Introduction
Uveal melanoma (UM) is the most frequently occurring
primary intraocular tumor in adults, and is associated with
significant mortality (1). Several
histologic prognostic factors have been described for this type of
cancer, such as large tumor diameter (LTD), location at onset, age
at time of diagnosis, presence of epitheloid cells and involvement
of the ciliary body (2). The cause
of UM is unknown, but several risk factors have been associated
with the development of the disease such as light irides, uveal
naevi, dysplastic naevus syndrome and oculodermal melanocytosis. UM
most commonly affects Caucasian males. Despite the early diagnosis,
the mortality due to UM has remained relatively unchanged. Specific
genetic alterations can predict the development of metastasis and
survival in patients with UM. Monosomy 3 strongly predicts
metastatic risk and other chromosomal abnormalities, also
correlated with metastatic diseases (3,4).
Approximately half of the patients develop metastases, most
frequently in the liver (5,6). Monosomy 3 correlates with epitheloid
histology, ciliary body involvement and poor outcome (6). Lack of chromosome 3 has been
demonstrated in 5–10% of all the patients, and the remaining copy
is duplicated (7). Occasionally,
partial deletions of chromosome 3 have been detected and a common
region of allelic loss on 3p25 and on 3q24-q26 could be defined.
Most likely these regions harbor putative tumor-suppressor genes,
but no specific genes have yet been identified (7). Monosomy 3 is present in 50–60% of
tumors, which is associated with isochromosome 8q and high level of
8q gain (8). The common region of
amplification was found to range from 8q24.1 to 8q24.3. A potential
metastasis-suppressor gene, LZTS1, is located in 8p21
(9). In UM, other recurrent
chromosome alterations, such as lack of 1p and 16q, have been
described (10). One of the
suggested tumor-suppressor genes, APITD1, in the 1p36 region
was shown to be negligible for survival rate and the common deleted
regions on chromosome 1 were found to range from 1p34.3 to 36.2
(10). Infrequently, abnormalities
of other chromosomes such as gain of 6p, loss of 6q, loss of 9p,
loss of chromosome 10, loss of 11q23-q25, and gain of chromosomes 7
and 10 have been reported (11). UM
can be classified into 2 groups based on the status of chromosome
3: class 1 tumors with 2 copies, and class 2 tumors, with monosomy
of chromosome 3. The characteristics of these tumors basically
differ; class 1 tumors have been characterized by gain of 6p and 8q
while class 2 tumors by monosomy 3 and gain of entire 8q (12). Class 1 tumors exhibit low
aneuploidy, and patients rarely have metastases whereas class 2
tumors have a higher chance of aneuploidy and patients have a high
risk to develop metastases (13).
Hypothalamic luteinizing hormone-releasing hormone (LH-RH) is the
primary link between the hypothalamus and the pituitary gland in
the regulation of gonadal functions and has a pivotal role in
vertebrate reproduction (14). The
effects of LH-RH and its analogs are mediated by high-affinity
G-protein-coupled receptors located on the membranes of the
pituitary gonadotrophs and several cancer cells (15–17).
Tumoral receptors for LH-RH have been detected on human breast,
prostatic, ovarian, endometrial and pancreatic cancers and in human
melanomas, non-Hodgkin's lymphomas and renal cell carcinomas
(14–20). Over the past decade, a direct
receptor-mediated antiproliferative effect of LH-RH-analogs on
tumor cells was proposed (14,16,17,19–21).
The receptors for LH-RH (LH-RH-R) on human tumors can also serve as
targets for LH-RH analogs linked to various cytotoxic agents
(15–17,22,23).
In our previous study, it was demonstrated that a high percentage
(47%) of human UMs express the type-I receptor for LH-RH (24). The gene encoding LH-RH-R is located
on chromosome 4q21.2; however, the numerical aberrations of
chromosome 4 have never been studied in UM.
In the present study, we aimed to investigate the
copy number of chromosome 3, particularly the monosomy of
chromosome 3 which has been extensively described in the aggressive
behavior of UM, and chromosome 4 in 46 human UM specimens using
fluorescence in situ hybridization (FISH). Furthermore,
chromosome index (CI) and ‘dominant’ cell population values for
chromosome 3 and 4 were determined. Additionally, we analyzed the
survival rate of the UM patients according to their CI. The
correlation between LH-RH-R expression and the copy number of
chromosome 3 and 4 was also investigated.
Materials and methods
Human UM tissues
Specimens of human UM were obtained from 46 patients
30–84 years of age at the time of enucleation at the Department of
Ophthalmology of the University of Debrecen, Debrecen, Hungary.
Normal lymphocyte samples, used as positive controls, were
collected at the Department of Pathology of the University of
Debrecen. Informed consent was obtained before enucleation, and the
present study was performed according to the tenets of the
Declaration of Helsinki and the Local Institutional Ethics
Committee. Fresh tumor tissue was obtained within 1 h after
enucleation, according to a standardized protocol. Briefly, an
incision was made through the tumor, leaving the optic nerve
intact. The quantity of tissue obtained (5–8 mm3)
depended on the size of the tumor. A sample was taken from the side
opposite the optic nerve and selected portions of the melanoma
tissues were flash frozen and stored at −80̊C. Conventional
histopathologic examination was performed on all tumors and the
origin of the tumor was confirmed. Follow-up data from the time of
diagnosis until the end of the study were obtained by reviewing the
charts of the patients (whether we had the availability) and/or by
contacting their general physicians. The clinicopathological data
of the 46 patients are summarized in Table I. UM samples were divided into 4
groups based on the CI: NN (normal CI3 and CI4), NP (normal CI3
pathological CI4), PN (pathological CI3 and normal CI4) and PP
(pathological CI3 and CI4). To simplify the evaluation, 2 major
groups were also created: N (including NN) and P (containing NP, PN
and PP).
 | Table I.Clinicopathological characteristics,
chromosome index (CI) results and survival data of the 46 uveal
melanoma patients. |
Table I.
Clinicopathological characteristics,
chromosome index (CI) results and survival data of the 46 uveal
melanoma patients.
| Sample ID | Gender | Age (years) | Type | Eye | Localization | Survival | CI3 | CI4 | Postoperative
days |
|---|
| 1 | F | 79 | ND | L | C | Deceased | 1.43 | 2.72 |
210 |
| 2 | M | 76 | Spindle | L | P | Alive | 2.00 | 2.65 | 1,559 |
| 3 | F | 44 | Spindle-B | L | Inferior temporal:
P | Alive | 2.19 | 3.39 | 1,770 |
| 4 | F | 50 | Spindle | R | Temporal: P | Alive | 2.41 | 3.00 | 1,497 |
| 5 | M | 76 | Spindle | R | P | Deceased
(liver) | 2.18 | 3.94 |
620 |
| 6 | F | 30 | Spindle-A | L | P | Alive | 2.17 | 3.34 | 1,770 |
| 7 | M | 66 | Epithelioid | L | Temporal: P | Alive | 2.04 | 4.01 |
333 |
| 8 | M | 61 | Spindle-B | L | Temporal: P | Alive | 2.21 | 2.81 | 1,505 |
| 9 | M | 53 | ND | L | Superior temporal:
P | Alive | 2.04 | 3.94 | 1,260 |
| 10 | M | 53 | Epithelioid | R | P | Alive | 1.48 | 2.79 | 1,442 |
| 11 | F | 79 | Epithelioid | R | P | Dead | 2.07 | 3.43 |
548 |
| 12 | M | 67 | Epithelioid | L | P | Alive | 2.10 | 2.53 | 1,630 |
| 13 | F | 72 | Epithelioid | L | Temporal: P | Deceased
(liver) | 1.37 | 5.39 |
317 |
| 14 | M | 35 | Spindle | L | Superior nasal:
P | Alive | 1.71 | 2.94 |
740 |
| 15 | M | 55 | Spindle-B | L | P | Alive | 2.68 | 3.03 | 1,545 |
| 16 | M | 65 | Spindle-B | R | Anterior temporal:
P | Dead | 2.53 | 1.91 |
467 |
| 17 | F | 68 | Spindle | L | P | Alive | 2.07 | 1.75 | 1,702 |
| 18 | M | 71 | Spindle-B | R | P | Alive | 2.28 | 3.43 | 1,006 |
| 19 | M | 69 | Mixed | R | Anterior nasal:
P | Alive | 1.37 | 2.31 |
958 |
| 20 | M | 64 | ND | L | Temporal: P | Deceased
(bone) | 1.79 | 2.39 |
312 |
| 21 | F | 75 | Epithelioid | L | Temporal: P | Alive | 2.26 | 3.04 |
846 |
| 22 | F | 79 | ND | R | C | Alive | 2.43 | 2.36 | 1,442 |
| 23 | F | 75 | Mixed | L | Anterior nasal:
P | Alive | 1.06 | 1.94 | 1,902 |
| 24 | M | 70 | Mixed | R | P | Alive | 1.99 | 2.06 | 1,022 |
| 25 | M | 47 | Epithelioid | L | C | Deceased
(liver) | 1.53 | 2.08 |
832 |
| 26 | M | 42 | Epithelioid | R | P | Alive | 2.05 | 2.48 |
947 |
| 27 | M | 72 | Epithelioid | L | P | Alive | 1.97 | 2.48 |
932 |
| 28 | F | 68 | Epithelioid | L | Juxtapapillary | Alive | 1.87 | 221 |
965 |
| 29 | M | 72 | Epithelioid | L | P | Deceased
(liver) | 1.88 | 2.27 |
29 |
| 30 | M | 64 | Spindle | L | Anterior retinal:
P | Alive | 1.23 | 2.22 | 2,021 |
| 31 | M | 42 | Epithelioid | R | P | Deceased
(orbita) | 2.01 | 2.82 |
303 |
| 32 | F | 68 | Epithelioid | R | P | Deceased
(liver/lung) | 1.66 | 2.55 |
439 |
| 33 | M | 51 | Spindle-B | L | C | Alive | 0.94 | 2.14 | 1,609 |
| 34 | F | 50 | Spindle-B | R | Juxtapapillary | Alive | 2.22 | 2.50 | 1,097 |
| 35 | M | 56 | ND | L | Anterior temporal:
P | Alive | 1.33 | 2.37 |
648 |
| 36 | F | 55 | Epithelioid | L | Anterior | Alive | 2.07 | 2.04 |
623 |
| 37 | F | 83 | Spindle-A | R | nasal: P | Deceased
(liver) | 1.40 | 1.80 |
261 |
| 38 | F | 63 | Spindle-A | R | C | Alive | 1.31 | 2.10 |
490 |
| 39 | M | 70 | Spindle-B | R | Temporal: P | Alive | 1.17 | 2.33 |
950 |
| 40 | F | 61 | Spindle | L | P | Alive | 1.88 | 2.04 |
740 |
| 41 | M | 70 | Epithelioid | L | P | Alive | 1.41 | 1.81 |
524 |
| 42 | F | 70 | Epithelioid | R | P | Alive | 1.35 | 2.26 |
582 |
| 43 | F | 71 | Mix | R | Anterior | Alive | 1.76 | 2.28 |
559 |
| 44 | F | 52 | Mix | R | Temporal: P | Alive | 1.93 | 2.52 |
560 |
| 45 | F | ND | Spindle | L | C | Alive | 1.93 | 2.99 |
592 |
| 46 | F | 54 | Spindle | R | Anterior temporal:
P | Alive | 1.84 | 1.91 |
613 |
Touch preparations
The tumor tissues were transferred from −80 to
−20̊C. The tissue samples were used for touch preparations, which
were obtained by pressing frozen tissue samples several times on
the surface of a silanized slide. The slides were fixed in
methanol:acetic acid (3:1), air dried, washed with 70% acetic acid
solution and distilled water, dehydrated with 70, 80 and 90%
ethanol and air dried. The slides were stored at −20̊C until
further use.
FISH
DNA FISH probes
Numerical aberrations of chromosome 3 and 4 were
studied by FISH with centromere-specific probes (CEP; Chromosome
Enumeration DNA FISH Probes, Vysis, Germany). The probes consisted
of chromosome 3- or 4-specific tandem-repeat DNA sequences. The CEP
probes were directly labeled with SpectrumOrange (chromosome 3) and
SpectrumGreen (chromosome 4) fluorophores. The centromeric probes
contain 7 µl CEP hybridization buffer, 1 µl probe and 1 µl
distilled water.
FISH hybridization
FISH was carried out according to a general protocol
with some modifications (25). The
slides containing the touch preparations were fixed in
methanol:acetic acid (3:1) at −20̊C, and then incubated in 15 µl
10% pepsin in 100 µl 1 M HCl. The slides were washed with 1X PBS
buffer, and then dehydrated in 70, 85 and 100% alcohol series and
air dried. DNA FISH probe was added, coverslips were applied and
sealed to the slide with rubber cement. The slides were denatured
at 75̊C for 5 min and hybridized overnight at 42̊C. After
hybridization, the slides were washed with 50% formamide/2X
standard saline citrate (SSC) solution at 42̊C for 7 min, and then
with 2X SSC solution at 42̊C for 7 min. The slides were then
counterstained with 4,6-diamidino-2-phenylindole (DAPI) in
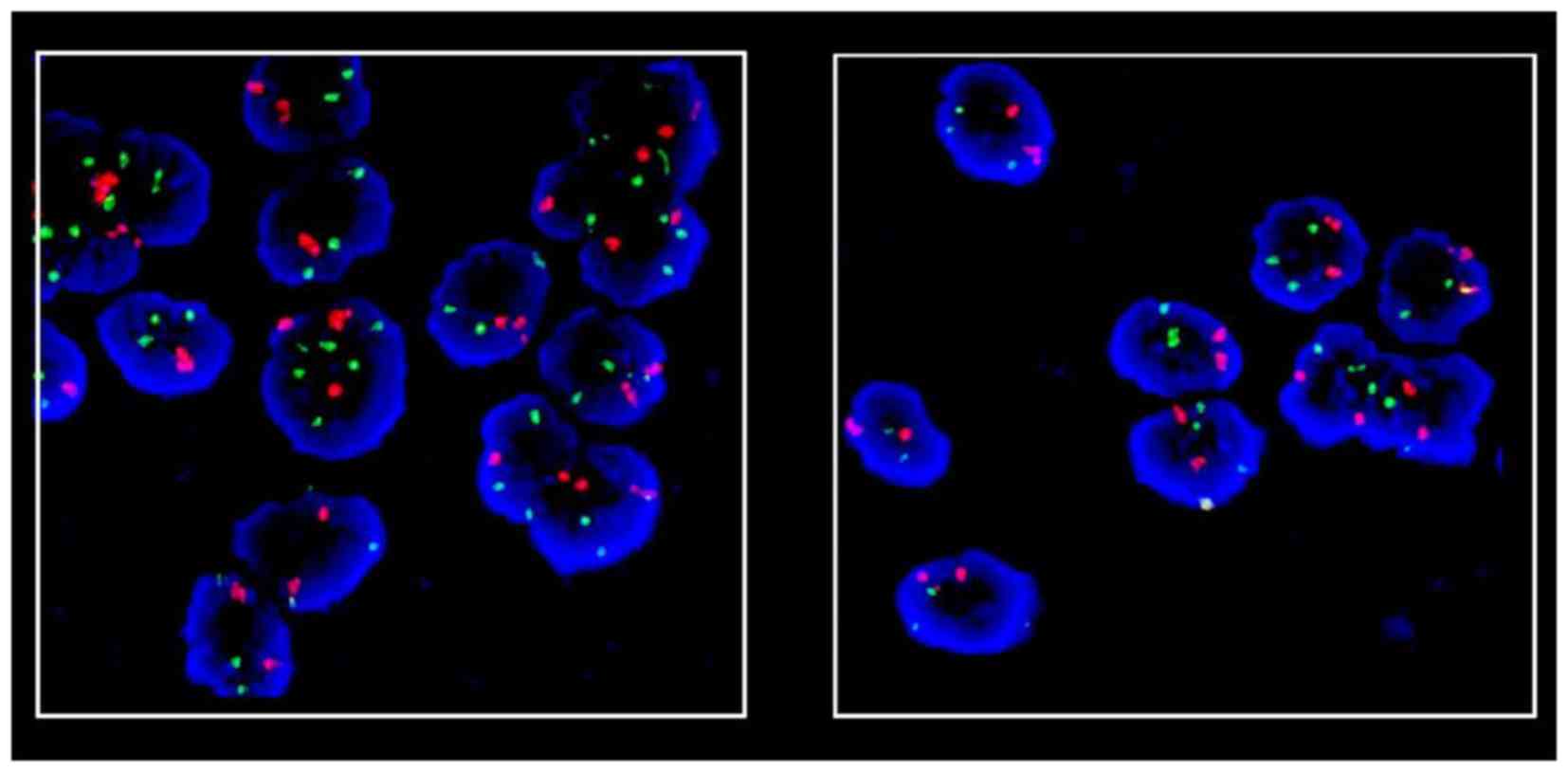
anti-fade solution (Fig. 1).
Fluorescence microscopy
Slides were evaluated using a fluorescence
microscope (Axio Imager Z2; Zeiss, Oberkochen, Germany). Image
capture was performed by a monochrome charge-coupled device camera
attached to the fluorescence microscope and ISIS software
(MetaSystems, Altlussheim, Germany).
FISH analysis
Numerical aberrations of chromosome 3 and 4 were
assessed by analyzing chromosome copy number on the basis of 100
relevant tumor cell nuclei. CI values for chromosome 3 and 4 were
determined for the ratio of the whole FISH signal in the sample and
the number of nuclei. Chromosome loss was stated as <1.75,
polysomy was stated as >2.25 chromosome copy numbers/nucleus.
‘Dominant’ cell population value was determined. A cell population
with a certain chromosome copy number was considered as ‘dominant’
cell population where the cut-off limit was 15% (26).
Statistical analysis
Indices for chromosome 3 and 4 were analyzed from
the UM samples. The two datasets were evaluated using
D'Agostino-Pearson omnibus normality test, and then Spearman
correlation analysis was performed. Chromosome results, receptor
findings and clinicopathological data were also analyzed.
Statistical analysis was carried out with the use of GraphPad Prism
6.03 (GraphPad, San Diego, CA, USA).
Survival in the groups was plotted against the
postoperative days (elapsed until death or the end of the follow-up
period), according to the Kaplan-Meier method. Differences among
the groups were investigated by means of Mantel-Cox log-rank and
Gehan-Breslow-Wilcoxon tests. Statistical analysis was carried out
with GraphPad Prism 6.03 software.
Results
Distribution of chromosome 3
Based on CI values, monosomy of chromosome 3 was
found in 16 (35%) samples. In 6 specimens (13%), >2 copies of
chromosome 3 were found. Normal biparental disomy was observed in
24 samples (52%). In 26 samples one signal/cell/‘dominant’ cell
population could be detected, whereas in 9 cases, clones containing
3 or more chromosome/nucleus were found. In 2 specimens, either
loss of chromosome or polysomy were observed. Normal distribution
of chromosome 3 was detected in 13 cases. In addition, the normal
tissue samples contained negligible abnormal cell population
(<15%) (Table II).
Representative distribution of chromosome 3 is shown in Fig. 1.
 | Table II.Distribution of chromosome 3 in the
human uveal melanoma specimens. |
Table II.
Distribution of chromosome 3 in the
human uveal melanoma specimens.
|
| Chromosome 3 |
|---|
|
|
|
|---|
|
| ‘Dominant’ cell
population 1 | ‘Dominant’ cell
population 2 |
|
|---|
|
|
|
|
|
|---|
| Sample ID | Signals/cell | % | Signals/cell | % | CI |
|---|
| 33 | 1 | 85 |
|
| 0.94 |
| 23 | 1 | 94 |
|
| 1.06 |
| 39 | 1 | 78 |
|
| 1.17 |
| 30 | 1 | 77 |
|
| 1.23 |
| 38 | 1 | 69 |
|
| 1.31 |
| 35 | 1 | 71 |
|
| 1.33 |
| 42 | 1 | 66 |
|
| 1.35 |
| 13 | 1 | 65 |
|
| 1.37 |
| 19 | 1 | 64 |
|
| 1.37 |
| 37 | 1 | 62 |
|
| 1.40 |
| 41 | 1 | 62 |
|
| 1.41 |
| 1 | 1 | 62 |
|
| 1.43 |
| 10 | 1 | 60 |
|
| 1.48 |
| 25 | 1 | 52 |
|
| 1.53 |
| 32 | 1 | 39 |
|
| 1.66 |
| 14 | 1 | 49 |
|
| 1.71 |
| 43 | 1 | 25 |
|
| 1.76 |
| 20 | 1 | 34 |
|
| 1.79 |
| 46 | 1 | 23 |
|
| 1.84 |
| 28 | 1 | 21 |
|
| 1.87 |
| 29 | 1 | 19 |
|
| 1.88 |
| 40 | 1 | 17 |
|
| 1.88 |
| 44 |
|
Normal |
|
| 1.93 |
| 45 |
|
Normal |
|
| 1.93 |
| 27 | 1 | 21 |
|
| 1.97 |
| 24 |
|
Normal |
|
| 1.99 |
| 2 | 1 | 35 |
|
| 2.00 |
| 31 |
|
Normal |
|
| 2.01 |
| 7 |
|
Normal |
|
| 2.04 |
| 9 |
|
Normal |
|
| 2.04 |
| 26 |
|
Normal |
|
| 2.05 |
| 11 |
|
Normal |
|
| 2.07 |
| 17 |
|
Normal |
|
| 2.07 |
| 36 | 3 | 17 |
|
| 2.07 |
| 12 | 1 | 24 | 3 | 18 | 2.10 |
| 6 |
|
Normal |
|
| 2.17 |
| 5 |
|
Normal |
|
| 2.18 |
| 3 |
|
Normal |
|
| 2.19 |
| 8 |
|
Normal |
|
| 2.21 |
| 34 | 3 | 21 |
|
| 2.22 |
| 21 | 3 | 19 |
|
| 2.26 |
| 18 | 1 | 42 | ≥4 | 27 | 2.28 |
| 4 |
|
| 3 | 18 | 2.41 |
| 22 | 3 | 20 |
|
| 2.43 |
| 16 | 3 | 18 | ≥4 | 18 | 2.53 |
| 15 | 3 | 29 | ≥4 | 22 | 2.68 |
Distribution of chromosome 4
Based on the CI values, chromosome 4 could be
detected in normal biparental disomy in 14 samples (30%), while 32
cases (70%) showed >2 signals/nucleus. In 8 samples one
signal/cell/‘dominant’ cell population was observed, whereas in 41
cases, clones containing 3 or more chromosome/nucleus were found.
In 6 specimens either loss of chromosome or polysomy was observed.
Normal distribution of chromosome 4 was detected only in 3 cases
(Table III). Representative
distribution of chromosome 4 is shown in Fig. 1.
 | Table III.Distribution of chromosome 4 in the
human uveal melanoma specimens. |
Table III.
Distribution of chromosome 4 in the
human uveal melanoma specimens.
|
| Chromosome 4 |
|---|
|
|
|
|---|
|
| ‘Dominant’ cell
population 1 | ‘Dominant’ cell
population 2 |
|
|---|
|
|
|
|
|
|---|
| Sample ID | Signals/cell | % | Signals/cell | % | CI |
|---|
| 17 | 1 | 22 |
|
| 1.75 |
| 37 | 1 | 44 | 3 | 24 | 1.80 |
| 41 | 1 | 39 | 3 | 20 | 1.81 |
| 16 | 1 | 15 |
|
| 1.91 |
| 46 | 1 | 26 | 3 | 17 | 1.91 |
| 23 |
|
Normal |
|
| 1.94 |
| 36 | 1 | 23 | 3 | 24 | 2.04 |
| 40 | 1 | 30 | 3 | 32 | 2.04 |
| 24 |
|
Normal |
|
| 2.06 |
| 25 |
|
Normal |
|
| 2.08 |
| 38 | 1 | 20 | 3 | 28 | 2.10 |
| 33 | 3 | 21 |
|
| 2.14 |
| 28 | 1 | 16 | 3 | 21 | 2.21 |
| 30 | 3 | 26 |
|
| 2.22 |
| 42 | 3 | 36 |
|
| 2.26 |
| 29 | 3 | 26 |
|
| 2.27 |
| 43 | 3 | 38 |
|
| 2.28 |
| 19 | 3 | 15 |
|
| 2.31 |
| 39 | 3 | 44 |
|
| 2.33 |
| 22 | 3 | 20 |
|
| 2.36 |
| 35 | 3 | 32 |
|
| 2.37 |
| 20 | 3 | 18 |
|
| 2.39 |
| 26 |
|
| ≥4 | 18 | 2.48 |
| 27 | 3 | 25 |
|
| 2.48 |
| 34 | 3 | 37 |
|
| 2.50 |
| 44 | 3 | 34 |
|
| 2.52 |
| 12 | 3 | 20 |
|
| 2.53 |
| 32 | 3 | 42 |
|
| 2.55 |
| 2 | 3 | 16 |
|
| 2.65 |
| 1 | ≥4 | 30 |
|
| 2.72 |
| 10 | 3 | 25 | ≥4 | 22 | 2.79 |
| 8 | 3 | 45 |
|
| 2.81 |
| 31 | 3 | 48 |
|
| 2.82 |
| 14 | 3 | 19 | ≥4 | 33 | 2.94 |
| 45 | 3 | 58 | ≥4 | 21 | 2.99 |
| 4 | 3 | 26 | ≥4 | 26 | 3.00 |
| 15 | 3 | 19 | ≥4 | 39 | 3.03 |
| 21 | 3 | 79 |
|
| 3.04 |
| 6 | 3 | 28 | ≥4 | 42 | 3.34 |
| 3 | 3 | 27 | ≥4 | 43 | 3.39 |
| 11 | 3 | 28 | ≥4 | 47 | 3.43 |
| 18 | 3 | 15 | ≥4 | 24 | 3.43 |
| 5 | 3 | 19 | ≥4 | 72 | 3.94 |
| 9 |
|
| ≥4 | 81 | 3.94 |
| 7 | 3 | 22 | ≥4 | 67 | 4.01 |
| 13 |
|
| ≥4 | 91 | 5.39 |
Statistical results
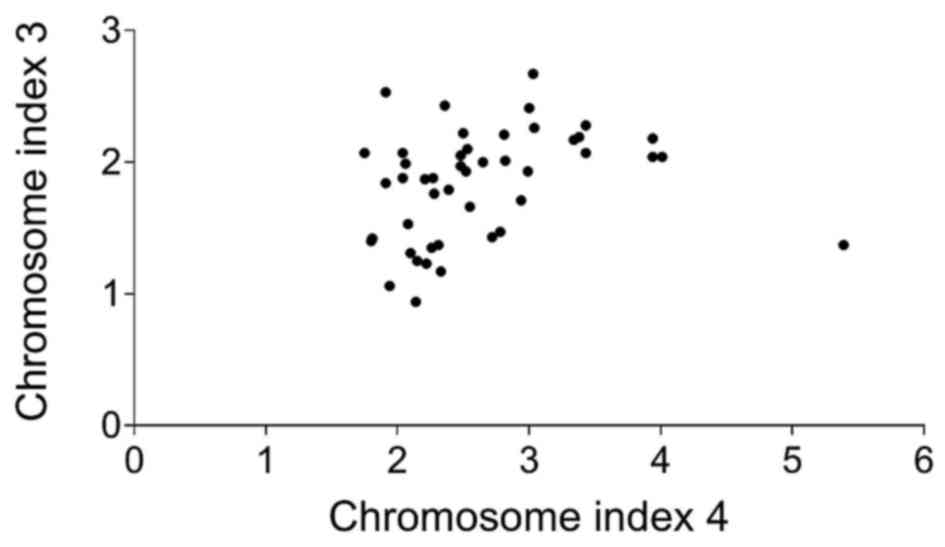
According to the statistical analysis, there was
(Spearman r=0.42; 0.139–0.639; CI, 0.95%) a statistically
significant (p<0.05) correlation between the copy number of
chromosome 3 and 4 (Fig. 2). CI
values for chromosomes 3 and 4 were determined for the samples and
were considered to be normal (N, 1.75–2.25) or pathological (P,
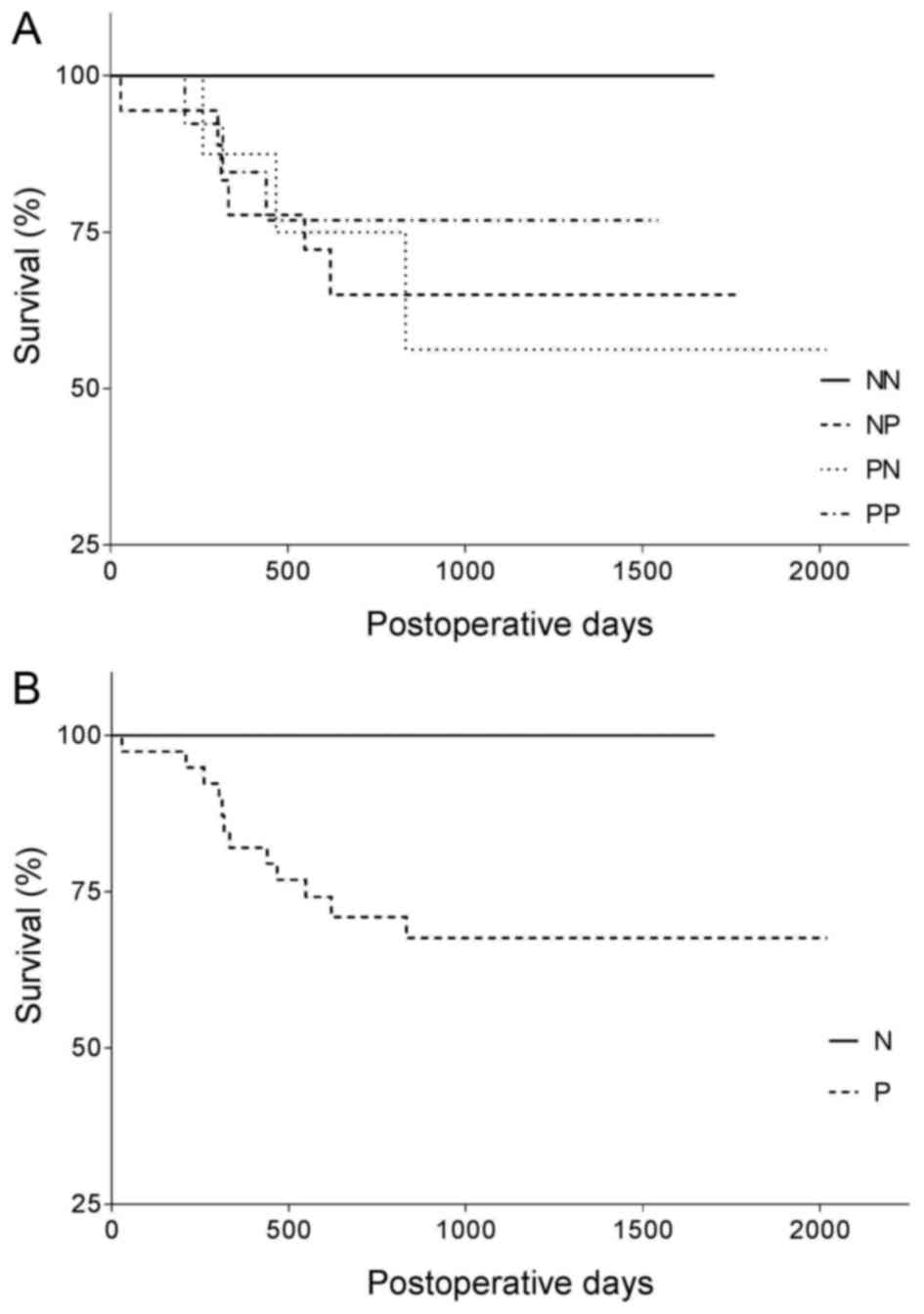
<1.75 or >2.25). Comparing the survival rate of the 4 groups
(NN, NP, PN and PP), an obvious difference was revealed, however
statistically significant differences could not be shown (p=0.38
for the Mantel-Cox test, and p=0.43 for the Gehan-Breslow-Wilcoxon
test). Even the 2 major groups (N and P) were not found to be
significantly different (p=0.12 by both the Mantel-Cox and
Gehan-Breslow-Wilcoxon tests), in spite of the considerable
difference between their survival curves (Fig. 3). The correlation of aberrations in
chromosome 3 and 4 with LH-RH-R findings was also investigated in
17 UM samples where receptor data were available (24). No significant correlation was found
among chromosome expression and LH-RH-R incidence and binding
characteristics. Furthermore based on our findings and the
clinicopathological data, no correlation was observed between
clinical outcome and chromosome 3 and 4 status (data not
shown).
Discussion
Uveal melanoma (UM) is the most common form of
primary ocular cancer in adults, with a mortality rate of 50% at
10–15 years after detection of the disease (27). Clinical treatment for the disease
includes photocoagulation, radiotherapy, local tumor incision and
eye removal. However, none of these treatments improves the
survival rate noticeably (28).
Adjuvant systemic therapy is mainly used in patients with high-risk
of metastasis or in patients already presenting with metastasis,
but the response rates to classical chemotherapeutic agents remain
low (29). We previously
demonstrated that LH-RH-R is expressed in approximately half (47%)
of human UMs (24). The effects of
LH-RH and its analogs are mediated by high-affinity
G-protein-coupled receptors for LH-RH located on the membranes of
the pituitary gonadotrophs and different human types of cancers
(16,17,19–21).
The presence of LH-RH-R in various types of cancers and cancer cell
lines originating from organs other than those of the reproductive
system has been shown in various studies (14,16–19,21,30).
Both agonists and antagonists of LH-RH may serve as potential
therapeutic agents, acting directly on the target cancer cells
(16,17,30–32).
LH-RH agonists inhibit the gonadotropin secretion after continuous
exposure (31). In contrast,
antagonists of LH-RH produce a competitive blockade of LH-RH-R
leading to an immediate cessation of the secretion of gonadotropins
and sex steroids, reducing the time of the onset of therapeutic
effects compared to the agonists (33). Agonistic analogs, such as
triptorelin, leuprolelin, goserelin and buserelin are extensively
applied in gynecology and oncology (14,16–18,30).
Potent antagonists of LH-RH, such as cetrorelix, ganirelix,
abarelix and degarelix, have also been developed and are now
available for clinical use (14,16–18,30,33).
The receptors for LH-RH on human tumors also serve as targets for
LH-RH analogs linked to cytotoxic agents (15–17,19,23,34).
In the analog AN-152 (AEZS-108) doxorubicin (DOX) is covalently
linked to the LH-RH agonist D-Lys6-LH-RH, that binds to
the receptors present on the surface of breast, prostatic, ovarian
and other cancer cells (15–17,23,34).
This analog has been extensively investigated in a large number of
experimental studies (14–19,23,30,34),
and also tested clinically in ovarian, endometrial, prostatic and
bladder cancer. It is in clinical phase III trials on endometrial
cancer (35). Generally, the
genetic background of different cancers is extensively
investigated. For example, aberrations of chromosome 4 have been
demonstrated in cervical cancer, small cell lung cancer,
glioblastoma and chronic lymphocytic leukemia (36–39).
Chromosome 4 hyperploidy is the most prominent alteration found in
Barrett's metaplasia and 89% of the patients display this
aberration (40). Notably, the gene
encoding LH-RH-R is located on chromosome 4q21.2. The numerical
aberrations of chromosome 4 have never been studied in UM.
It was reported that monosomy 3 strongly predicts
metastatic risk and other chromosomal abnormalities correlate with
metastatic disease (3,4). Monosomy 3 in choroidal melanoma is a
significant predictor of metastasis-related death and has been
associated with a 70% decrease in 5-year survival. Infrequently,
abnormalities of other chromosomes such as losses of 1p, 6q, 9p,
10, 11q23-q25, and gain of chromosomes 6p, 7, 8q and 10 have been
reported (3,11). Recently, several potential genes
were proposed in UM, such as GNAQ, DDEF1,
NBS1, HDM2, BCL-2 and CCND1; however, a
significant role for most of these genes must be further
investigated in tumorigenesis and progression towards metastasis
must be confirmed (41,42).
In the present study, one of our aims was to
investigate the copy number of chromosome 3 due to the fact that it
has been implicated in the aggressive behavior of UM. More
importantly, copy number of chromosome 4 was also studied in the
same human UM specimens using fluorescence in situ
hybridization (FISH). The correlation between LH-RH-R expression,
clinicopathological findings and numerical aberrations of
chromosome 3 and 4 was similarly analyzed.
FISH can detect chromosomal alterations that are
consistent with a diagnosis of neoplasia. Several studies have
shown that FISH has significantly higher sensitivity for the
detection of tumor cells than conventional cytology (43–45).
FISH is also able to detect various types of cytogenetic
alterations including aneusomy, duplication, amplification,
deletion and translocation (8). In
general, 3 basic types of DNA probes are used: centromeric
(chromosome enumeration probes), whole chromosome (whole chromosome
paints) and locus-specific probes (46).
We demonstrated in the present study, for the first
time, that chromosome 4 is present in an abnormal copy number in
the majority of UMs. Based on the chromosome index (CI) values, in
70% of samples of chromosome 4, more than 2 signals/nucleus were
detected while the normal 2 copies were found only in 30% of the
cases. The monosomy of chromosome 3 was detected in 35% of the
samples while in 13% of the cases polysomy was observed. Our
results are somewhat different from previous studies concerning the
frequency of the monosomy of chromosome 3 (50%) (47–50).
This slight difference may be partially explained by the possibly
diverse genetic background of the Hungarian population.
In case of chromosome 3, based on ‘dominant’ cell
population values, one signal/cell/‘dominant’ cell population was
observed in 26 samples whereas we found clones containing 3 or more
chromosomes/nucleus in 9 cases. In 2 specimens either loss of the
chromosome or polysomy was observed.
In the case of chromosome 4, one
signal/cell/‘dominant’ was observed in 8 samples whereas in 41
cases clones containing 3 or more chromosomes/nucleus were
detected. In 6 specimens either loss of the chromosome or polysomy
was observed.
According to our statistical analysis, there is a
moderate, statistically significant correlation between the copy
numbers of chromosome 3 and 4, but no correlation was found with
LH-RH-R expression and chromosome aberrations.
We also determined the survival rate of the UM
patients according to their CI. Comparison of the survival rate of
the 4 groups (NN, NP, PN and PP) and the 2 major groups (N and P),
a moderate difference was revealed, although statistically
significant differences could not be proven in spite of the
considerable difference between their survival curves. As mentioned
above, the diverse genetic background of the Hungarian population
as well as the limited number of human UM specimens may have
contributed to the limitation of the present study. Our research is
in the early phase of the investigation of chromosome 4 status;
therefore, multivariate statistical analysis may not be a proper
statistical test at this moment. However, investigation of a larger
population may be important which may indeed require a more
powerful statistical test, such as multivariate statistical
analysis. In conclusion, our results provide new informations
concerning the genetic background of UM and may lead to a more
precise prognosis and novel therapeutic approaches for cancer of
the eye.
Acknowledgements
The present study was supported by the Hungarian
Scientific Research Fund (OTKA) K 81596 (to G.H.), TAMOP
4.2.2.A-11/1/KONV-2012-0025 Project (to G.H.),
TAMOP-4.2.2/B-10/1-2010-0024 (to E.S), and the Gedeon Richter's
Talentum Foundation (to E.S.). The present study was co-financed by
the European Union and the European Social Fund. The present study
is dedicated to the late Dr Andrea Treszl who recently died from
metastatic breast cancer. Her intellectual, spiritual and personal
contributions provided a great inspiration for our research on UM.
We thank Dr Rudolf Gesztelyi for his excellent assistance in the
statistical part of the present study.
References
|
1
|
Perry JD and Singh AD: Uveal melanoma:
Epidemiologic aspects. Clin Ophthalmic Oncol. 75–87. 2014.
|
|
2
|
Mooy CM and De Jong PT: Prognostic
parameters in uveal melanoma: A review. Surv Ophthalmol.
41:215–228. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dopierala J, Damato BE, Lake SL, Taktak
AFG and Coupland SE: Genetic heterogeneity in uveal melanoma
assessed by multiplex ligation-dependent probe amplification.
Invest Ophthalmol Vis Sci. 51:4898–4905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mensink HW, Vaarwater J, Kiliç E, Naus NC,
Mooy N, Luyten G, Brüggenwirth HT, Paridaens D and de Klein A:
Chromosome 3 intratumor heterogeneity in uveal melanoma. Invest
Ophthalmol Vis Sci. 50:500–504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Damato B: Does ocular treatment of uveal
melanoma influence survival? Br J Cancer. 103:285–290. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abildgaard SKO and Vorum H: Proteomics of
uveal melanoma: A minireview. J Oncol. 2013:8209532013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hughes S, Damato BE, Giddings I, Hiscott
PS, Humphreys J and Houlston RS: Microarray comparative genomic
hybridisation analysis of intraocular uveal melanomas identifies
distinctive imbalances associated with loss of chromosome 3. Br J
Cancer. 93:1191–1196. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kilic E, van Gils W, Lodder E, Beverloo
HB, van Til ME, Mooy CM, Paridaens D, de Klein A and Luyten GP:
Clinical and cytogenetic analyses in uveal melanoma. Invest
Ophthalmol Vis Sci. 47:3703–3707. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Onken MD, Worley LA and Harbour JW: A
metastasis modifier locus on human chromosome 8p in uveal melanoma
identified by integrative genomic analysis. Clin Cancer Res.
14:3737–3745. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Häusler T, Stang A, Anastassiou G, Jöckel
KH, Mrzyk S, Horsthemke B, Lohmann DR and Zeschnigk M: Loss of
heterozygosity of 1p in uveal melanomas with monosomy 3. Int J
Cancer. 116:909–913. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van den Bosch T, Kilic E, Paridaens D and
de Klein A: Genetics of uveal melanoma and cutaneous melanoma: Two
of a kind? Dermatol Res Pract. 2010:3601362010.PubMed/NCBI
|
|
12
|
Onken MD, Worley LA, Ehlers JP and Harbour
JW: Gene expression profiling in uveal melanoma reveals two
molecular classes and predicts metastatic death. Cancer Res.
64:7205–7209. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ehlers JP, Worley L, Onken MD and Harbour
JW: Integrative genomic analysis of aneuploidy in uveal melanoma.
Clin Cancer Res. 14:115–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schally AV: Luteinizing hormone-releasing
hormone analogs: Their impact on the control of tumorigenesis.
Peptides. 20:1247–1262. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schally AV and Nagy A: Chemotherapy
targeted to cancers through tumoral hormone receptors. Trends
Endocrinol Metab. 15:300–310. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schally AV and Halmos G: Targeting to
Peptide Receptors. Drug Delivery in Oncology. Felix K, Peter S and
Henning S: Wiley-VCH; Weinheim: pp. 1219–1261. 2012
|
|
17
|
Schally AV, Comaru-Schally AM, Nagy A,
Kovacs M, Szepeshazi K, Plonowski A, Varga JL and Halmos G:
Hypothalamic hormones and cancer. Front Neuroendocrinol.
22:248–291. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schally AV, Szepeshazi K, Nagy A,
Comaru-Schally AM and Halmos G: New approaches to therapy of
cancers of the stomach, colon and pancreas based on peptide
analogs. Cell Mol Life Sci. 61:1042–1068. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szepeshazi K, Schally AV and Halmos G:
LH-RH receptors in human colorectal cancers: Unexpected molecular
targets for experimental therapy. Int J Oncol. 30:1485–1492.
2007.PubMed/NCBI
|
|
20
|
Halmos G, Arencibia JM, Schally AV, Davis
R and Bostwick DG: High incidence of receptors for luteinizing
hormone-releasing hormone (LHRH) and LHRH receptor gene expression
in human prostate cancers. J Urol. 163:623–629. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moretti RM, Marelli Montagnani M, Van
Groeninghen JC and Limonta P: Locally expressed LHRH receptors
mediate the oncostatic and antimetastatic activity of LHRH agonists
on melanoma cells. J Clin Endocrinol Metab. 87:3791–3797. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu SV, Schally AV, Hawes D, Xiong S,
Fazli L, Gleave M, Cai J, Groshen S, Brands F, Engel J, et al:
Expression of receptors for luteinizing hormone-releasing hormone
(LH-RH) in prostate cancers following therapy with LH-RH agonists.
Clin Cancer Res. 16:4675–4680. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seitz S, Buchholz S, Schally AV, Weber F,
Klinkhammer-Schalke M, Inwald EC, Perez R, Rick FG, Szalontay L,
Hohla F, et al: Triple negative breast cancers express receptors
for LHRH and are potential therapeutic targets for cytotoxic
LHRH-analogs, AEZS 108 and AEZS 125. BMC Cancer. 14:8472014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Treszl A, Steiber Z, Schally AV, Block NL,
Dezso B, Olah G, Rozsa B, Fodor K, Buglyo A, Gardi J, et al:
Substantial expression of luteinizing hormone-releasing hormone
(LHRH) receptor type I in human uveal melanoma. Oncotarget.
4:1721–1728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balázs M, Adám Z, Treszl A, Bégány A,
Hunyadi J and Adány R: Chromosomal imbalances in primary and
metastatic melanomas revealed by comparative genomic hybridization.
Cytometry. 46:222–232. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bonaldi L, Midena E, Filippi B, Tebaldi E,
Marcato R, Parrozzani R and Amadori A: FISH analysis of chromosomes
3 and 6 on fine needle aspiration biopsy samples identifies
distinct subgroups of uveal melanomas. J Cancer Res Clin Oncol.
134:1123–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kilic E, Naus NC, van Gils W, Klaver CC,
van Til ME, Verbiest MM, Stijnen T, Mooy CM, Paridaens D, Beverloo
HB, et al: Concurrent loss of chromosome arm 1p and chromosome 3
predicts a decreased disease-free survival in uveal melanoma
patients. Invest Ophthalmol Vis Sci. 46:2253–2257. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang C and Wei W: The miRNA expression
profile of the uveal melanoma. Sci China Life Sci. 54:351–358.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bedikian AY, Legha SS, Mavligit G,
Carrasco CH, Khorana S, Plager C, Papadopoulos N and Benjamin RS:
Treatment of uveal melanoma metastatic to the liver: a review of
the M. D. Anderson Cancer Center experience and prognostic factors.
Cancer. 76:1665–1670. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Engel JB and Schally AV: Drug Insight:
Clinical use of agonists and antagonists of
luteinizing-hormone-releasing hormone. Nat Clin Pract Endocrinol
Metab. 3:157–167. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kovacs M and Schally AV: Comparison of
mechanisms of action of luteinizing hormone-releasing hormone
(LHRH) antagonist cetrorelix and LHRH agonist triptorelin on the
gene expression of pituitary LHRH receptors in rats. Proc Natl Acad
Sci USA. 98:12197–12202. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tolkach Y, Joniau S and Van Poppel H:
Luteinizing hormone-releasing hormone (LHRH) receptor agonists vs
antagonists: A matter of the receptors? BJU Int. 111:1021–1030.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kovacs M, Schally AV, Csernus B and Rekasi
Z: Luteinizing hormone-releasing hormone (LH-RH) antagonist
Cetrorelix down-regulates the mRNA expression of pituitary
receptors for LH-RH by counteracting the stimulatory effect of
endogenous LH-RH. Proc Natl Acad Sci USA. 98:1829–1834. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Keller G, Schally AV, Gaiser T, Nagy A,
Baker B, Westphal G, Halmos G and Engel JB: Human malignant
melanomas express receptors for luteinizing hormone releasing
hormone allowing targeted therapy with cytotoxic luteinizing
hormone releasing hormone analogue. Cancer Res. 65:5857–5863. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu SV, Tsao-Wei DD, Xiong S, Groshen S,
Dorff TB, Quinn DI, Tai YC, Engel J, Hawes D, Schally AV, et al:
Phase I, dose-escalation study of the targeted cytotoxic LHRH
analog AEZS-108 in patients with castration- and taxane-resistant
prostate cancer. Clin Cancer Res. 20:6277–6283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Singh RK, Indra D, Mitra S, Mondal RK,
Basu PS, Roy A, Roychowdhury S and Panda CK: Deletions in
chromosome 4 differentially associated with the development of
cervical cancer: Evidence of slit2 as a candidate tumor suppressor
gene. Hum Genet. 122:71–81. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shivapurkar N, Virmani AK, Wistuba II,
Milchgrub S, Mackay B, Minna JD and Gazdar AF: Deletions of
chromosome 4 at multiple sites are frequent in malignant
mesothelioma and small cell lung carcinoma. Clin Cancer Res.
5:17–23. 1999.PubMed/NCBI
|
|
38
|
Burford A, Little SE, Jury A, Popov S,
Laxton R, Doey L, Al-Sarraj S, Jürgensmeier JM and Jones C:
Distinct phenotypic differences associated with differential
amplification of receptor tyrosine kinase genes at 4q12 in
glioblastoma. PLoS One. 8:e717772013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Geller MD, Pei Y, Spurgeon SE, Durum C and
Leeborg NJ: Chronic lymphocytic leukemia with a FGFR3
translocation: Case report and literature review of an uncommon
cytogenetic event. Cancer Genet. 207:340–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Doak SH, Jenkins GJ, Parry EM, D'Souza FR,
Griffiths AP, Toffazal N, Shah V, Baxter JN and Parry JM:
Chromosome 4 hyperploidy represents an early genetic aberration in
premalignant Barrett's oesophagus. Gut. 52:623–628. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Landreville S, Agapova OA, Harbour JW and
Manuscript A: Emerging insights into the molecular pathogenesis of
uveal melanoma. Future Oncol. 4:629–636. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bauer J, Kilic E, Vaarwater J, Bastian BC,
Garbe C and de Klein A: Oncogenic GNAQ mutations are not
correlated with disease-free survival in uveal melanoma. Br J
Cancer. 101:813–815. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Halling KC and Kipp BR: Bladder cancer
detection using FISH (UroVysion assay). Adv Anat Pathol.
15:279–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Coleman JF, Theil KS, Tubbs RR and Cook
JR: Diagnostic yield of bone marrow and peripheral blood FISH panel
testing in clinically suspected myelodysplastic syndromes and/or
acute myeloid leukemia: A prospective analysis of 433 cases. Am J
Clin Pathol. 135:915–920. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Göhring G, Giagounidis A, Büsche G,
Hofmann W, Kreipe HH, Fenaux P, Hellström-Lindberg E and
Schlegelberger B: Cytogenetic follow-up by karyotyping and
fluorescence in situ hybridization: Implications for monitoring
patients with myelodysplastic syndrome and deletion 5q treated with
lenalidomide. Haematologica. 96:319–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bishop R: Applications of fluorescence in
situ hybridization (FISH) in detecting genetic aberrations of
medical significance. Biosci Horiz. 3:85–95. 2010. View Article : Google Scholar
|
|
47
|
Horsman DE, Sroka H, Rootman J and White
VA: Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer
Genet Cytogenet. 45:249–253. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Prescher G, Bornfeld N and Becher R:
Nonrandom chromosomal abnormalities in primary uveal melanoma. J
Natl Cancer Inst. 82:1765–1769. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Höglund M, Gisselsson D, Hansen GB, White
VA, Säll T, Mitelman F and Horsman D: Dissecting karyotypic
patterns in malignant melanomas: Temporal clustering of losses and
gains in melanoma karyotypic evolution. Int J Cancer. 108:57–65.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Scholes AGM, Damato BE, Nunn J, Hiscott P,
Grierson I and Field JK: Monosomy 3 in uveal melanoma: Correlation
with clinical and histologic predictors of survival. Invest
Ophthalmol Vis Sci. 44:1008–1011. 2003. View Article : Google Scholar : PubMed/NCBI
|