Introduction
Oral squamous cell carcinoma (OSCC) represents the
sixth most common solid cancer worldwide and tongue squamous
carcinoma (TSCC) is one of the major oral malignant tumor subtypes
(1–3). TSCC has the worst prognosis for early
stage disease compared with any other head and neck cancer subsite
(4). Despite treatment with the
combination of surgery, radiotherapy and chemotherapy, which lead
to significant morbidity, failure rates remain unacceptably high
(4,5). One of the reasons for the dismal
prognosis is that current treatment cannot eliminate
cancer-initiating cells (CICs) (6,7). A
comprehensive understanding of the molecular basis of CICs in TSCC
may contribute to the identification of novel therapeutic targets
to improve patient outcome.
Tripartite motif containing 14 (TRIM14), a newly
identified gene located on chromosome 9q22, contains a B-box, a
coiled-coil domain, and a C-terminal PRYSPRY domain, but lacks the
N-terminal RING domain found in most TRIM family proteins (8). TRIM14 is markedly upregulated in TSCC
cell lines and clinical tissues and its expression was found to be
significantly correlated with the TNM classification in patients
with TSCC (9). TRIM14 expression
may be an independent prognostic indicator for the survival of
patients with TSCC (9). Ectopic
expression of TRIM14 in TSCC cells promoted proliferation,
angiogenesis and increased resistance to cisplatin-induced
apoptosis of TSCC cells (9).
Furthermore, TRIM14 overexpression significantly promoted the
tumorigenicity of TSCC cells in vivo whereas silencing of
endogenous TRIM14 caused an opposite outcome (9). It has been proposed that TRIM14 may
represent a novel therapeutic target for the treatment of TSCC
(9).
MicroRNAs (miRNAs/miRs) are regulatory, non-coding
RNAs ~18-25 nucleotides in length and are expressed at specific
stages of tissue development or cell differentiation, and have
large-scale effects on the expression of a variety of genes at the
post-transcriptional level (10–15).
miRNAs induce mRNA degradation or translational suppression of
targeted transcripts by base-pairing with the 3 untranslated region
3′UTR of its targeted mRNAs (10–15).
Dysregulated miRNAs have an important role in the development of
TSCC (16). However, elucidating
the roles of miRNAs in cancer biology, particularly in TSCC,
remains an ongoing process.
In the present study, we showed that TRIM14 induced
the formation of CICs and epithelial-mesenchymal transition (EMT)
in TSCC SCC25 cells. Its overexpression promoted
cisplatin-resistance in the SCC25 cells. We found that
overexpression of miR-15b suppressed TRIM14 and inhibited
cancer-initiating cell phenotypes in the SCC25 cells. Moreover,
overexpression of miR-15b promoted mesenchymal-epithelial
transition (MET) in the SCC25 cells and sensitized
cisplatin-resistant SCC25 (SCC25-res) cells to cisplatin. Thus, we
conclude that miR-15b inhibited cancer stem cell phenotypes and its
restoration reversed chemoresistance of cisplatin by targeting
TRIM14 in TSCC. Elucidating the molecular mechanism of EMT and
cancer-initiating cells in TSCC may further help us to understand
the pathogenesis and progression of the disease, and offer novel
targets for the discovery of new drugs.
Materials and methods
Human TSCC cell line SCC25 and
SCC25-res cells
SCC25 cells were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA). To obtain
cisplatin-resistant tongue cancer cells, we treated SCC25 cells
with escalating concentrations of cisplatin from 107 to
105 M. The established SCC25-res cells grew at a similar
rate in the presence or absence of 105 M cisplatin for 3
days (data not shown). IC50 is the cisplatin
concentration that reduces proliferating cells by 50%. The
IC50 value of SCC25-res cells increased by 12-fold,
respectively, as compared with the SCC25 cells (data not
shown).
Western blotting
Protein extracts were resolved through
SDS-polyacrylamide gel electrophoresis, transferred to
polyvinylidene difluoride membranes (Bio-Rad, Berkeley, CA, USA),
and probed with antibodies against human ERCC1, YAP, TRIM14, CD44,
CD133, OCT4, Nanog (all from Abcam, Cambridge, MA, UK) or β-actin
(Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Sphere forming assay
Cells were seeded on 0.5% agar pre-coated 6-well
plates and cultured for 1 week. Single spheres were selected and
counted.
Soft agar assay
The soft agar assay was carried out as previously
described (17).
MTT assay
To monitor resistance to cisplatin, SCC25 and
SCC25-res cells were treated with cisplatin at different
concentrations for 24 h. The MTT assay was performed as previously
described (18).
Real-time PCR for miRNAs
Total RNA was isolated from cultured cells using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Detection of the
mature form of miRNAs and the mRNA expression of TRIM14 were
performed using specific qRT-PCR primers. The primer sequences of
GAPDH were: forward, 5′-ATTCAACGGCACAGTCAAGG-3′ and reverse,
5′-GCAGAAGGGGCGGAGATGA-3′. The primer sequences of TRIM14 were:
forward, 5′-GCAGAGACAGAGCTAGACTGTAAAGGT-3′ and reverse,
5′-CCTGGTCACACAATTGATATGGA-3′.
Immunofluorescence staining
The cells after treatment were fixed with 4%
paraformaldehyde and permeabilized with 1% Triton X. After
blocking, the cells were incubated with the primary antibodies
against TRIM14, E-cadherin or vimentin (Abcam) overnight at 4°C. On
the following day, the cells were incubated with the secondary
antibodies and 4,6-diamidine-2-phenylindole dihydrochloride (DAPI),
and viewed under a fluorescence microscope.
Methods of bioinformatics
The analysis of potential miRNA target sites was
carried out using the commonly used prediction algorithms miRanda
(http://www.microrna.org/).
Statistical analysis
Data are presented as mean ± SEM. Student's t-test
(two-tailed) was used to compare two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
TRIM14 induces the formation of the
CIC phenotypes in SCC25 cells
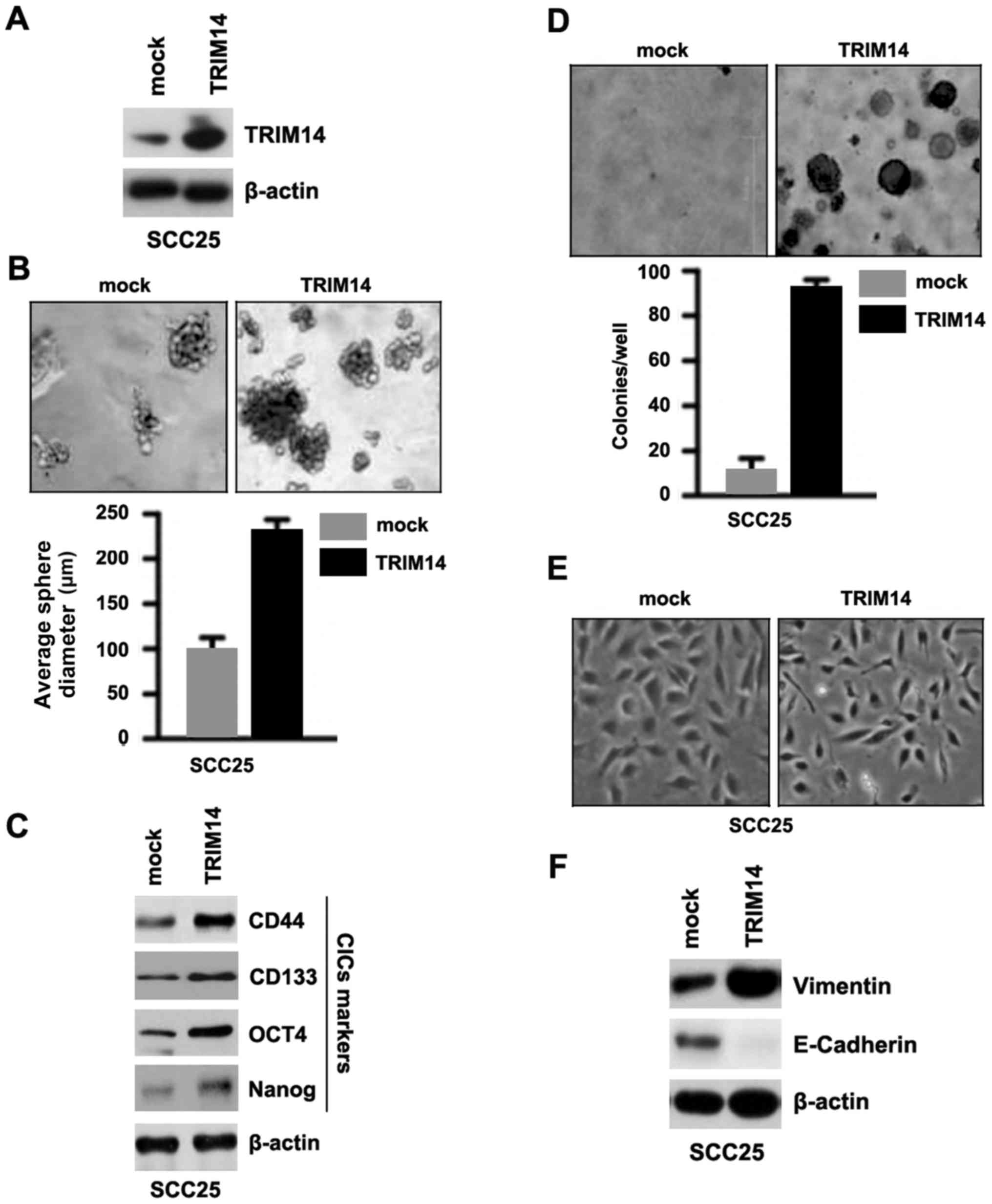
In order to ascertain whether TRIM14 affects CIC
traits in SCC25 cells, we performed a sphere forming assay to
assess the capacity of CIC or CIC-like self renewal in SCC25 cells.
We tested whether TRIM14-expressing plasmids could stably express
TRIM14 protein in the SCC25 cells. The results showed that TRIM14
protein was significantly increased by the TRIM14-expressing
plasmids in the cells (Fig. 1A).
Sphere forming assay showed that TRIM14-overexpressing cells formed
much larger spheres after 14 days of culture as compared with the
control cells, indicating markedly increased CIC traits by
TRIM14-expressing plasmids (Fig.
1B). CD44, CD133, OCT4 and Nanog are positively associated with
stem cell-like characteristics in cancer (19–22).
To identify whether TRIM14 can regulate CD44, CD133, OCT4 and Nanog
protein expression, we performed western blotting of the SCC25
cells transfected with the TRIM14-expressing plasmids and empty
vectors. The results showed that CD44, CD133, OCT4 and Nanog
protein were upregulated in the SCC25 cells transfected with the
TRIM14-expressing plasmids (Fig.
1C). To determine whether cells with CIC characteristics could
have increased clonogenic ability, we performed clonogenic assay.
We found that clonogenic ability was significantly increased in the
SCC25 cells transfected with the TRIM14-expressing plasmids
(Fig. 1D). In addition, the results
from the cell morphology demonstrated that overexpression of TRIM14
induced EMT phenotype in the SCC25 cells (Fig. 1E). We also performed western
blotting to detect E-cadherin (epithelial marker) and vimentin
(mesenchymal marker) protein in the SCC25 cells transfected with
the TRIM14-expressing plasmids and empty vectors. We found that
E-cadherin protein was downregulated and vimentin was upregulated
by TRIM14 in the SCC25 cells transfected with the TRIM14-expressing
plasmids (Fig. 1F).
TRIM14 overexpression leads to
cisplatin-resistance in TSCC
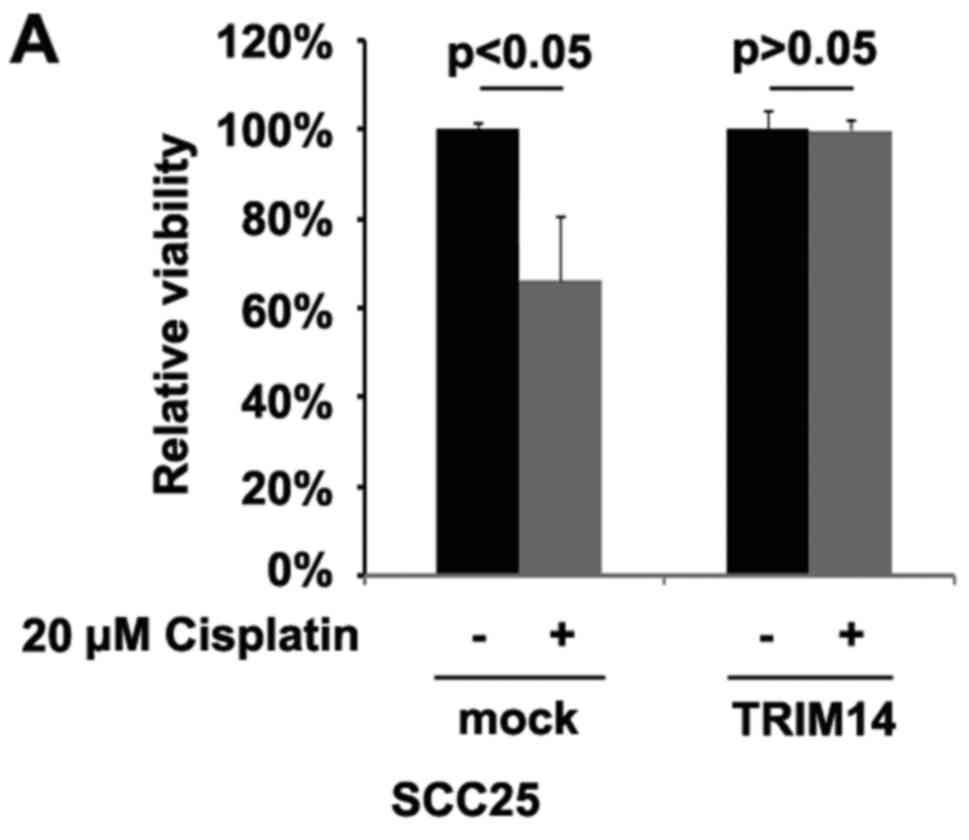
To identify whether TRIM14 can affect cisplatin
resistance in SCC25 cells, we performed MTT assay. We overexpressed
TRIM14 in cisplatin-sensitive SCC25 cells and its overexpression
transformed the cisplatin-sensitive SCC25 cells to
cisplantin-resistant cells (Fig.
2A), suggesting that its overexpression is involved in
cisplatin resistance. High expression of ERCC1 and YAP are
associated with cispatin-resistance in locally advanced squamous
cell carcinoma of the head and neck (23,24).
Our results showed that ERCC1 and YAP protein were upregulated by
TRIM14 (Fig. 2B).
miR-15b degrades TRIM14 in TSCC
Having demonstrated that overexpression of TRIM14
induced the formation of CIC phenotypes and EMT, next we studied
the mechanisms regulating TRIM14 expression in TSCC. miRs are a
class of small non-coding RNAs (~22 nucleotides) and negatively
regulate protein-coding gene expression by targeting mRNA
degradation or translation inhibition (11–13).
To further confirm whether TRIM14 could be regulated
by miRNA, we used the commonly used prediction algorithm miRanda
(http://www.microrna.org/microrna/home.do) to analyze
3′UTR of TRIM14. A dozen miRNAs were identified by the algorithm.
However, we were interested in miR-15b, since it is one of the most
significantly downregulated miRNAs in SCC25-res cells and its
restoration effectively reversed the phenotype of EMT in SCC25-res
cells, and sensitized them to chemotherapy (25).
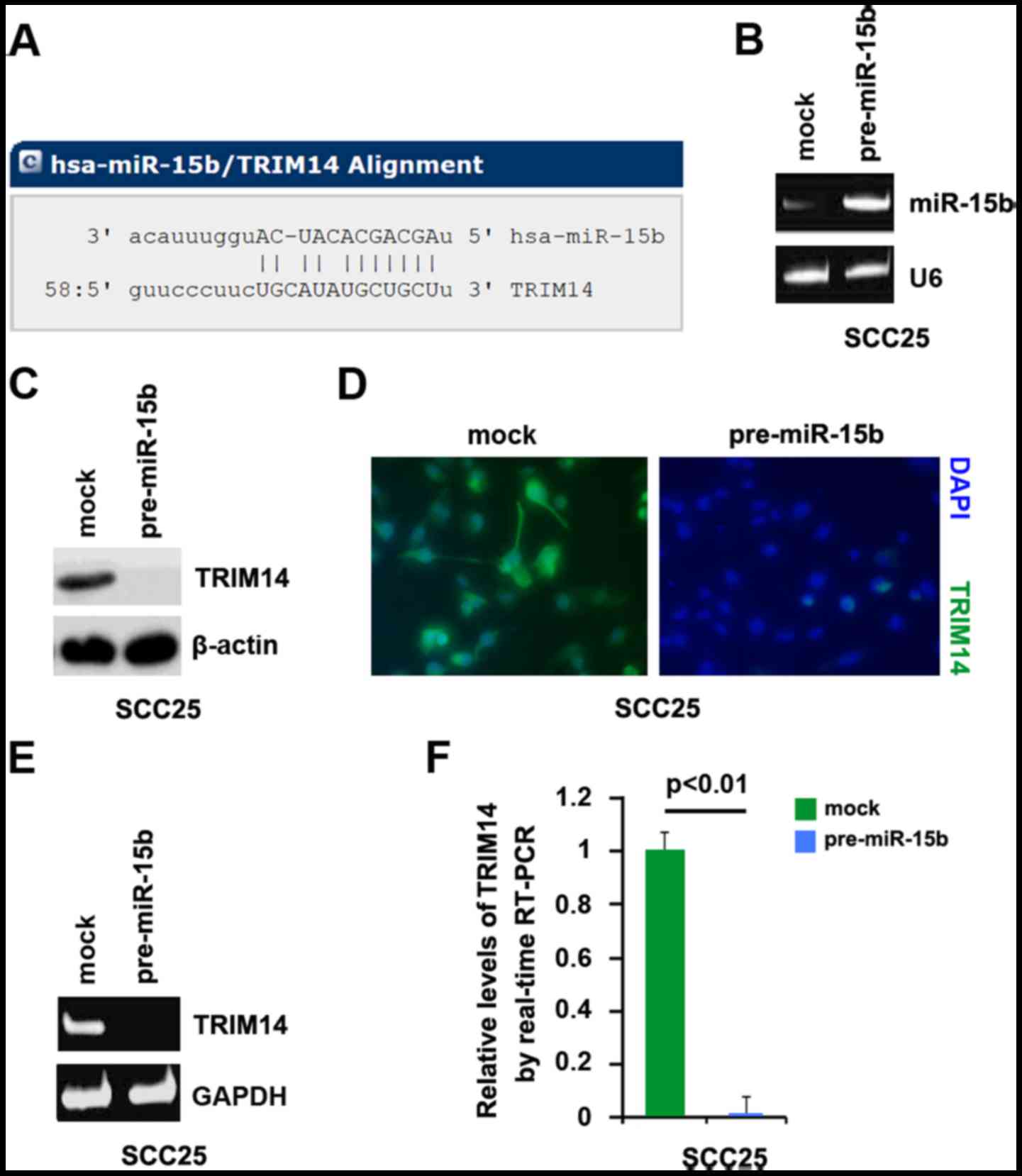
Target sites on 3′UTR of TRIM14 are shown in
Fig. 3A. We reasoned that miR-15b
downregulates TRIM14 expression by targeting its 3′UTR in TSCC.
Downregulation of miR-15b can contribute to upregulation of TRIM14
and cisplatin resistance in TSCC. In an attempt to identify the
role of miR-15b in regulating TRIM14 expression in TSCC, we
transfected SCC25 cells with pre-miR-15b and control miR. After
transfection, miR-15b expression was detected by real-time PCR and
the results showed that miR-15b was significantly increased by
pre-miR-15b in the cells (Fig.
3B).
To confirm the reason, we performed western blotting
to detect TRIM14 expression in the SCC25 cells transfected with
pre-miR-15b and control miR. The results showed that TRIM14 protein
was significantly inhibited by miR-15b (Fig. 3C). We next performed
immunofluorescence analyses in the SCC25 cells transfected with
pre-miR-15b and control miR. The results showed that TRIM14 protein
was evidently inhibited in the cells transfected with pre-miR-15b
(Fig. 3D). To identify whether
miR-15b can degrade TRIM14 mRNA, we performed RT-PCR and real-time
PCR and we found that TRIM14 mRNA was degraded by miR-15b (Fig. 3E and F).
miR-15b inhibits CIC phenotypes in the
SCC25-res cells
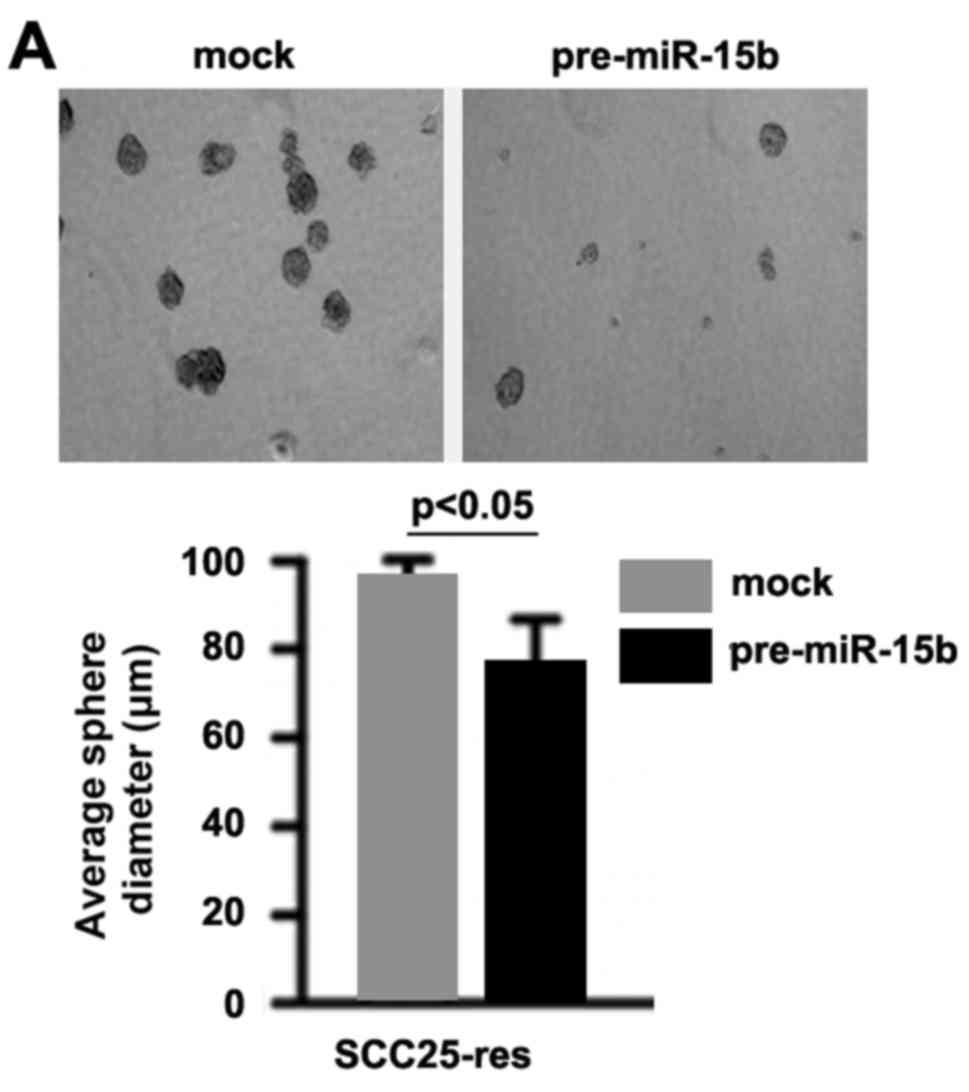
In order to ascertain whether miR-15b affects CIC
traits in the SCC25-res cells, we performed sphere forming assay to
assess the capacity of CIC or CIC-like self renewal in the
SCC25-res cells. Sphere forming assay showed that
miR-15b-overexpressing cells formed much smaller spheres after 14
days of culture as compared with control cells, indicating markedly
decreased CIC traits by pre-miR-15b (Fig. 4A). To identify whether TRIM14 can
regulate CD44, CD133, OCT4 and Nanog protein expression, we
performed western blotting in SCC25-res cells transfected with
pre-miR-15b and control miR. The results showed that CD44, CD133,
OCT4 and Nanog protein were downregulated in the SCC25-res cells
transfected with pre-miR-15b (Fig.
4B).
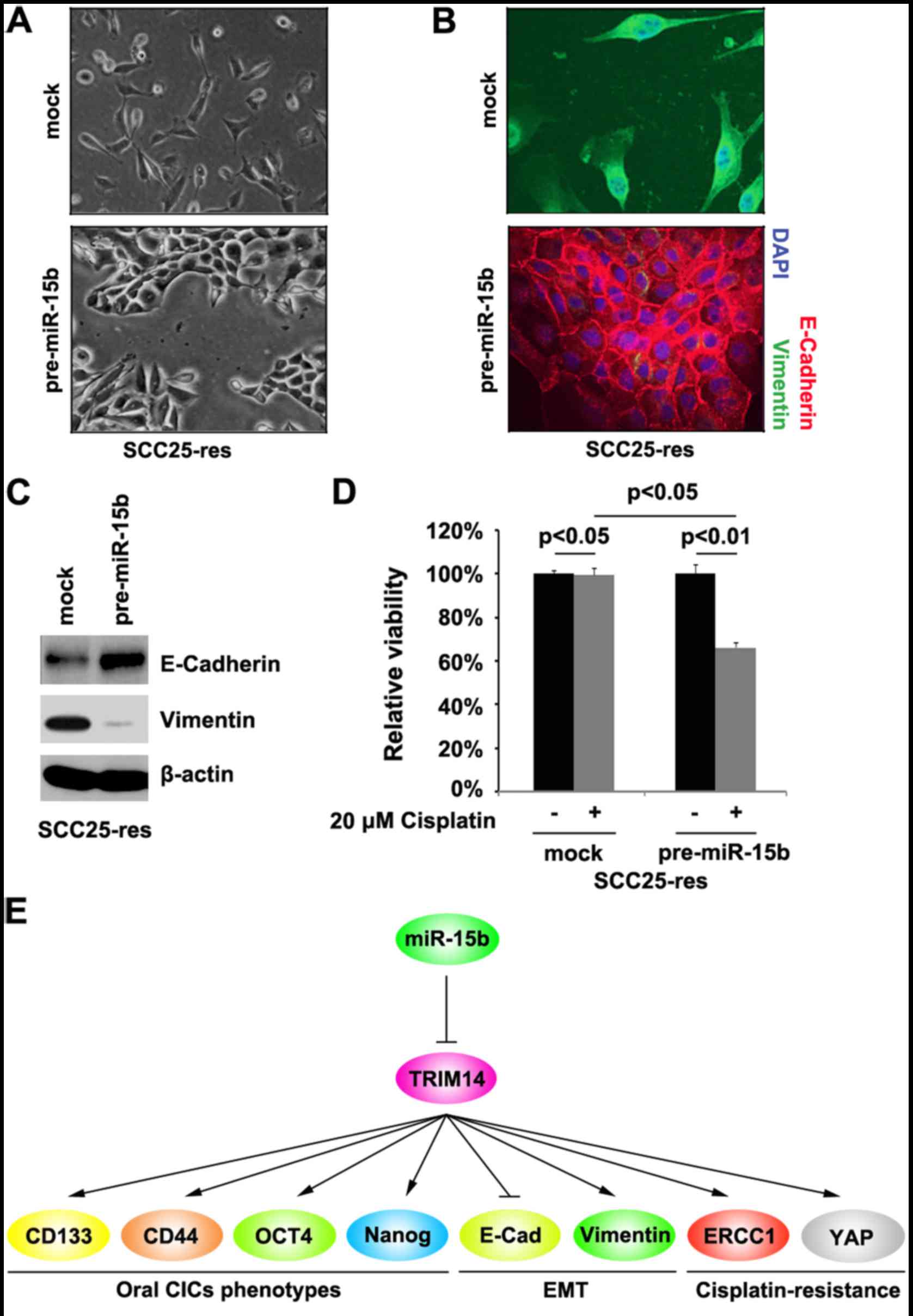
miR-15b induces MET and sensitizes
SCC25-res cells to cisplatin
Cell morphology demonstrated that overexpression of
miR-15b induced MET phenotypes in SCC25-res cells (Fig. 5A). In order to detect whether
miR-15b can affect E-cadherin (epithelial marker) and vimentin
(mesenchymal marker) protein, we performed immunofluorescence
analyses in the SCC25-res cells transfected with pre-miR-15b and
control miR. We found that restoration of miR-15b promoted
E-cadherin and inhibited vimentin in the SCC25-res cells (Fig. 5B). We also performed western
blotting to detect E-cadherin and vimentin protein in the SCC25-res
cells transfected with pre-miR-15b and control miR. We found that
E-cadherin protein was upregulated and vimentin was downregulated
by miR-15b in the SCC25-res cells (Fig.
5C).
To study the role of miR-15b in cisplatin
resistance, we performed MTT assay in the SCC25-res cells. The
results showed that restoration of miR-15b sensitized SCC25-res
cells to cisplatin (Fig. 5D).
Discussion
miR-15b is the one of most significantly
downregulated microRNAs (miRNAs/miRs) in cisplatin-resistant SCC25
(SCC25-res) cells and its restoration effectively reversed EMT in
the SCC25-res cells, and sensitized them to chemotherapy, while
inhibition of miR-15b in the sensitive lines induced EMT and
conferred chemoresistance (25).
Moreover, EMT cells can have cancer-initiating cell-like features
and CICs exhibit a mesenchymal phenotype under most circumstances.
Consequently, malignant tumors tend to relapse after surgical
resection. This character is believed to be largely attributable to
the stem cell-like properties of a fraction of cells (26). In the present study, we found that
miR-15b may play an important role in the formation of CICs and MET
of TSCC and in the regulation of cisplatin (cisplatin is widely
used to treat oral squamous cell carcinoma, however, many patients
exhibit acquired drug resistance). These findings provide novel
insights into the potential role of miR-15b deregulation in
promoting the formation of CICs and conferring chemoresistance in
TSCC (Fig. 5E).
Ectopic expression of TRIM14 in TSCC cells was found
to promote proliferation, angiogenesis and increased resistance to
cisplatin-induced apoptosis in TSCC cells in vitro.
Furthermore, TRIM14 overexpression significantly promoted the
tumorigenicity of TSCC cells in vivo whereas silencing of
endogenous TRIM14 caused an opposite outcome (9). In line with this report, we showed
that overexpression of TRIM14 induced the formation of CIC traits
and EMT (Fig. 5E). The results
further confirmed the idea that CICs exhibiting a mesenchymal
phenotype frequently can promote chemotherapy resistance in cancer.
Contrary to miR-15b, TRIM14 is markedly upregulated in TSCC cell
lines and clinical tissues (9,25).
Restoration of miR-15b to inhibit TRIM14 may represent a novel
therapeutic target for the treatment of TSCC.
Low expression of ERCC1 is an independent predictor
for prolonged survival (23).
Moreover, it has been proposed that ERCC1 expression may be a
useful predictive marker of locally advanced squamous cell
carcinoma of the head and neck in patients treated with
cisplatin-based concurrent chemoradiotherapy (23). We demonstrated that TRIM14
overexpression significantly promoted ERCC1 expression in the TSCC
cells. Deregulation of the miR-15b/TRIM14/ERCC1 axis may play an
important role in the progression of TSCC. YAP overexpression
correlates with epithelial-mesenchymal transition and nodal
metastasis, resulting in cisplatin-resistance. Thus, targeting YAP
could be a new therapeutic strategy for the treatment of patients
with oral squamous cell carcinoma that are resistant to cisplatin
(24). We found that TRIM14
significantly upregulated YAP protein level in the SCC25 cells
(Fig. 5E). Deregulation of the
miR-15b/TRIM14/YAP axis may promote the formation of
cisplatin-resistant TSCC.
Acknowledgements
The present study was supported by the Shandong
Province Science and Technology Research Project
(2013G0021812).
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Argiris A, Karamouzis MV, Raben D and
Ferris RL: Head and neck cancer. Lancet. 371:1695–1709. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rusthoven K, Ballonoff A, Raben D and Chen
C: Poor prognosis in patients with stage I and II oral tongue
squamous cell carcinoma. Cancer. 112:345–351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brennan S, Corry J, Kleid S, Porceddu S,
Yuen K, Rischin D and Peters LJ: Prospective trial to evaluate
staged neck dissection or elective neck radiotherapy in patients
with CT-staged T1-2 N0 squamous cell carcinoma of the oral tongue.
Head Neck. 32:191–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Q, Shi S, Yen Y, Brown J, Ta JQ and
Le AD: A subpopulation of CD133+ cancer stem-like cells
characterized in human oral squamous cell carcinoma confer
resistance to chemotherapy. Cancer Lett. 289:151–160. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meroni G and Diez-Roux G: TRIM/RBCC, a
novel class of ‘single protein RING finger’ E3 ubiquitin ligases.
BioEssays. 27:1147–1157. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Su X, Wang J, Chen W, Li Z, Fu X and Yang
A: Overexpression of TRIM14 promotes tongue squamous cell carcinoma
aggressiveness by activating the NF-κB signaling pathway.
Oncotarget. 7:9939–9950. 2016.PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small
RNAs with antisense complementarity to lin-14. Cell.
75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing
in Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duz MB, Karatas OF, Guzel E, Turgut NF,
Yilmaz M, Creighton CJ and Ozen M: Identification of miR-139-5p as
a saliva biomarker for tongue squamous cell carcinoma: A pilot
study. Cell Oncol. 39:187–193. 2016. View Article : Google Scholar
|
|
17
|
Kong D, Banerjee S, Ahmad A, Li Y, Wang Z,
Sethi S and Sarkar FH: Epithelial to mesenchymal transition is
mechanistically linked with stem cell signatures in prostate cancer
cells. PLoS One. 5:e124452010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ren ZG, Dong SX, Han P and Qi J: miR-203
promotes proliferation, migration and invasion by degrading SIK1 in
pancreatic cancer. Oncol Rep. 35:1365–1374. 2016.PubMed/NCBI
|
|
19
|
Park SY, Lee HE, Li H, Shipitsin M, Gelman
R and Polyak K: Heterogeneity for stem cell-related markers
according to tumor subtype and histologic stage in breast cancer.
Clin Cancer Res. 16:876–887. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mansour SF and Atwa MM:
Clinicopathological significance of CD133 and ALDH1 cancer stem
cell marker expression in invasive ductal breast carcinoma. Asian
Pac J Cancer Prev. 16:7491–7496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsai LL, Hu FW, Lee SS, Yu CH, Yu CC and
Chang YC: Oct4 mediates tumor initiating properties in oral
squamous cell carcinomas through the regulation of
epithelial-mesenchymal transition. PLoS One. 9:e872072014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct-4 and Nanog in oral cancer stem-like cells and high-grade
oral squamous cell carcinoma. Cancer Res. 14:4085–4095. 2008.
|
|
23
|
Jun HJ, Ahn MJ, Kim HS, Yi SY, Han J, Lee
SK, Ahn YC, Jeong HS, Son YI, Baek JH, et al: ERCC1 expression as a
predictive marker of squamous cell carcinoma of the head and neck
treated with cisplatin-based concurrent chemoradiation. Br J
Cancer. 99:167–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshikawa K, Noguchi K, Nakano Y, Yamamura
M, Takaoka K, Hashimoto-Tamaoki T and Kishimoto H: The Hippo
pathway transcriptional co-activator, YAP, confers resistance to
cisplatin in human oral squamous cell carcinoma. Int J Oncol.
46:2364–2370. 2015.PubMed/NCBI
|
|
25
|
Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M,
Zhang W, Chen W, Pan C, Liu Q, et al: MiR-200b and miR-15b regulate
chemotherapy-induced epithelial-mesenchymal transition in human
tongue cancer cells by targeting BMI1. Oncogene. 31:432–445. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sanai N, Alvarez-Buylla A and Berger MS:
Neural stem cells and the origin of gliomas. N Engl J Med.
353:811–822. 2005. View Article : Google Scholar : PubMed/NCBI
|