|
| HPV infection |
|
|
| 0.329 |
|
| 0.136 |
| No | 24 | 11 | 13 |
| 14 | 10 |
|
|
Yes | 41 | 24 | 17 |
| 16 | 25 |
|
| Diameter of tumor
(cm) |
|
|
| 0.427 |
|
| 0.427 |
|
<4 | 29 | 14 | 15 |
| 15 | 14 |
|
| ≥4 | 36 | 21 | 15 |
| 15 | 21 |
|
| Histological
type |
|
|
| 0.087 |
|
| 0.263 |
| Well,
moderate | 50 | 24 | 26 |
| 25 | 25 |
|
|
Poor | 15 | 11 | 4 |
| 5 | 10 |
|
| FIGO stage |
|
|
| 0.015 |
|
| 0.056 |
| Ib,
IIa | 35 | 14 | 21 |
| 20 | 15 |
|
| IIb,
IIIa | 30 | 21 | 9 |
| 10 | 20 |
|
| Lymph node
metastasis |
|
|
| 0.004 |
|
| 0.019 |
| No | 31 | 11 | 20 |
| 19 | 12 |
|
|
Yes | 34 | 24 | 10 |
| 11 | 23 |
|
The complete clinical follow-up data were analyzed
regarding the overall survival rate. We found that both low
methylation of WHSC1 and increased mRNA expression were correlated
with a poor clinical outcome (p=0.003 and 0.013, respectively;
Fig. 2D and E). Cox proportional
hazard regression analysis showed the prognostic significance of
the histological type (poor), the International Federation of
Gynecology and Obstetrics (FIGO) stage (IIb/IIIa), and lymph node
metastasis (p=0.001, 0.000 and 0.000, respectively; Table II). The low levels of methylation
of WHSC1 were associated with a relative risk of death of 3.164
(95% CI, 1.406–7.120; p=0.005), and the high mRNA expression of
WHSC1 was correlated with a relative risk of death of 2.589 (95%
CI, 1.183–5.664; p=0.017). However, the results of the multivariate
analysis revealed that poor differentiation (HR, 3.126; p=0.004)
and high tumor-node-metastasis (TNM) stage (HR, 3.787; p=0.001)
were independently correlated with a significantly increased risk
of death (Table II). These data
suggested that overexpression of WHSC1 may be involved in cervical
carcinogenesis.
 | Table II.Clinical characteristics of the
cervical cancer patients and the correlatation with overall
survival by Cox proportional hazard regression analysis. |
Table II.
Clinical characteristics of the
cervical cancer patients and the correlatation with overall
survival by Cox proportional hazard regression analysis.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years
(<55/≥55) | 1.264 | 0.613–2.607 | 0.525 |
|
|
|
| HPV infection
(no/yes) | 0.747 | 0.362–1.540 | 0.429 |
|
|
|
| Diameter of tumor
(<4/≥4 cm) | 1.104 | 0.525–2.323 | 0.794 |
|
|
|
| Histologic type
(well, moderate/poor) | 3.642 | 1.629–7.356 | 0.001 | 3.126 | 1.447–6.753 | 0.004 |
| FIGO stage (Ib,
IIa/IIb, IIIa) | 4.046 | 1.869–8.763 | 0.000 | 3.787 | 1.731–8.285 | 0.001 |
| LNM (no/yes) | 4.819 | 2.052–11.317 | 0.000 | 2.305 | 0.832–6.382 | 0.108 |
| WHSC1 methylation
(high/low) | 3.164 | 1.406–7.120 | 0.005 | 1.363 | 0.522–3.559 | 0.527 |
| WHSC1 mRNA level
(low/high) | 2.589 | 1.183–5.664 | 0.017 | 1.607 | 0.721–3.580 | 0.246 |
Overexpression of WHSC1 promotes cell
proliferation, migration and invasion
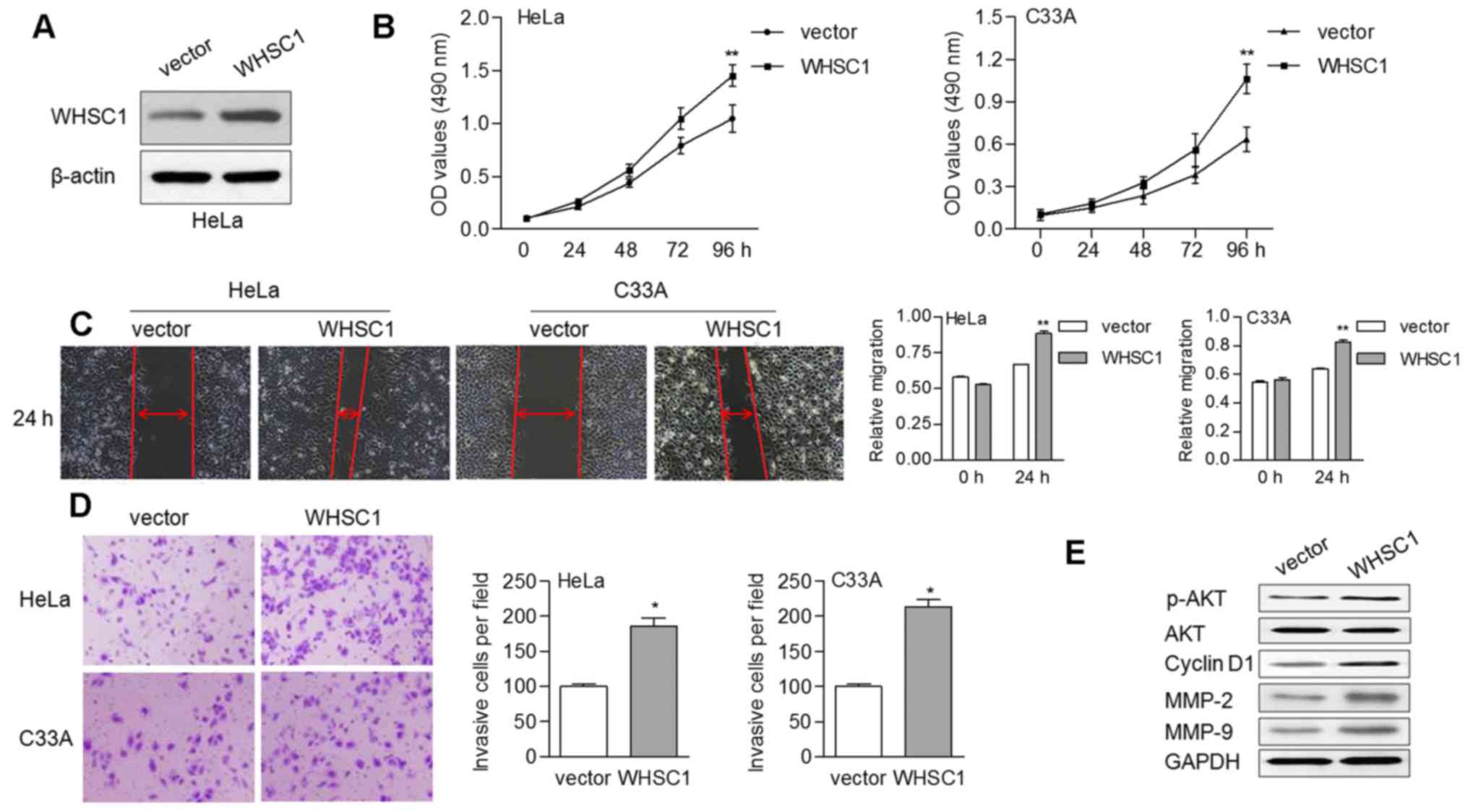
To ascertain the oncogenic role of WHSC1 in cervical
cancer, we performed in vitro experiments to reveal the
effects of WHSC1 on cell growth and motility. By transducing a
vector overexpressing WHSC1, WHSC1 levels were enhanced in the HeLa
cells (Fig. 3A). We found that
overexpression of WHSC1 contributed to cell proliferation in both
the HeLa and C33A cells (Fig. 3B).
Upregulation of WHSC1 also enhanced the migratory ability of HeLa
and C33A cells based on a wound-healing assay (Fig. 3C). Similarly, the invasive ability
of cells was increased by ectopic WHSC1 expression (Fig. 3D). Furthermore, the protein levels
of phosphorylated AKT (p-AKT), cyclin D1 (CCND1), MMP-2 and MMP-9
were increased in cells overexpressing WHSC1 (Fig. 3E). There were no alterations
observed regarding the expression of total AKT, irrespective of the
presence of WHSC1.
Silencing of WHSC1 inhibits cell
growth and motility
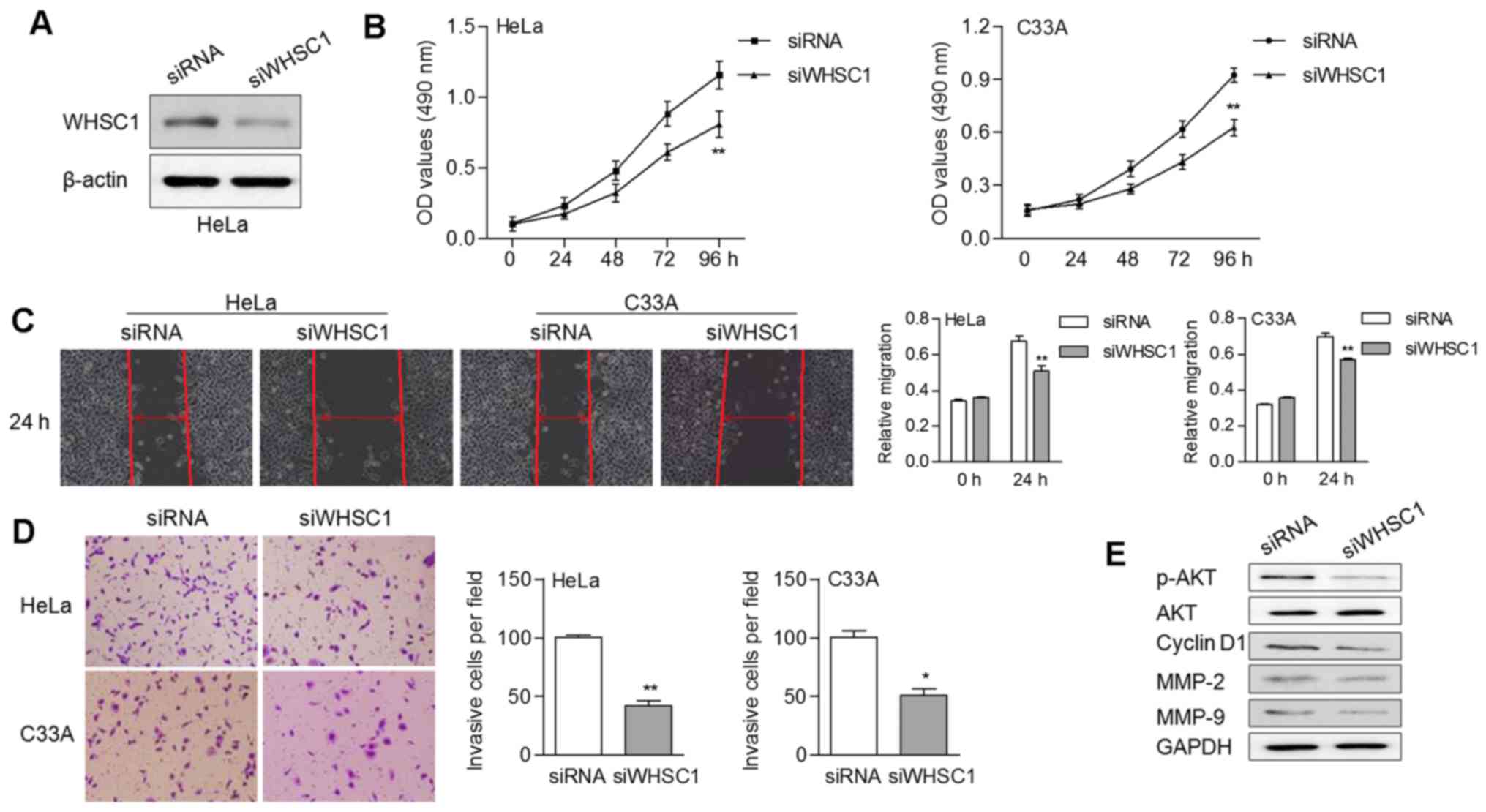
We also knocked down WHSC1 in cancer cells by RNA
interference using siRNA (Fig. 4A).
Stable expression of WHSC1 siRNA led to an obvious decrease of cell
proliferation (Fig. 4B). In
addition, knockdown of WHSC1 suppressed the migratory and invasive
abilities of cervical cancer cells (Fig. 4C and D). The protein expression
levels of p-AKT, CCND1, MMP-2 and MMP-9 were noticeably decreased
in cells with WHSC1 knockdown (Fig.
4E). These data suggested that WHSC1 probably contributed to
tumor progression by activating AKT/MMP-2 signaling in cervical
cancer.
Knockdown of WHSC1 inhibits tumor
growth in vivo
Based on the oncogenic effect of WHSC1 observed in
the in vitro experiments, a xenograft model of human
cervical cancer cells was applied in nude mice. There was a
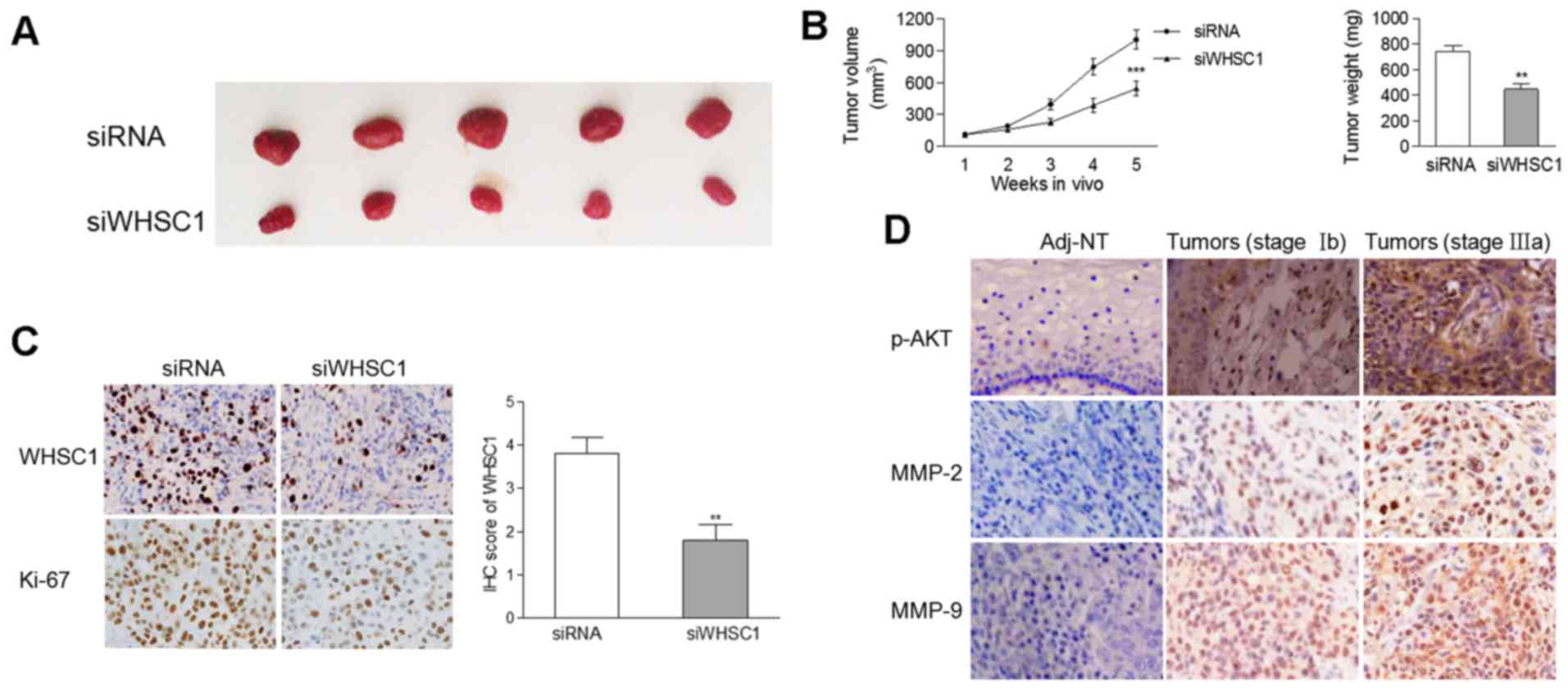
significant decrease in the tumor size and weight of tumors
containing cells with WHSC1 knockdown compared to tumors comprised
of cells from the control group (Fig.
5A and B). IHC revealed that the protein levels of WHSC1 and
Ki-67 were decreased in tumors stably-expressing siWHSC1 (Fig. 5C), suggesting an oncogenic role of
WHSC1 in vivo.
WHSC1 is positively correlated with
p-AKT, MMP-2 and MMP-9 expression in clinical samples
Considering that WHSC1 activated AKT/MMP-2 signaling
in vitro, we determined the protein levels of p-AKT, MMP-2
and MMP-9 by IHC in 65 pairs of cervical cancer samples and matched
normal tissues. As the degree of malignancy increased, there was a
gradual increase in the protein levels of p-AKT, MMP-2 and MMP-9 in
clinical samples (Fig. 5D),
suggesting that AKT/MMP-2 signaling was activated during cervical
tumorigenesis. We found that the mRNA expression of WHSC1 was
positively correlated with the protein levels of p-AKT (Spearman
r=0.292, p=0.018) and MMP-9 (Spearman r=0.304, p=0.014), but not
with MMP-2 (Spearman r=0.231, p=0.064).
Discussion
Epigenetic modification and the consequent
dysregulation of genes are emerging as promising novel targets in
the treatment and prognostic prediction of cervical cancer
(14,15). In the present study, we described a
novel mechanism in which epigenetic activation of WHSC1 is induced
by hypomethylation of its promoter and provided direct evidence
that WHSC1 is a critical oncogene in human cervical cancer.
First, we preliminarily determined the endogenous
WHSC1 levels in cervical cancer cells. Our data revealed that WHSC1
is overexpressed in tumors cells, which was similar with findings
from previous studies (13,16). Further MSP and BSP analyses
ascertained the hypomethylation status of the CpG island in the
promoter region of WHSC1, suggesting that hypomethylation of this
CpG island leads to WHSC1 activation during the development of
cervical cancer. Using the publicly available database MethHC
(methhc.mbc.nctu.edu.tw), we found that
the methylation level of the CpG island near the WHSC1 promoter was
markedly decreased in cervical cancer (p<0.005) and inversely
associated with its mRNA expression (correlation r=−0.113,
p=1.11×10−16; data not shown). In addition, the results
from this database showed that the methylation levels of WHSC1 were
also decreased in colorectal adenocarcinoma, head and neck
squamous, lung squamous and renal clear cell carcinoma.
Collectively, all these observations indicated that promoter
hypomethylation may be an important mechanism responsible for the
activation of WHSC1 in various human cancers. We reviewed previous
studies and found that WHSC1 was also reported as a hypomethylated
gene in head and neck cancer by genome-wide methylation profiling
(17). Our findings provide the
direct evidence that hypomethylation of WHSC1 is involved in the
pathogenesis of cervical cancer.
Regarding the aforementioned findings, we observed
that WHSC1, a specific histone methyltransferase, induced the
dimethylation of histone 3 at lysine 36 (H3K36 me2) and decreased
the trimethylation of histone 3 at lysine 27 (H3K27 me3) (18,19).
WHSC1 regulated the expression of TWIST1 and NEK7 through H3K36 me2
(19,20). Presumably, genomic disorders of
H3K36 me2 and tumor-related genes mediated by WHSC1 may contribute
to malignant programming.
We assessed the WHSC1 methylation level in 65 pairs
of tumors and corresponding normal tissues, and found that the
increased hypomethylation of WHSC1 in tumors indicated tumor
progression, suggesting the clinical importance of epigenetic
modification. Furthermore, increased expression of WHSC1 was
associated with lymph node metastasis and a worse survival rate.
Similar observations were also documented in previous studies
(12,13,21).
By comparing the mRNA levels of WHSC1 across 3 analyses from
Oncomine (www.oncomine.org), we found that the
expression of WHSC1 was significantly upregulated in cervical
cancer tissues (p=9.88×10−11, data not shown). Our data
were consistent with these results, thus supporting the oncogenic
role of WHSC1 in the development of cervical tumorigenesis.
Upregulated WHSC1 mRNA was not an independent prognostic factor in
the present study, while opposing observations were noted in other
studies regarding the protein expression of WHSC1 (22,23).
Tissue-specific expression and the types of samples assessed may
explain these differences. Further analysis based on a larger
cohort may provide more accurate information regarding the
efficiency of WHSC1 as a prognostic factor.
Finally, the suppressive effect of WHSC1 on cell
growth and motility was ascertained, supporting the findings that
dysregulated WHSC1 is correlated with lymph node metastasis in
clinical cases. In addition, overexpression of WHSC1 led to
activation of the AKT signaling pathway and upregulation of MMP-2
and MMP-9, revealing the possible mechanism responsible for the
alterations of tumor cell behavior. In prostate cancer, TWIST1 was
identified as a target of WHSC1, which induced the migration and
invasion of immortalized RWPE-1 cells (19). WHSC1 knockdown resulted in
significantly induced apoptosis rates in cells from head and neck
squamous cell carcinoma (20).
Several tumor-related signaling pathways, such as the Wnt and
NF-κB, were also reported to be activated by WHSC1 (10,11).
Furthermore, we found that the protein levels of p-AKT and MMP-9
were correlated with WHSC1 in clinical samples, suggesting that the
AKT/MMP-2 signaling pathway is at least partially activated in
WHSC1-related cervical carcinogenesis. The matrix
metalloproteinases (MMPs) is a large family of proteases known as
zinc-containing metalloproteins (24). These enzymes are capable of
degrading extracellular matrix proteins (25). MMPs were reported to be critical
during advanced stages of tumor progression as regulators of tumor
cell migration, invasion and metastasis (26). In addition, both MMP-2 and MMP-9 act
as positive regulators of angiogenesis (27,28).
Our data expanded the knowledge of the role of WHSC1 in human
malignancies, suggesting an oncogenic function of WHSC1 in cervical
cancer.
In conclusion, our present findings describe WHSC1
as a novel hypomethylated gene in cervical cancer and provide
evidence that this gene is a promising prognostic marker.
Furthermore, we suggest that WHSC1 promotes more aggressive types
of cervical cancer through the modulation of the AKT/MMP-2
signaling pathway.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eskander RN and Tewari KS: Targeting
angiogenesis in advanced cervical cancer. Ther Adv Med Oncol.
6:280–292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alonso S, González B, Ruiz-Larroya T,
Durán Domínguez M, Kato T, Matsunaga A, Suzuki K, Strongin AY,
Gimènez-Bonafé P and Perucho M: Epigenetic inactivation of the
extracellular matrix metallopeptidase ADAMTS19 gene and the
metastatic spread in colorectal cancer. Clin Epigenetics.
7:1242015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tseng RC, Chang JW, Mao JS, Tsai CD, Wu
PC, Lin CJ, Lu YL, Liao SY, Cheng HC, Hsu HS, et al:
Growth-arrest-specific 7C protein inhibits tumor metastasis via the
N-WASP/FAK/F-actin and hnRNP U/β-TrCP/β-catenin pathways in lung
cancer. Oncotarget. 6:44207–44221. 2015.PubMed/NCBI
|
|
5
|
Søes S, Daugaard IL, Sørensen BS, Carus A,
Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL and
Kristensen LS: Hypomethylation and increased expression of the
putative oncogene ELMO3 are associated with lung cancer
development and metastases formation. Oncoscience. 1:367–374. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun W, Liu Y, Glazer CA, Shao C, Bhan S,
Demokan S, Zhao M, Rudek MA, Ha PK and Califano JA: TKTL1 is
activated by promoter hypomethylation and contributes to head and
neck squamous cell carcinoma carcinogenesis through increased
aerobic glycolysis and HIF1alpha stabilization. Clin Cancer Res.
16:857–866. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chesi M, Nardini E, Lim RS, Smith KD,
Kuehl WM and Bergsagel PL: The t(4;14) translocation in myeloma
dysregulates both FGFR3 and a novel gene, MMSET,
resulting in IgH/MMSET hybrid transcripts. Blood. 92:3025–3034.
1998.PubMed/NCBI
|
|
8
|
Lauring J, Abukhdeir AM, Konishi H, Garay
JP, Gustin JP, Wang Q, Arceci RJ, Matsui W and Park BH: The
multiple myeloma associated MMSET gene contributes to
cellular adhesion, clonogenic growth, and tumorigenicity. Blood.
111:856–864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brito JL, Walker B, Jenner M, Dickens NJ,
Brown NJ, Ross FM, Avramidou A, Irving JA, Gonzalez D, Davies FE,
et al: MMSET deregulation affects cell cycle progression and
adhesion regulons in t(4;14) myeloma plasma cells. Haematologica.
94:78–86. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toyokawa G, Cho HS, Masuda K, Yamane Y,
Yoshimatsu M, Hayami S, Takawa M, Iwai Y, Daigo Y, Tsuchiya E, et
al: Histone lysine methyltransferase Wolf-Hirschhorn syndrome
candidate 1 is involved in human carcinogenesis through regulation
of the Wnt pathway. Neoplasia. 13:887–898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang P, Guo L, Duan ZJ, Tepper CG, Xue L,
Chen X, Kung HJ, Gao AC, Zou JX and Chen HW: Histone
methyltransferase NSD2/MMSET mediates constitutive NF-κB signaling
for cancer cell proliferation, survival, and tumor growth via a
feed-forward loop. Mol Cell Biol. 32:3121–3131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kassambara A, Klein B and Moreaux J: MMSET
is overexpressed in cancers: Link with tumor aggressiveness.
Biochem Biophys Res Commun. 379:840–845. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hudlebusch HR, Santoni-Rugiu E, Simon R,
Ralfkiær E, Rossing HH, Johansen JV, Jørgensen M, Sauter G and
Helin K: The histone methyltransferase and putative oncoprotein
MMSET is overexpressed in a large variety of human tumors. Clin
Cancer Res. 17:2919–2933. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin A, Zhang Q, Kong X, Jia L, Yang Z,
Meng L, Li L, Wang X, Qiao Y, Lu N, et al: JAM3 methylation
status as a biomarker for diagnosis of preneoplastic and neoplastic
lesions of the cervix. Oncotarget. 6:44373–44387. 2015.PubMed/NCBI
|
|
15
|
Sood S, Patel FD, Ghosh S, Arora A,
Dhaliwal LK and Srinivasan R: Epigenetic alteration by DNA
methylation of ESR1, MYOD1 and hTERT gene
promoters is useful for prediction of response in patients of
locally advanced invasive cervical carcinoma treated by
chemoradiation. Clin Oncol. 27:720–727. 2015. View Article : Google Scholar
|
|
16
|
Gu C, Feng L, Peng H, Yang H, Feng Z and
Yang Y: MTDH is an oncogene in multiple myeloma, which is
suppressed by Bortezomib treatment. Oncotarget. 7:4559–4569.
2016.PubMed/NCBI
|
|
17
|
Zhang XY, Li M, Sun K, Chen XJ, Meng J, Wu
L, Zhang P, Tong X and Jiang WW: Decreased expression of GRIM-19 by
DNA hypermethylation promotes aerobic glycolysis and cell
proliferation in head and neck squamous cell carcinoma. Oncotarget.
6:101–115. 2015.PubMed/NCBI
|
|
18
|
Popovic R, Martinez-Garcia E, Giannopoulou
EG, Zhang Q, Zhang Q, Ezponda T, Shah MY, Zheng Y, Will CM, Small
EC, et al: Histone methyltransferase MMSET/NSD2 alters EZH2 binding
and reprograms the myeloma epigenome through global and focal
changes in H3K36 and H3K27 methylation. PLoS Genet.
10:e10045662014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ezponda T, Popovic R, Shah MY,
Martinez-Garcia E, Zheng Y, Min DJ, Will C, Neri A, Kelleher NL, Yu
J, et al: The histone methyltransferase MMSET/WHSC1 activates
TWIST1 to promote an epithelial-mesenchymal transition and invasive
properties of prostate cancer. Oncogene. 32:2882–2890. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saloura V, Cho HS, Kiyotani K, Alachkar H,
Zuo Z, Nakakido M, Tsunoda T, Seiwert T, Lingen M, Licht J, et al:
WHSC1 promotes oncogenesis through regulation of NIMA-related
kinase-7 in squamous cell carcinoma of the head and neck. Mol
Cancer Res. 13:293–304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang S, Zhang Y, Meng F, Liu Y, Xia B,
Xiao M, Xu Y, Ning X, Li H and Lou G: Overexpression of multiple
myeloma SET domain (MMSET) is associated with advanced tumor
aggressiveness and poor prognosis in serous ovarian carcinoma.
Biomarkers. 18:257–263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou P, Wu LL, Wu KM, Jiang W, Li JD, Zhou
LD, Li XY, Chang S, Huang Y, Tan H, et al: Overexpression of MMSET
is correlation with poor prognosis in hepatocellular carcinoma.
Pathol Oncol Res. 19:303–309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiao M, Yang S, Chen J, Ning X, Guo L,
Huang K and Sui L: Overexpression of MMSET in endometrial cancer: A
clinicopathologic study. J Surg Oncol. 107:428–432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hrabeta J, Eckschlager T, Stiborova M,
Heger Z, Krizkova S and Adam V: Zinc and zinc-containing
biomolecules in childhood brain tumors. J Mol Med (Berl).
94:1199–1215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Basset P, Bellocq JP, Wolf C, Stoll I,
Hutin P, Limacher JM, Podhajcer OL, Chenard MP, Rio MC and Chambon
P: A novel metalloproteinase gene specifically expressed in stromal
cells of breast carcinomas. Nature. 348:699–704. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chabottaux V and Noel A: Breast cancer
progression: Insights into multifaceted matrix metalloproteinases.
Clin Exp Metastasis. 24:647–656. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nelson AR, Fingleton B, Rothenberg ML and
Matrisian LM: Matrix metalloproteinases: Biologic activity and
clinical implications. J Clin Oncol. 18:1135–1149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Raithatha SA, Muzik H, Muzik H, Rewcastle
NB, Johnston RN, Edwards DR and Forsyth PA: Localization of
gelatinase-A and gelatinase-B mRNA and protein in human gliomas.
Neuro Oncol. 2:145–150. 2000. View Article : Google Scholar : PubMed/NCBI
|