Introduction
Human and mouse genomes generate many transcripts
that lack the capacity to produce functional proteins. These
products include non-coding RNAs (ncRNA) (1). Small (<200 nucleotides) ncRNAs are
involved in multiple biological processes such as neuronal
development, and are potential key regulators of human diseases,
including cancer (2,3). The functions and mechanisms of action
of long ncRNA (lncRNA), transcripts ranging from 200 nucleotides to
100 kb, are also being uncovered, in applications ranging from
cancer to epigenetics (4–8). Some of these lncRNA are expressed in a
cellular compartment- or tissue-specific manner and at
substantially lower levels than mRNAs (9), which suggests key regulatory roles in
gene expression both at transcriptional and post-transcriptional
levels (10–12).
Malat1, also known as non-coding nuclear-enriched
abundant transcript 2 (Neat2), was originally identified as an
lncRNA whose expression was increased in early-stage non-small cell
lung cancers that later metastasized compared with those that did
not progress into tumors (13).
Malat1 lacks open reading frames of significant length, is highly
conserved and is located in the nucleus, specifically in nuclear
speckles (14), where it regulates
alternative splicing (15). In
addition, post-transcriptional modifications of Malat1 yield a
tRNA-like 61 nt transcript (termed mascRNA) that is exported to the
cytoplasm (16) for as yet elusive
functions.
High expression levels of Malat1 within tumors have
been associated with poor prognosis and severity of various types
of cancer, including bladder (17),
lung (18), gallbladder (19) and liver cancers (20). In addition, circulating Malat1
levels have also been recently shown to predict development of
hepatocellular carcinoma (21,22),
suggesting its possible use as a reliable biomarker. Moreover,
Malat1 may be directly implicated in the development of cancer
through its effect on cellular proliferation and its facilitating
role in cell invasion and metastasis, two important hallmarks of
cancer (18,23,24).
In cultured cells, Malat1 promotes proliferation and cell motility
by influencing the expression of oncogenic transcription factors
(e.g., B-MYB) or motility-related genes through transcriptional
and/or post-transcriptional regulation (23,25,26).
Nevertheless, the direct causal contribution of Malat1 to
carcinogenesis in vivo remains poorly investigated. In this
study, we tested the hypothesis that absence of Malat1 impairs the
development of liver carcinogenesis induced by the genotoxic agent
DEN. Our results rather indicate that Malat1 ablation does not
modify the susceptibility to DEN-associated hetapocarcinoma.
Materials and methods
Animals
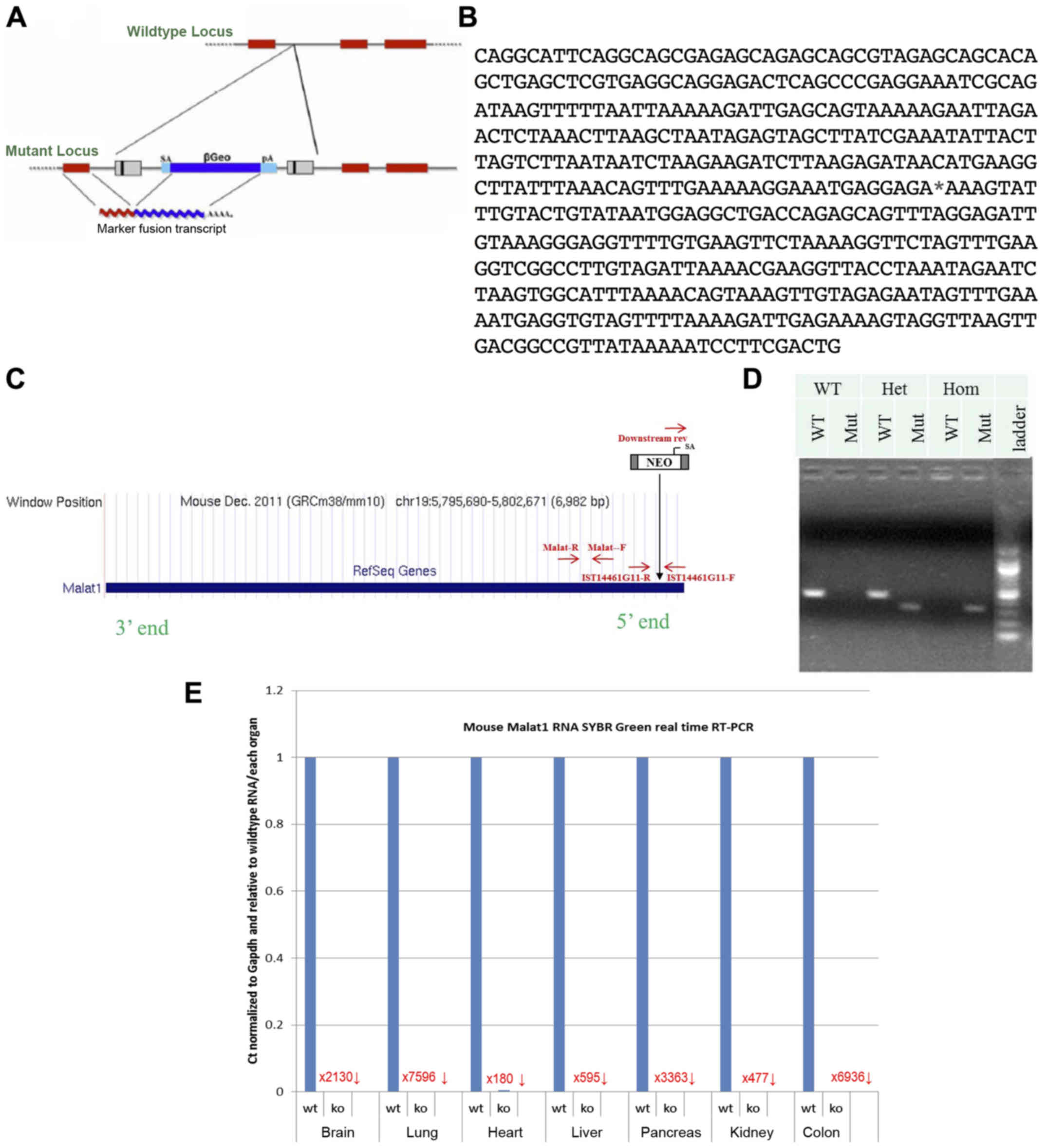
Mutant Malat1 mice were generated using a
gene-trapping technique (27)
(Fig. 1A). Mice (strain C57BL/6)
were cloned from an ES cell line (IST14461G11; Texas A&M
Institute for Genomic Medicine, TIGM). The ES cell clone contained
a retroviral insertion in the Malat1 gene identified from
the TIGM gene trap database (Fig.
1B), and was microinjected into C57BL/6 host blastocysts to
generate germline chimeras using standard procedures (28). The retroviral OmniBank Vector 74
contained a splice acceptor sequence (SA) followed by a 5′
selectable marker neomycin resistance genes, for identification of
successful gene trap events followed by a polyadenylation signal
(pA) (Fig. 1C). Insertion of the
retroviral vector into the Malat1 gene led to the splicing
of the endogenous upstream exons into this cassette to produce a
fusion transcript and terminate expression of the RNA downstream.
Chimeric males were bred to C57BL/6 females for germline
transmission of the mutant Malat1 allele. The correct
mutation was confirmed using PCR-based genotyping protocol using
primers specific for genomic insertion site and for the vector
5′-AGAGCAGAGCAGCGTAGAGC-3′, 5′-TAACGGCCGTCAACTTAACC-3′,
5′-CCAATAAACCCTCTTGCAGTTGC-3′ (Fig.
1D). Malat1 expression levels were quantified in several
tissues to confirm gene knockout (Fig.
1E). Compared to wild-type littermates, Malat1−/−
mice showed an absence of Malat1 expression in all tissues studied
(Fig. 1E). Notably, expression in
the liver was reduced by 595-fold (Fig.
1E).
At 15-day-old, wild-type (WT) and
Malat1−/− male littermates were injected with 5 µg/g of
the potent genotoxic agent N-nitrosodiethylamine (DEN) (Sigma no.
N0258) to induce liver tumors (29). Mice were weighed every day for the
first 15 days and then weekly for the remaining of the protocol.
Mice were exposed to a 12:12-h dark-light cycle and kept at ambient
temperature of ± 2°C. Animals were euthanized by exsanguination
(cardiac puncture) under ketamine/xylazine anesthesia. Tissues were
collected, weighed, and snap frozen or fixed in 4% paraformaldehyde
(PFA) until further processing. All mice were cared for and handled
in conformance with the Canadian Guide for the Care and Use of
Laboratory Animals, and protocols were approved by our
institutional animal care committee.
Histology
Scoring of histologic parameters was performed by an
anatomic pathologist with experience in pulmonary pathology,
independently and blinded to experimental data, using an Olympus
BX53 microscope. A semiquantitative scale was used to score
bronchial/endobronchial, peribronchial, perivascular, interstitial,
pleural and intra alveolar inflammation, capillary vascular
congestion and pulmonary edema. When present, metastases were
measured and photographed.
Plasma biochemistry
At sacrifice, blood was collected by intracardiac
puncture and placed into a tube containing EDTA. Plasma was stored
at −80°C for further biochemical analyses. Plasma cholesterol and
triglycerides were measured using colorimetric assays (Thermo
Fisher Scientific and Wako, respectively).
Glucose tolerance test
To evaluate glucose tolerance, mice were fasted for
12 h starting at 8 pm with free access to water. The following
morning, mice were weighed, baseline glycemia was measured and mice
were injected intraperitoneally with 2 g/kg of D-glucose. Glycemia
was measured in blood from the tail vein at different intervals
following glucose injection using an Accu-Chek performa glucometer
(Roche).
RNA extraction and real-time
quantitative PCR analysis
Total RNA was extracted using Aurum total RNA fatty
and fibrous tissue kit (Bio-Rad). Purity, degradation state and
concentration of the RNA samples were analyzed by the Experion
automated electrophoresis system (Bio-Rad). cDNA was synthetized
from 1 µg of RNA using qScript reverse transcriptase (Quanta
Bioscience, USA) according to the manufacturer's instructions.
Semi-quantitative PCR was carried using an ABI 7900. Chemical
detection of the PCR products was achieved with SYBR Green
Jumpstart Taq ReadyMix without MgCl2 (Sigma, Oakville,
ON, USA) (30). All reactions were
performed in duplicate and relative level of gene expression was
determined by the standard curve method. Results were normalized to
the expression level of the reference gene hypoxanthine-guanine
phosphoribosyltransferase (HPRT), which did not differ between
groups. PCR primers used are listed in Tables I and II.
 | Table I.Primers used for the quantification
of genes detailed in Fig. 5. |
Table I.
Primers used for the quantification
of genes detailed in Fig. 5.
| Gene name | Forward | Reverse |
|---|
| HPRT |
AAACTTTGCTTTCCCTG |
AGGCTTTGTATTTGGCT |
| Ki67 |
AGGAGGCAGCTAAGGACACA |
ACACTTCCTTGGGGTCCTCT |
| P53 |
AGAGACCGCCGTACAGAAGAAG |
TTTTTATGGCGGGAAGTAGAC |
| HDAC3 |
CACCCGCATCGAGAATCAGAAC |
CAGCGTCGGCCTCGTCAGTC |
| TERT |
AGGGTAAGCTGGTGGAGGTT |
GATGCTCTGCTCGATGACAA |
| HDAC1 |
ACGGGAGGCTCTGTCGCAAGTG |
CCAGCCCCAATGTCCCGTAGG |
| NFκB |
GCTCAGCGGGCAGTATTCCT |
AGTCCCCGCGCTGCTCCTCTAT |
| Foxo3 |
GGCTCCCCAACCGGCTCCTTCAA |
CACGTTCCGGCGGGCATTCTGG |
| FoxoA1 |
CTCCCGGTACTTCTCTGCTG |
GTGGTCGAGTTGGACTGGTT |
| P27 |
CGAGCCTGGAGCGGATGGAC |
GCGCGGGGGCCTGTAGTAGAAC |
| P21 |
CAGGTCGGCAGGAGGCATATCTAG |
ATCCCAGATAAGCCCACCCC |
| TNFα |
AACTAGTGGTGCCAGCCGATG |
CGGACTCCGCAAAGTCTAAG |
| c-myc |
CTCGCCGCCGCTGGGAAACTT |
AGGGGCATCGTCGTGGCTGTCTG |
| CDK4 |
GTCAGTTTCTAAGCGGCCTG |
CACGGGTGTTGCGTATGTAG |
| IL-6 |
AGTTGCCTTCTTGGGACTGA |
CAGAATTGCCATTGCACAAC |
 | Table II.Primer used to assess the reduction
in Malat1 expression in Malat1 knockout animals (Fig. 1E). |
Table II.
Primer used to assess the reduction
in Malat1 expression in Malat1 knockout animals (Fig. 1E).
| Gene name | Forward | Reverse |
|---|
| Malat1 |
GGCAGAATGCCTTTGAAGAG (named Malat1-F in
Fig. 1C) |
GGTCAGCTGCCAATGCTAGT (named Malat1-R in
Fig. 1C) |
| GAPDH |
GGCATTGCTCTCAATGACAAC |
GCCATGTAGGCCATGAGGT |
Data analysis
Data are presented as mean ± SEM. Statistical
differences were analyzed by Chi-square, ANOVA, ANOVA repeated
measures, and Fisher's tests (ad hoc) when appropriate. A
p-value <0.05 was considered statistically significant.
Results
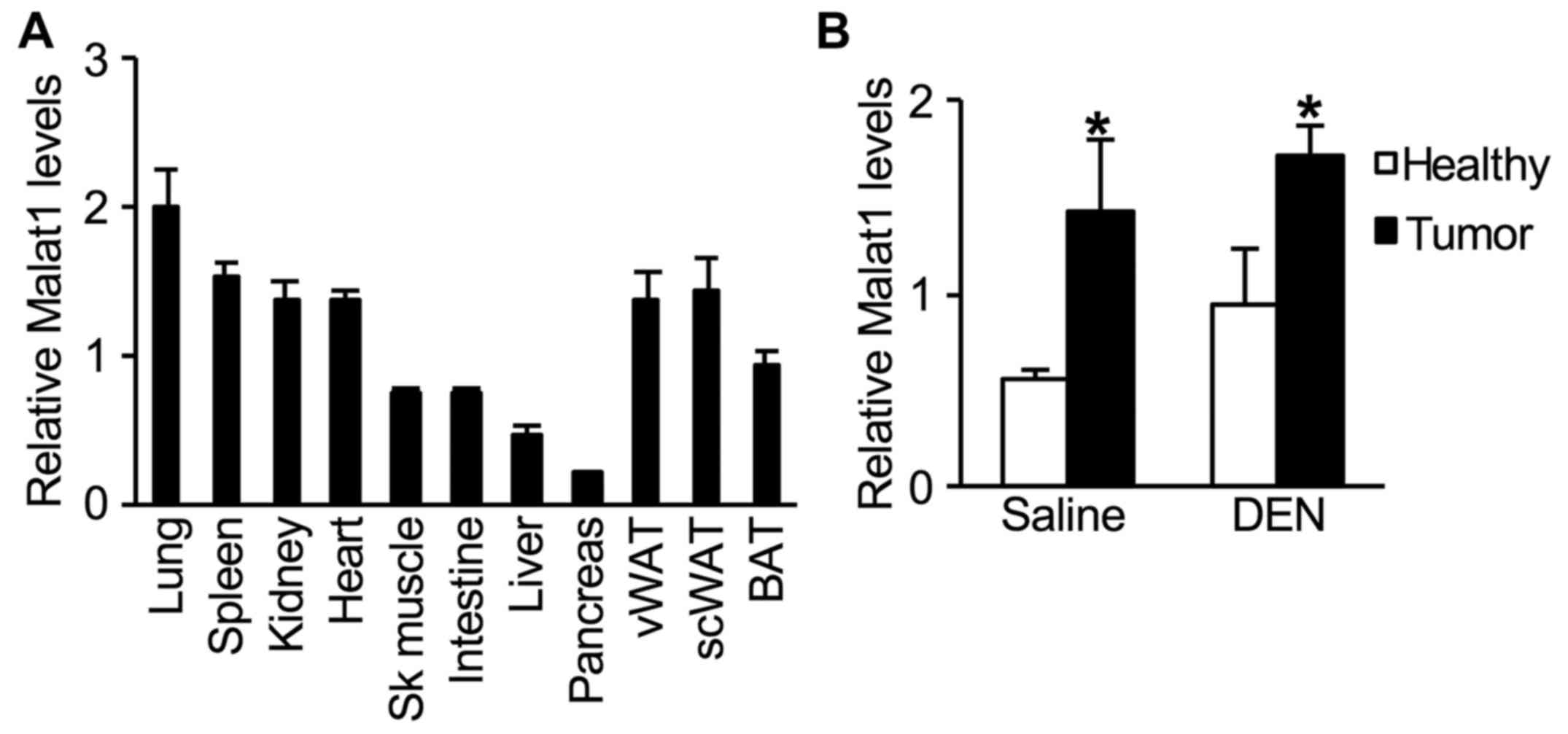
In untreated 2-month-old WT male mice, hepatic
expression levels of Malat1 were lower when compared to those in
other tissues (Fig. 2A). One year
after DEN injection, an increase in Malat1 expression was observed
in tumors when compared to that in healthy liver tissue (Fig. 2B). However, this increase in Malat1
levels was similar whether tumors developed spontaneously as
function of age, or induced by DEN administration (Fig. 2B).
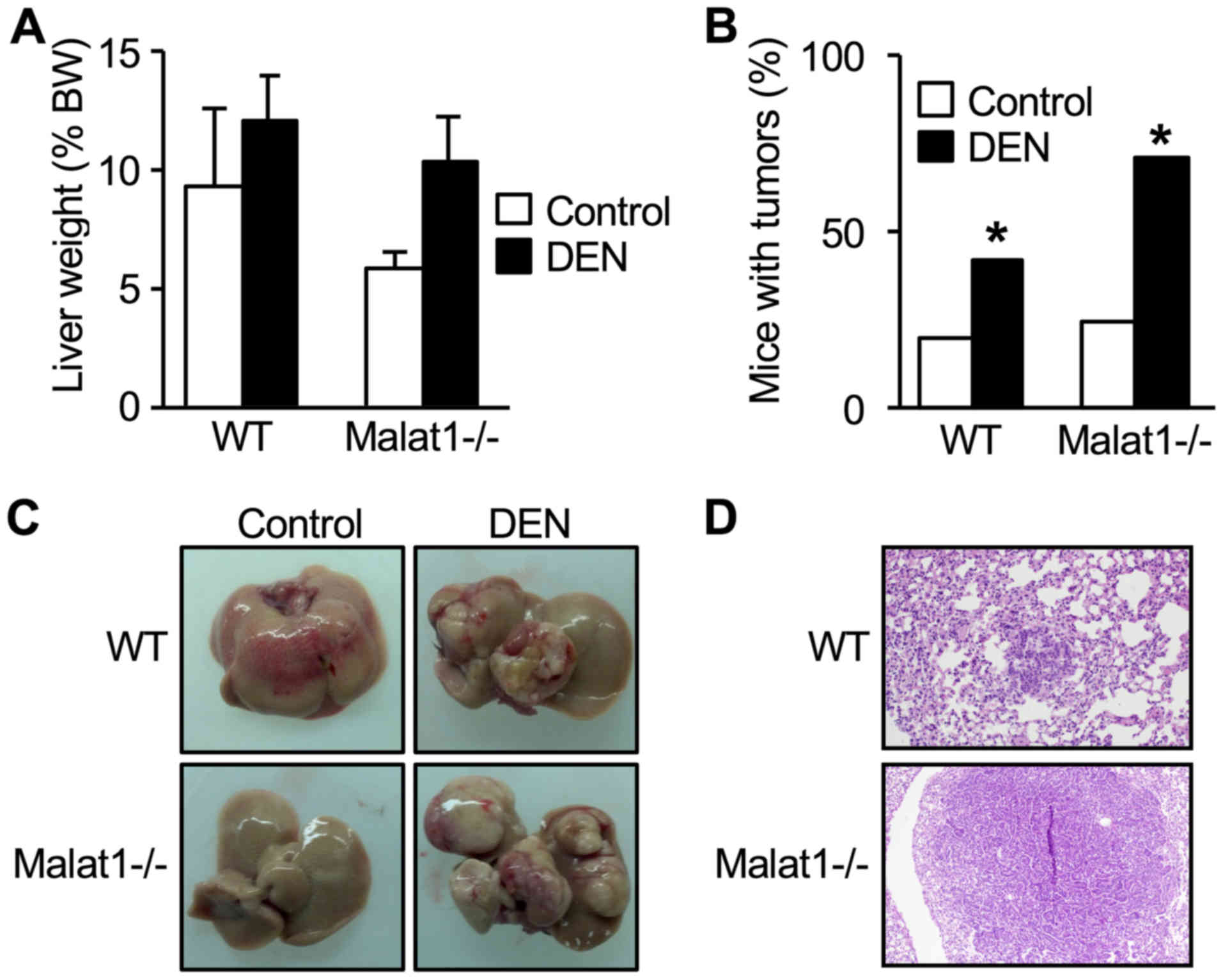
In WT mice, DEN injection did not change total liver
weight (Fig. 3A), but resulted in
an increased number and prevalence of tumors (Fig. 3B). This resulted in a distorted
tissue with increased number of lobes containing at least one tumor
(Fig. 3C). These features equally
developed in DEN-treated Malat1−/− animals (Fig. 3A-C) (no statistical difference
between DEN-treated WT and Malat1−/− mice), indicating
that absence of Malat1 had no impact on the incidence and severity
of DEN-induced hepatocarcinoma.
Histopathology examination of the lung and
semi-quantitative histologic scaling between DEN-treated WT mice
and Malat1−/− littermates revealed comparable prevalence
of peribronchial (2 mice vs 0, respectively), perivascular (2 vs 4)
interstitial (0 vs 0), pleural (0 vs 0), and intra-alveolar
inflammation (1 vs 0). One DEN-treated WT mouse had atypical cell
foci suggesting a lung metastasis, whereas one Malat1−/−
mouse had well-defined, differentiated lung metastasis (Fig. 3D).
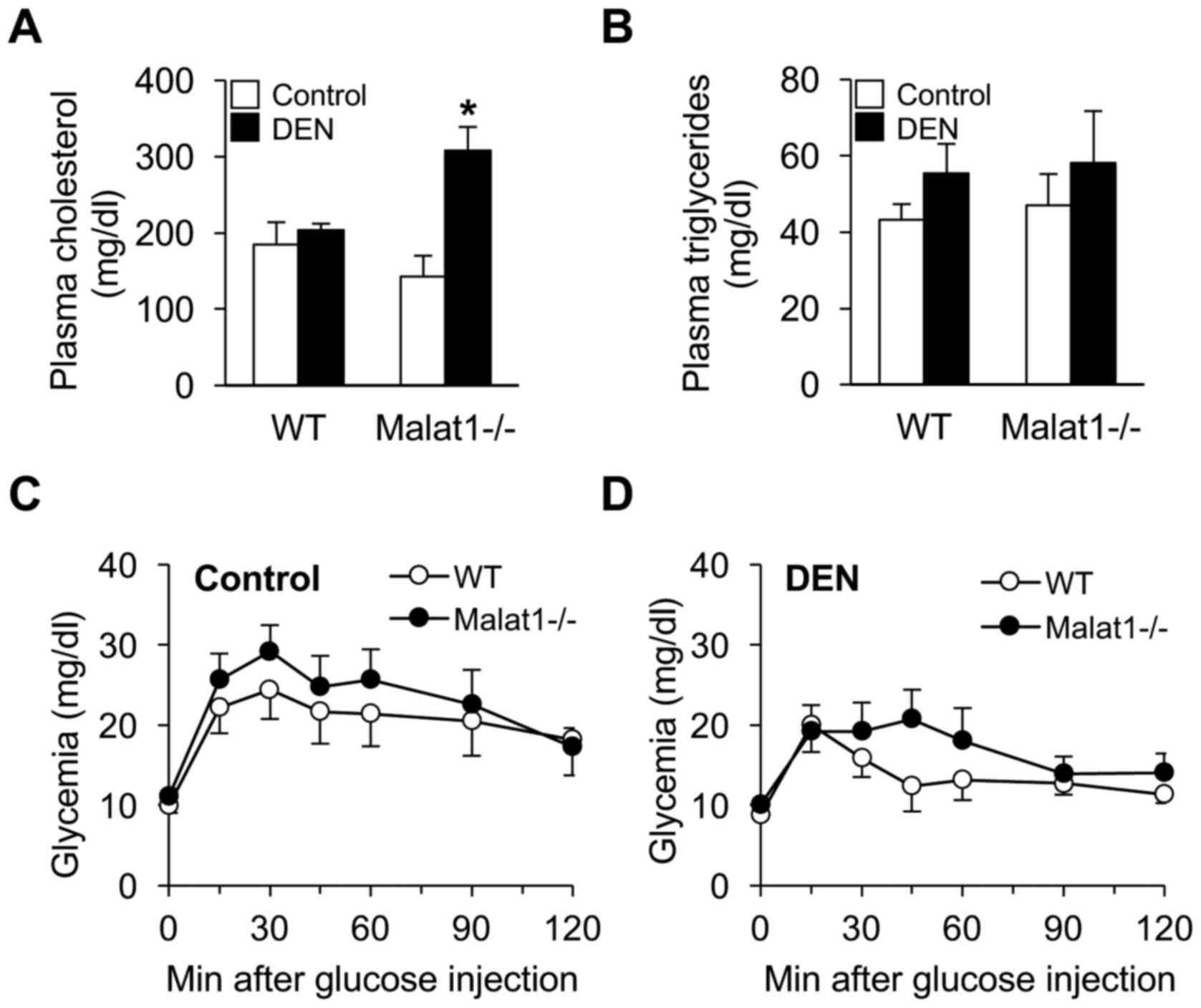
Beyond structural damage, deletion of Malat1 may
have modulated hepatic metabolic functions upon DEN administration.
Although DEN had no impact on plasma cholesterol levels in WT mice,
it unexpectedly increased 2-fold those of Malat1−/−
littermates (Fig. 4A). In contrast,
no difference in triglyceride levels (Fig. 4B) or glucose tolerance (Fig. 4C) was observed between groups.
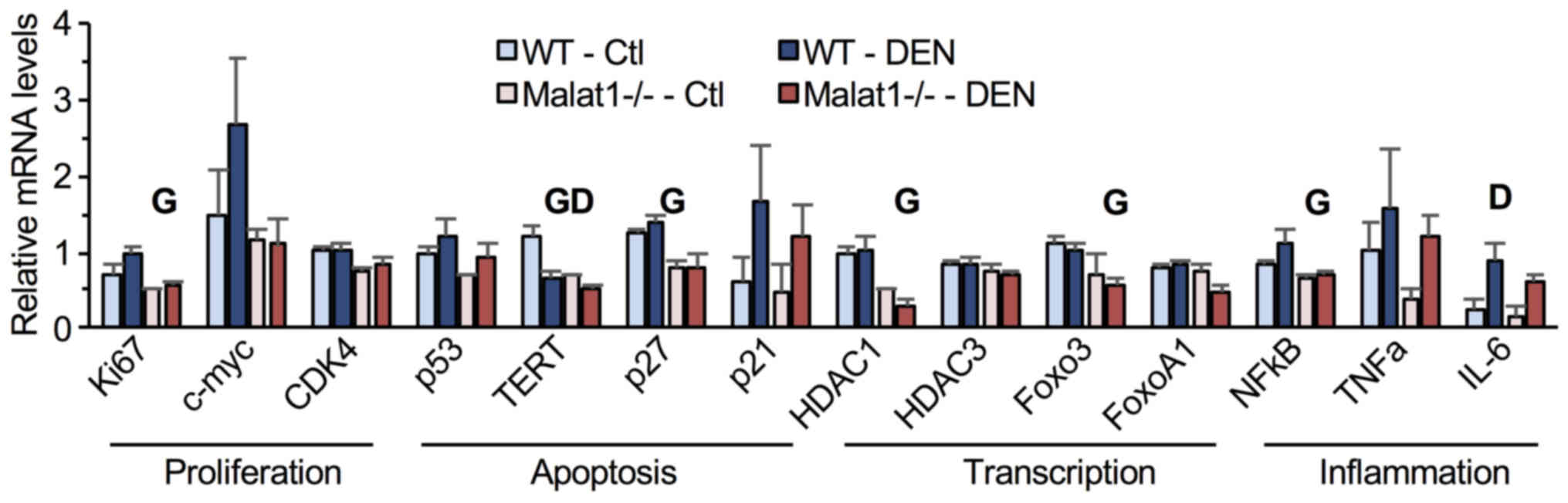
Finally, the expression of genes involved in cell
cycle and inflammation was quantified in liver tumors. Compared to
their WT littermates, Malat1−/− mice showed
significantly lower hepatic mRNA levels of Ki67, TERT, HDAC1, NFκB,
Foxo3, p27, and IL-6 (Fig. 5). DEN
treatment diminished TERT mRNA levels but increased those of IL-6
(Fig. 5). No significant
interaction between genotype and DEN administration was observed,
indicating that absence of Malat1 did not modify the
transcriptional response to DEN in hepatic tumors.
Discussion
Many studies performed in cultured cells have
demonstrated a stimulating role of Malat1 on cancer cell
proliferation (20,31,32).
Malat1 expression is increased during normal cell cycle progression
in normal diploid human fibroblasts and has been found important
for G1/S and mitotic division (23). Accordingly, Malat1 is overexpressed
in various cancer types including lung, liver and breast cancer,
and is associated with a poor prognostic (26). Moreover, Malat1 overexpression has
been shown to predict tumor recurrence of hepatocellular carcinoma
after liver transplantation (20).
Despite these findings, the role of Malat1 in tumorigenesis in
vivo is still not established. This study tested whether
genetic deletion of Malat1 in mice impaired the development of
hepatocarcinoma induced by the genotoxic agent DEN, a well-known
regimen for the induction of hepatocellular carcinoma (29).
Compared to its levels in many tissues, Malat1
expression was low in liver of young and healthy WT mice. In
tumors, Malat1 was 2- to 3-fold higher than in healthy liver
tissue; however, no further increase was observed upon DEN
treatment. This indicates that Malat1 expression may have reached a
plateau in actively proliferating hepatocytes, perhaps due to
mutual inhibition by YAP and SRSF1 (33). In this context, it was thus expected
that genetic ablation of Malat1 would in theory translate into
large, physiologically relevant impacts in a model in which
increased expression of Malat1 normally occurs. However, the main
finding of this study is that Malat1−/− mice are as
susceptible to DEN-induced liver cancer as WT mice. Beyond this,
Malat1 may be implicated in the development of cancer through its
involvement in facilitating invasion and metastasis (23,24,34).
However, we found no significant difference in the prevalence of
pulmonary inflammation or lung metastases between WT and
Malat1−/− mice. This could be attributable to the
experimental regimen, as DEN is a powerful genotoxic agent that may
bypass some of the tumorigenic pathways triggered by Malat1. In
this view, it would be interesting to test the influence of Malat1
in other tumor-inducing contexts, such as tobacco-induced lung
cancer.
Genetic deletion of Malat1 (complete germline
knockout) in mice has very little impact on pre- and post-natal
development and on adult phenotypes when tested in normal
conditions, as shown by three independent groups (35–38).
The findings observed in the mouse line used herein also show that
the basal modulation of glucose and lipid metabolism, functions
regulated by the liver, were not affected by the absence of Malat1,
at least in the fasted state. Although triglyceridemia and glucose
tolerance were similar between WT and Malat1−/−
littermates in response to DEN, plasma cholesterol levels were
robustly increased in DEN-treated Malat1−/− mice. Thus,
Malat1 may play important roles in the regulation of cholesterol
homeostasis. Interestingly, a recent report showed that, in
contrast, Malat1 expression is high in the fatty liver of obese
ob/ob mice, and that it promotes cholesterol accumulation in HepG2
hepatocytes by increasing SREBP1-c protein stability (39). Thus, these observations suggest that
Malat1 may exert beneficial impacts on cholesterol metabolism in a
manner dependent upon conditions in which liver functions are
perturbed. The mechanisms for these divergent effects could be
independent from obesity-associated, IL-6-induced liver
inflammation and tumorigenesis (40), since Malat1 deficiency did not
modify the increase in IL-6 expression triggered by DEN treatment.
Therefore, the role of Malat1 on cholesterol homeostasis remains to
be established in vivo in different settings, including
non-alcoholic fatty liver disease (NAFLD).
In vitro (17, 25, 32) and in a mouse model of
xenografts (18), Malat1 was shown
to stimulate cytoskeleton components, cell proliferation, and
cellular motility of cancer cells through impacting gene
transcription, not alternative splicing per se (18). Consistent with these findings, our
study shows that absence of Malat1 results in a significant
downregulation of many genes involved in cell cycle and
inflammation. Nonetheless, the observed changes in gene expression
were not sufficient to induce a robust and specific phenotype. It
remains to be investigated whether knockout of Malat1 modified the
expression of genes through cis-regulatory mechanism as
previously reported in other cell types (38).
A study performed in metastatic renal cell carcinoma
has suggested Malat1 as a putative FoxP3 target gene (41). Interestingly, this family of
transcription factors has been shown to be involved in the sexual
dimorphism observed in the development of liver cancer (42). Since female mice are not affected by
DEN exposure (29), it would be
interesting to test whether this dimorphism exists in
Malat1−/− mice using other types of carcinogens or by
crossing Malat1−/− mice with susceptible transgenic
strains.
In conclusion, we hypothesized that absence of
Malat1 would confer resistance to liver carcinogenesis induced by
DEN. As expected, DEN treatment stimulated an increase in liver
tumors, and number of lobes with at least one tumor. However, these
changes were completely similar to those observed in
Malat1−/− mice, despite differences in their
transcriptional mRNA profiles. In conclusion, gene deletion of
Malat1 does not impact cell proliferation upon DEN-induced
hepatocarcinoma in vivo. Thus, in this model, the role of
Malat1 in the regulation of hepatocyte proliferation is either
minimal or masked by redundant and/or overwhelming mechanisms, not
present in in vitro settings, including hormonal cues. Since
Malat1 has been found to be highly upregulated in many other types
of cancer, the impact of Malat1 deficiency to the in vivo
development of these diseases remains to be investigated.
Acknowledgements
This study was supported by a grant from the Natural
Sciences and Engineering Research Council (NSERC) of Canada to F.P.
M.J.G. was a recipient of an MSc studenship from the Fonds
d'enseignement et de recherche - FER from Laval University/Faculty
of Pharmacy. S.C. is the recipient of a PhD studentship award from
the Fonds de Recherche du Québec-Santé (FRQS). F.P. holds a Senior
Scholar Award from the FRQS.
References
|
1
|
Ankö ML and Neugebauer KM: Long non-coding
RNAs add another layer to pre-mRNA splicing regulation. Mol Cell.
39:833–834. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li J, Meng H, Bai Y and Wang K: Regulation
of lncRNA and its role in cancer metastasis. Oncol Res. 23:205–217.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dhamija S and Diederichs S: From junk to
master regulators of invasion: lncRNA functions in migration, EMT
and metastasis. Int J Cancer. 139:269–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Betancur JG: Pervasive lncRNA binding by
epigenetic modifying complexes - The challenges ahead. Biochim
Biophys Acta. 1859:93–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blythe AJ, Fox AH and Bond CS: The ins and
outs of lncRNA structure: How, why and what comes next? Biochim
Biophys Acta. 1859:46–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun M, Nie FQ, Wang ZX and De W:
Involvement of lncRNA dysregulation in gastric cancer. Histol
Histopathol. 31:33–39. 2016.PubMed/NCBI
|
|
9
|
Kaikkonen MU, Lam MT and Glass CK:
Non-coding RNAs as regulators of gene expression and epigenetics.
Cardiovasc Res. 90:430–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rinn JL and Chang HY: Genome regulation by
long non-coding RNAs. Annu Rev Biochem. 81:145–166. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long non-coding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilusz JE, Sunwoo H and Spector DL: Long
non-coding RNAs: Functional surprises from the RNA world. Genes
Dev. 23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel non-coding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hutchinson JN, Ensminger AW, Clemson CM,
Lynch CR, Lawrence JB and Chess A: A screen for nuclear transcripts
identifies two linked non-coding RNAs associated with SC35 splicing
domains. BMC Genomics. 8:392007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained non-coding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilusz JE, Freier SM and Spector DL: 3′
end processing of a long nuclear-retained non-coding RNA yields a
tRNA-like cytoplasmic RNA. Cell. 135:919–932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The non-coding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng
MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA Malat1
promotes gallbladder cancer development by acting as a molecular
sponge to regulate miR-206. Oncotarget. 7:37857–37867.
2016.PubMed/NCBI
|
|
20
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo F, Sun B, Li H, Xu Y, Liu Y, Liu X, Lu
L, Li J, Wang Q, Wei S, et al: A MALAT1/HIF-2α feedback loop
contributes to arsenite carcinogenesis. Oncotarget. 7:5769–5787.
2016.PubMed/NCBI
|
|
22
|
Konishi H, Ichikawa D, Yamamoto Y, Arita
T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et
al: Plasma level of metastasis-associated lung adenocarcinoma
transcript 1 is associated with liver damage and predicts
development of hepatocellular carcinoma. Cancer Sci. 107:149–154.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al:
Long non-coding RNA MALAT1 controls cell cycle progression by
regulating the expression of oncogenic transcription factor B-MYB.
PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tano K, Mizuno R, Okada T, Rakwal R,
Shibato J, Masuo Y, Ijiri K and Akimitsu N: MALAT-1 enhances cell
motility of lung adenocarcinoma cells by influencing the expression
of motility-related genes. FEBS Lett. 584:4575–4580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1 - a paradigm for long non-coding RNA function in cancer. J
Mol Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hansen GM, Markesich DC, Burnett MB, Zhu
Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP and
Abuin A: Large-scale gene trapping in C57BL/6N mouse embryonic stem
cells. Genome Res. 18:1670–1679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hogan B, Beddington R, Costantini F and
Lacy E: Manipulating the Mouse Embryo: A Laboratory Manual. Cold
Spring Harbor Laboratory Press; New York, NY: 1994
|
|
29
|
Fausto N and Campbell JS: Mouse models of
hepatocellular carcinoma. Semin Liver Dis. 30:87–98. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miard S, Dombrowski L, Carter S, Boivin L
and Picard F: Aging alters PPARgamma in rodent and human adipose
tissue by modulating the balance in steroid receptor coactivator-1.
Aging Cell. 8:449–459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo F, Li Y, Liu Y, Wang J, Li Y and Li G:
Inhibition of metastasis-associated lung adenocarcinoma transcript
1 in CaSki human cervical cancer cells suppresses cell
proliferation and invasion. Acta Biochim Biophys Sin (Shanghai).
42:224–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han Y, Liu Y, Nie L, Gui Y and Cai Z:
Inducing cell proliferation inhibition, apoptosis, and motility
reduction by silencing long non-coding ribonucleic acid
metastasis-associated lung adenocarcinoma transcript 1 in
urothelial carcinoma of the bladder. Urology. 81:209.e201–207.
2013. View Article : Google Scholar
|
|
33
|
Wang J, Wang H, Zhang Y, Zhen N, Zhang L,
Qiao Y, Weng W, Liu X, Ma L, Xiao W, et al: Mutual inhibition
between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced
tumourigenesis in liver cancer. Cell Signal. 26:1048–1059. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nakagawa S, Naganuma T, Shioi G and Hirose
T: Paraspeckles are subpopulation-specific nuclear bodies that are
not essential in mice. J Cell Biol. 193:31–39. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakagawa S, Ip JY, Shioi G, Tripathi V,
Zong X, Hirose T and Prasanth KV: Malat1 is not an essential
component of nuclear speckles in mice. RNA. 18:1487–1499. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eissmann M, Gutschner T, Hämmerle M,
Günther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun
T, Zörnig M, et al: Loss of the abundant nuclear non-coding RNA
MALAT1 is compatible with life and development. RNA Biol.
9:1076–1087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang B, Arun G, Mao YS, Lazar Z, Hung G,
Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al: The lncRNA
Malat1 is dispensable for mouse development but its transcription
plays a cis-regulatory role in the adult. Cell Rep. 2:111–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yan C, Chen J and Chen N: Long non-coding
RNA MALAT1 promotes hepatic steatosis and insulin resistance by
increasing nuclear SREBP-1c protein stability. Sci Rep.
6:226402016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park EJ, Lee JH, Yu GY, He G, Ali SR,
Holzer RG, Osterreicher CH, Takahashi H and Karin M: Dietary and
genetic obesity promote liver inflammation and tumorigenesis by
enhancing IL-6 and TNF expression. Cell. 140:197–208. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwarzer A, Wolf B, Fisher JL, Schwaab T,
Olek S, Baron U, Tomlinson CR, Seigne JD, Crosby NA, Gui J, et al:
Regulatory T-cells and associated pathways in metastatic renal cell
carcinoma (mRCC) patients undergoing DC-vaccination and
cytokine-therapy. PLoS One. 7:e466002012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Z, Tuteja G, Schug J and Kaestner KH:
Foxa1 and Foxa2 are essential for sexual dimorphism in liver
cancer. Cell. 148:72–83. 2012. View Article : Google Scholar : PubMed/NCBI
|