Introduction
Primary liver cancer, particularly hepatocellular
carcinoma (HCC), is the sixth most common and aggressive malignancy
worldwide, accounting for over 800,000 deaths per year (1,2). In
China, the mortality rate of HCC is the second highest, and the
5-year survival rate is below 5% (3). The major cause of mortality in HCC
patients is tumor cell invasion and metastasis, which is a complex
process that involves changes in the extracellular matrix, leading
to translocation from the primary tumor to distant organs and
growth at new sites. Multiple reports have shown that various
signaling pathways are involved in tumor metastasis, but the
details of the molecular mechanisms underlying tumor invasion and
metastasis remain largely unclear. Therefore, it is crucial to
elucidate the mechanisms of tumor metastasis and to develop a novel
strategy for HCC treatment.
MicroRNAs (miRNAs) are a class of endogenous
noncoding RNAs that are 21–23 nucleotides in length (in mammals).
They are evolutionarily conserved and bind primarily to
complementary sequences in the 3′-untranslated region (UTR) of
their target mRNAs, inhibiting translation or destabilizing the
mRNAs (4). miRNAs can function as
oncogenes or tumor suppressors, and miRNA dysfunction is involved
in many biological processes, including the development and
progression of various human cancers (4,5). The
aberrant expression of specific miRNAs is directly implicated in
tumorigenesis, including processes of growth, apoptosis, and
metastasis (6,7). For example, miR-199a suppresses
tumorigenicity and multidrug resistance of ovarian
cancer-initiating cells (8). Thus,
miRNAs act as potentially important tumor hallmarks, and their
deregulation has been studied to identify tools for early
diagnosis, prognosis, monitoring, and treatment (9,10).
However, the functions and mechanisms of miRNAs involved in HCC
metastasis are largely unknown.
In this study, we examined the effect of miR-539 on
HCC cells to determine its potential contribution to HCC migration
and invasion. We evaluated the expression of miR-539 in HCC cell
lines and clinical specimens. Additionally, we investigated the
functional role of miR-539 in the migration and invasion of HCC
cell lines and the contribution of miR-539 to HCC cancer malignancy
and the underlying molecular mechanisms. Our findings elucidated
important roles of miR-539 in HCC and suggested novel therapeutic
strategies for HCC treatment.
Materials and methods
Human tissue samples
The HCC samples and corresponding adjacent
non-tumorous liver tissues were obtained from 30 patients with
primary HCC who underwent surgery at the First Affiliated Hospital
of Xi'an Jiaotong University (Xi'an, China) from July 2012 to June
2014. The clinicopathological characteristics of the patients were
independently determined by two experienced pathologists. All
tissue samples were immediately frozen in liquid nitrogen and
stored at −80°C until RNA extraction. None of the patients
recruited in this study had undergone preoperative chemotherapy,
radiotherapy, or other therapy.
Lentiviral production, plasmid
construction, and transfection
Viral vectors containing the miR-539 (GenBank ID:
664612) construct and the green fluorescent protein (GFP) construct
were purchased from Takara (Shiga, Japan). The miR-539 sequence was
sub-cloned into the pLVX-IRES-Hyg vector (Takara) to generate
pLV-miR-539. The viral particles were harvested 48 h after
transfection of pLV-miR-539 into HEK293T cells using the Lenti-HT
Packaging Mix (Takara). HCCLM3 and SK-HEP1 cells were infected with
the harvested recombinant lentivirus in the presence of 6 µg/ml
Polybrene (Sigma, St. Louis, MO, USA). The empty pLV-GFP vector
encoding GFP was used as the control (Fig. 1). The viral particles were harvested
as previously described (11) and
used in all subsequent studies.
The full-length 3′-UTR of the FSCN1 gene containing
the putative miR-539 binding sites was amplified by PCR and was
inserted into the pGL3 vector under the control of the CMV promoter
(Promega, Madison, WI, USA). The coding sequences of FSCN1 were
amplified by PCR and cloned into the pCDNA3.1(+) vector
(Invitrogen, Carlsbad, CA, USA) to generate pCDNA-FSCN1. The primer
sequences used to amplify the target genes are listed in Table I. The correct insertion of the
PCR-amplified sequences was confirmed by sequencing. The plasmid
was transfected using Lipofectamine 2000 (Invitrogen) according to
the manufacturer's instructions.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
|
| Primer sequences
(5′-3′) | Length (bp) |
|---|
| GAPDH-F |
TCATGGGTGTGAACCATGAGAA |
146 |
| GAPDH-R |
GGCATGGACTGTGGTCATGAG |
|
| FSCN1-F |
CACAGGCAAATACTGGACGGT |
101 |
| FSCN1-R |
CCACCTTGTTATAGTCGCAGAAC |
|
| miR-539-F |
GGAGAAATTATCCTTGGTGTG | / |
| U6-F |
ATTGGAACGATACAGAGAAGATT | / |
| FSCN1-F |
GGAATTCATGACCGCCAACGGCACAGCCGA | 1497 |
| FSCN1-R |
CCCTCGAGTTAGTACTCCCAGAGCGAGGCGGG |
|
| FSCN1-wtUTR-F |
GCTCTAGATGCTAACCCCTTCTCCGCCAGGT |
491 |
| FSCN1-wtUTR-R |
GCTCTAGAAAGGGTCAGTCCTAGCCCACCCGA |
|
| FSCN1-mutUTR-F |
ACCTCAGATGGCTAAGTCACGCAGACACATGGAAGCG |
491 |
| FSCN1-mutUTR-R |
CGCTTCCATGTGTCTGCGTGACTTAGCCATCTGAGGT |
|
RNA extraction and quantitative
polymerase chain reaction (qRT-PCR)
Total RNA from cultured cells and frozen tissues was
extracted using the RNAiso reagent (Takara) according to
manufacturer's instruction. To measure the miR-539 levels, we used
a miRNA reverse transcription kit (Invitrogen) and SYBR Green
Master Mix kit (Roche Diagnostics, Indianapolis, IN, USA) to obtain
the cDNA using qRT-PCR. Aliquots (1 µg) of RNA were reverse
transcribed to cDNA (20 µl) with oligo(dT) and M-MuLV reverse
transcriptase (Fermentas, Glen Burnie, MD, USA) in accordance with
the manufacturer's instructions. One-fifth of the resulting cDNA
was used as a template for PCR using a SYBR Green PCR kit (Takara)
in a LightCycler 480 Real-time PCR System (Roche Diagnostics,
Mannheim, Germany). The housekeeping gene
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an
internal control for each mRNA expression experiment. U6 was used
for miRNA normalization. The primer sequences for the target genes
are listed in Table I. The cycling
conditions were as follows: an initial 10 min of pre-denaturation
at 95°C, followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec,
and 72°C for 15 sec. The specificity of the amplification products
was confirmed by melting curve analysis. Each RT reaction was run
in triplicate. The relative quantification of miR-539 or FSCN1 was
performed using the 2−∆∆Ct method to analyze the qRT-PCR
data (12).
Cell proliferation studies
Cells were plated in 96-well plates at
2×103 per well. At the indicated time points, 100 µl of
3-(4–5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT,
0.5 mg/ml, Roche Diagnostics) was added to each well, and the cells
were incubated for 4 h at 37°C, followed by removal of the culture
medium and addition of 150 µl dimethyl sulfoxide (DMSO, Sigma) to
dissolve the formazan crystals. The OD value at 450 nm was then
determined and used to construct a growth curve with time to assess
cell proliferation. Each experiment was repeated at least
thrice.
Colony formation assay
For the soft-agar assay, 2,000 cells were suspended
in complete medium containing 0.6% agar (Sigma) and then added on
top of a layer of complete medium containing 1% agar in 6-well
plates. Colonies were counted under an inverted microscope after 20
days. The colonies were counted in 10 randomly chosen microscope
fields.
Wound healing assay
To further investigate cell migration, we used wound
healing assays as previously described (13). Cells (2×105) were seeded
in 6-well plates, incubated overnight, and then infected or
transfected with a test vector or vehicle. At 90% confluency, the
cell monolayer was scratched with a sterile 200-µl pipette tip.
Then, the plate was washed with PBS 5–6 times to remove the
suspended cells, and the cells were maintained in DMEM containing
1% FBS. Images were taken at 0 and 24 h along the scrape line under
a microscope to observe the migrated cells and wound healing, and
the cells in three wells from each group were quantified. The
results are expressed as the relative scratch width based on the
distance migrated relative to the original scratched distance.
ImageJ Plus was used to quantify the images of the wound healing
assays.
Transwell cell migration and invasion
assay
As previously described (14), cell migration and invasion were
assayed using Transwells with an 8 µm pore size (Corning Costar,
Cambridge, MA, USA). The Transwells were placed into 24-well
plates. The following day, the medium was removed, and 100 µl fresh
medium was added to the upper chambers coated without or with
Matrigel (BD Biosciences) in each insert. Then, 500 µl DMEM with
10% FBS was added to the lower chamber. After incubation for 24–48
h at 37°C in a 5% CO2 humidified incubator, the cells on
the upper surface were removed with a cotton swab, and the cells
that migrated or invaded to the bottom side of the membrane were
fixed with 95% absolute alcohol and stained with crystal violet for
20 min at room temperature. Four random fields of each membrane
were imaged under an inverted microscope (Olympus Corp., Tokyo,
Japan) at ×100 magnification, and the cells were counted for
statistical analysis. Each experiment was performed in
triplicate.
Bioinformatics and dual luciferase
reporter gene assay
Three generally accepted online bioinformatics
programs, TargetScan, RNAhybrid, and microRNA.org,
were used to predict the interactions between miR-539 and target
mRNAs. The predicted candidates were further investigated.
Luciferase reporter assays were performed using a construct that
contained the regions of interest that were sub-cloned into the
XbaI restriction site of the luciferase reporter vector pGL3
Basic Vector (Promega). The PCR products were amplified from the
full-length 3′-UTR of FSCN1 using the primers listed in Table I. Site-directed mutagenesis was
performed using the QuikChange Site-Directed Mutagenesis kit
(Agilent Technologies, Inc., Santa Clara, CA, USA) to alter the
miR-539 binding site sequence in the 3′-UTR FSCN1-amplified PCR
product. For luciferase assays, SK-HEP1 cells were seeded at
1.0×105/well in 24-well plates, cultured until they
reached 70% confluence, and co-transfected with LV-miR-con or
LV-miR-539; pGL3-control (400 ng; Promega), pGL3-FSCN1-WT (400 ng)
or pGL3-FSCN1-Mut; and pRL-TK (50 ng, Promega) using the
Lipofectamine 2000 reagent. Cells were lysed using cell lysis
buffer (Cell Signaling, Boston, MA, USA), and luciferase activity
was measured using the Dual-Luciferase Reporter Assay System
(Promega) according to the manufacturer's protocol. Three
independent experiments were performed, and the data are presented
as the mean ± SD.
Eukaryotic expression vector
construction
To construct a plasmid that expresses FSCN1 in
cells, we cloned the open reading frame (ORF) of FSCN1 into the
mammalian expression vector pcDNA3.1(+) (Invitrogen) using the
primers in Table I. The constructs
were confirmed by sequencing.
Preparation of cell extracts and
western blot analysis
Western blot analyses were performed as previously
described (15). Protein lysates
were prepared on ice in RIPA buffer [50 mM Tris-HCl (pH 8.0), 150
mM NaCl, 0.1% SDS, 1% NP40 and 0.5% sodium deoxycholate] for 30 min
with freshly added 0.1 mg/ml phenylmethylsulfonyl fluoride (PMSF),
1 mM sodium orthovanadate, and 1 mg/ml aprotinin. The supernatants
were collected by centrifugation at 12,000 × g at 4°C for 20 min.
Protein concentrations were determined using a BCA assay kit
(Pierce, Rockford, IL, USA). Aliquots of cell extracts containing
20–50 µg of total protein were resolved using 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to a 0.45 mm polyvinylidene fluoride (PVDF) membrane
(Millipore, Temecula, CA, USA). Membranes were blocked in 5%
non-fat dry milk in TBS-T [10 mM Tris-HCl (pH 8.0), 150 mM NaCl,
0.05% Tween-20] for 2 h at room temperature (RT) and then incubated
for 4 h at RT in Blotto A containing a 1:1000 dilution of rabbit
anti-FSCN1 and anti-GAPDH monoclonal antibodies. After a wash in
TBS-T buffer (5 min, RT), the membranes were incubated for 1 h at
RT in Blotto A containing a 1:10,000 dilution of
peroxidase-conjugated anti-mouse secondary antibody (Abcam,
Cambridge, UK). After a wash in TBS-T, the enhanced
chemiluminescence (ECL) luminol reagent (Pierce) was used to
develop the membrane according to the manufacturer's
recommendation. GAPDH was used as a loading control. All procedures
were performed three times.
Tumor studies in vivo
Athymic nude and BALB/c mice were obtained from the
Shanghai Experimental Animal Center (Shanghai, China) and
maintained according to the Animal Research Committee's guidelines
at Xi'an Jiaotong University. Briefly, a total of 2×106
SK-HEP1 cells were injected into the right leg of each mouse (seven
animals for each treatment) and then monitored every other day for
tumor growth. When the tumors grew to 40–50 mm3, the
animals were randomly divided into two treatment groups. Each mouse
was treated with LV-miR-con (100 µl, 2×108 TU/ml) or
LV-miR-539 (100 µl, 2×108 TU/ml) via intratumoral
injection. All treatments were administered every other day for a
total of five doses. Tumor growth was monitored by measuring
perpendicular tumor diameters using an electronic digital caliper.
Tumor volumes were calculated by the following formula: tumor
size=ab2/2, where a is the larger and b is the
smaller of the two dimensions. The tumor-bearing mice were
sacrificed 24 days after injection, and the tumors were removed,
weighed, and examined by immunohistochemistry (IHC) staining
assays.
Immunohistochemistry (IHC)
staining
Solid tumors were removed from the sacrificed mice
and were fixed with 4% formaldehyde. Paraffin-embedded tumor
tissues were sectioned to 5-µm thickness and mounted on positively
charged microscope slides, and 1 mM EDTA (pH 8.0) was used for
antigen retrieval. Endogenous peroxidase activity was quenched by
incubating the slides in methanol containing 3% hydrogen peroxide,
and the slides were then washed in PBS for 6 min. Next, the
sections were incubated for 2 h at RT with normal goat serum and
subsequently incubated at 4°C overnight with a primary antibody
(Abcam: 1:100, Ki67). Then, the sections were rinsed with PBS and
incubated with a horseradish peroxidase-linked goat anti-rabbit
antibody, followed by reaction with Mayer's hematoxylin. Ki-67 was
determined as the ratio of the number of cells positive for brown
staining to the total number of cells in a given field of view at
×400 magnification. Five fields of view were analyzed per
section.
Statistical analyses
The results represent the average of three
experiments, and the data are presented as the mean ± SD. Each
experiment was performed at least three times unless otherwise
specified. Statistical significance was determined using an
unpaired Student's t-test, and a value of P<0.05 was considered
to indicate a statistically significant difference.
Results
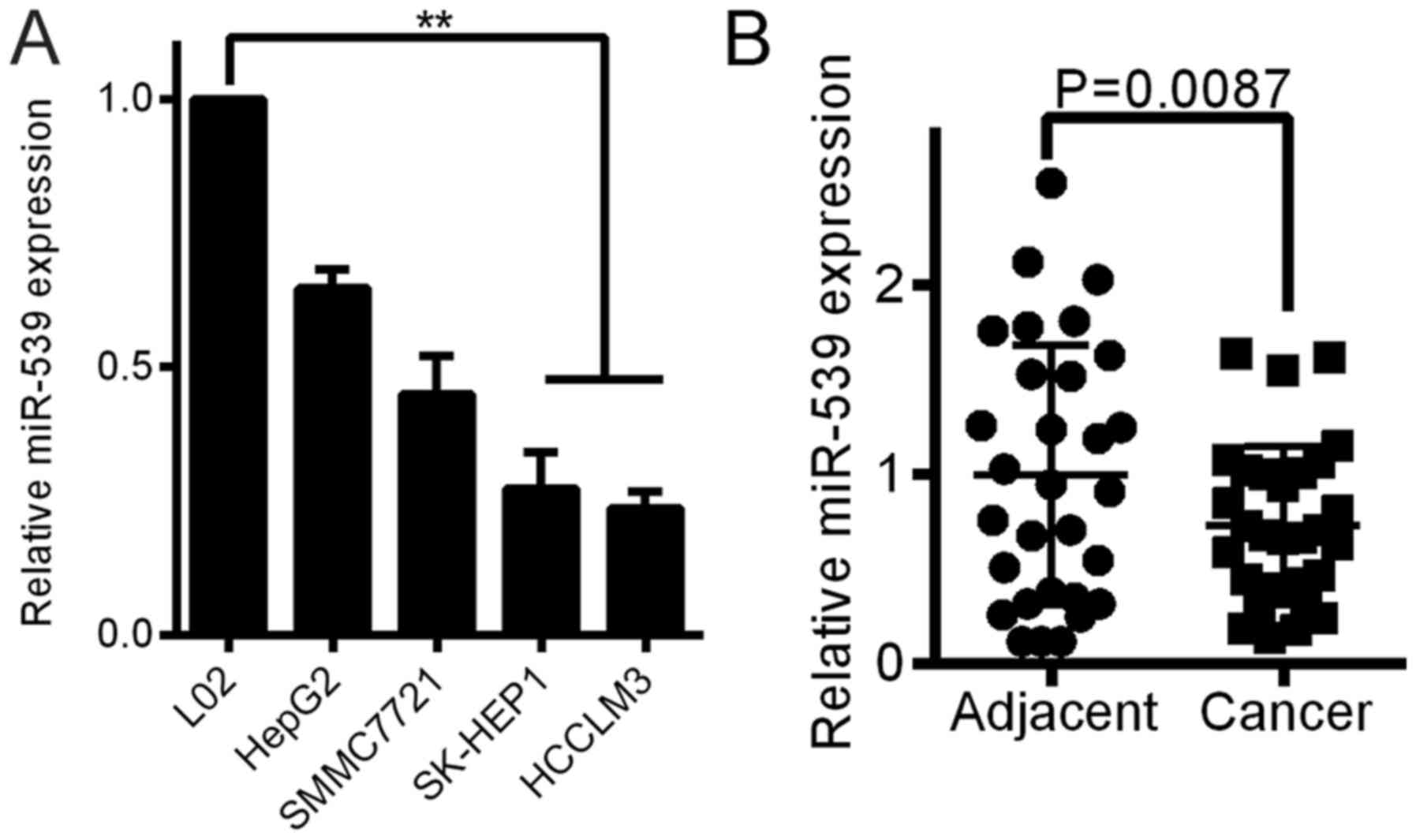
miR-539 is downregulated in HCC cell
lines and tissue samples
QRT-PCR analyses were performed to determine the
expression level of miR-539 in 4 HCC cell lines and the normal
liver cell line L02. The levels of miR-539 were downregulated in
all HCC cell lines compared with the miR-539 level in the L02 cells
(Fig. 1A). The miR-539 expression
was also measured in 30 freshly frozen HCC and adjacent normal
tissue samples. The expression of miR-539 was significantly
decreased in 19 HCC tissues compared to the expression in the
adjacent tissues (Fig. 1B,
P<0.01). Five HCC tissues were expressed to the high level of
the adjacent tissues, while 6 HCC tissues were the same with the
adjacent tissues.
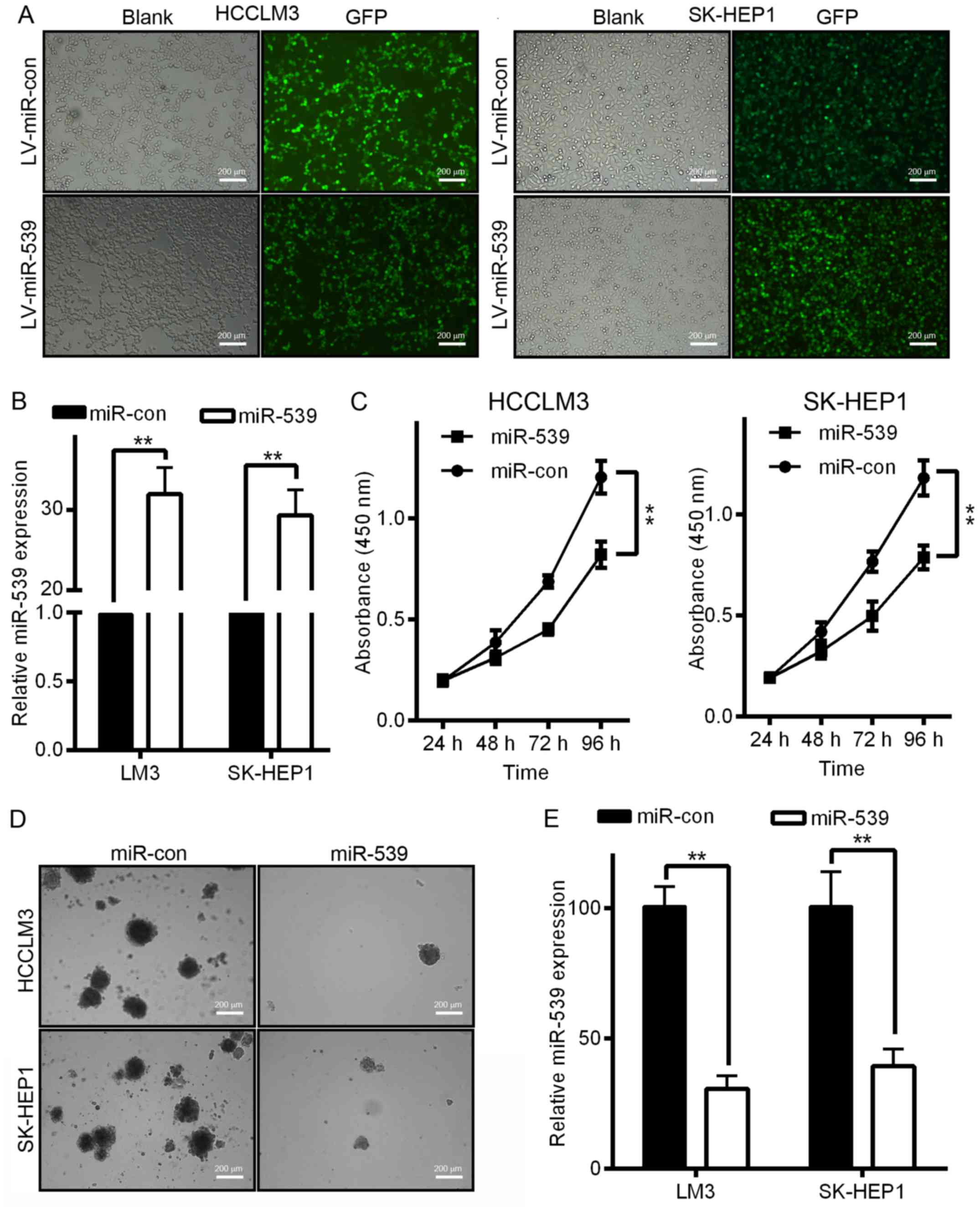
miR-539 inhibits HCC cell growth and
proliferation in vitro
The downregulation of miR-539 in both HCC tissues
and HCC cell lines suggested that miR-539 may play a role in HCC
tumorigenesis. To investigate the biological function of miR-539 in
the development and progression of HCC, we performed MTT and
soft-agar assays after the infection of HCC cells with LV-miR-539
or LV-miR-con. We first generated LV-miR-539 or LV-miR-con and then
used these constructs to infect HCCLM3 and SK-HEP1 cells. The
increased miR-539 expression was confirmed by qRT-PCR (Fig. 2B). As shown in Fig. 2C, HCCLM3 and SK-HEP1 cells infected
with LV-miR-539 displayed significant growth inhibition compared to
that of cells infected with LV-miR-con (P<0.01). Moreover, cells
infected with LV-miR-539 formed fewer and smaller colonies compared
with cells infected with LV-miR-con in an anchorage-independent
manner (Fig. 2D and E,
P<0.05).
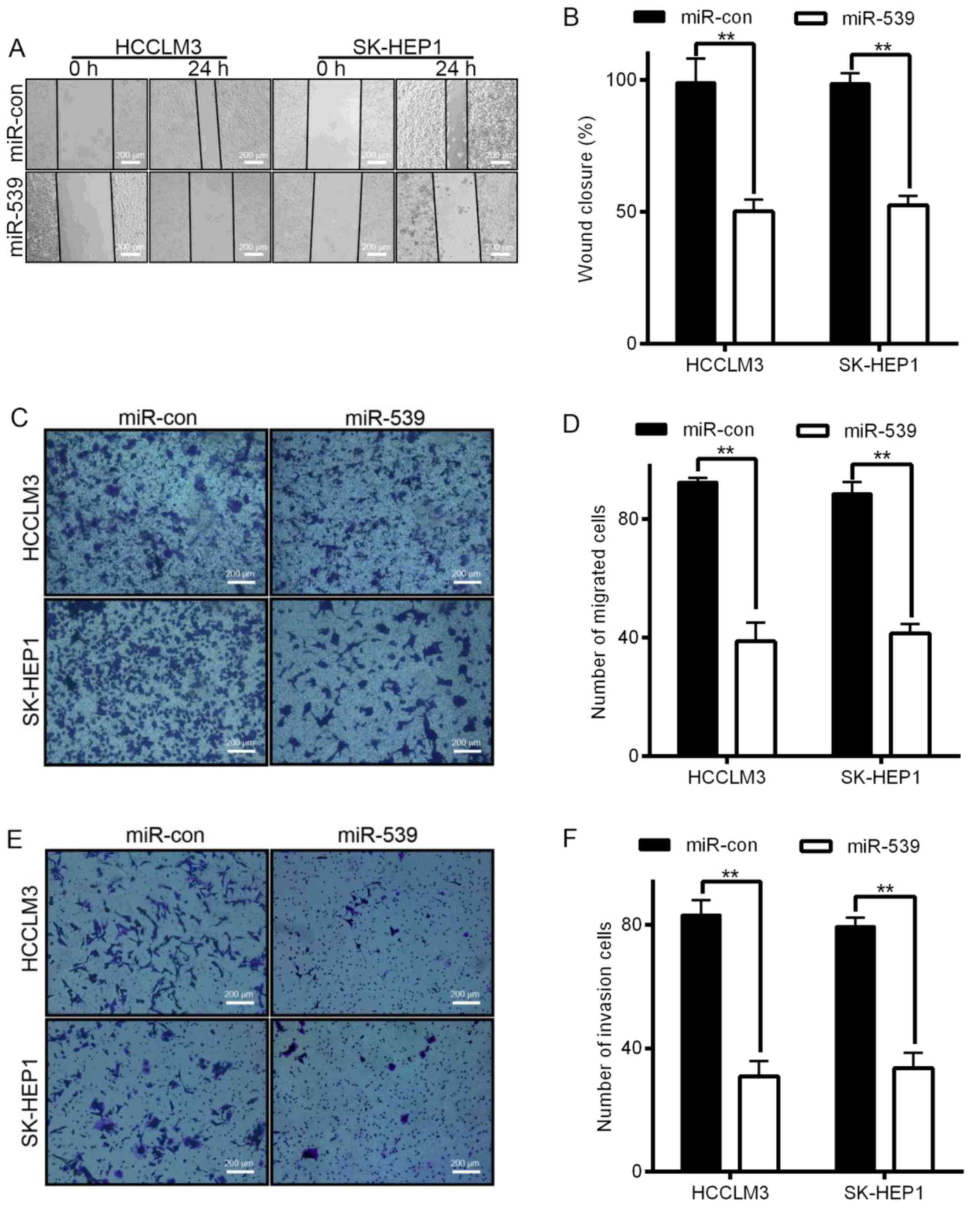
miR-539 inhibits HCC cell migration
and invasion in vitro
Invasion and migration through the basement membrane
is a characteristic property of metastatic cancer cells. We used
two different approaches to assess the role of miR-539 in the
migration of HCCLM3 and SK-HEP1 cells. The wound healing assays
showed that the migratory ability of HCCLM3 and SK-HEP1 cells
infected with LV-miR-539 was much weaker than that of cells
transfected with miR controls (Fig. 3A
and B). Transwell migration assays also demonstrated that the
overexpression of LV-miR-539 significantly suppressed the migratory
ability of HCCLM3 and SK-HEP1 cells (Fig. 3C and D, P<0.01). In addition,
Transwell invasion assays indicated that invasion was inhibited by
infection with LV-miR-539 (Fig. 3E and
F, P<0.05). These results, taken together, clearly
demonstrate that overexpression of miR-539 markedly reduces the
migration and invasion of HCC cancer cells.
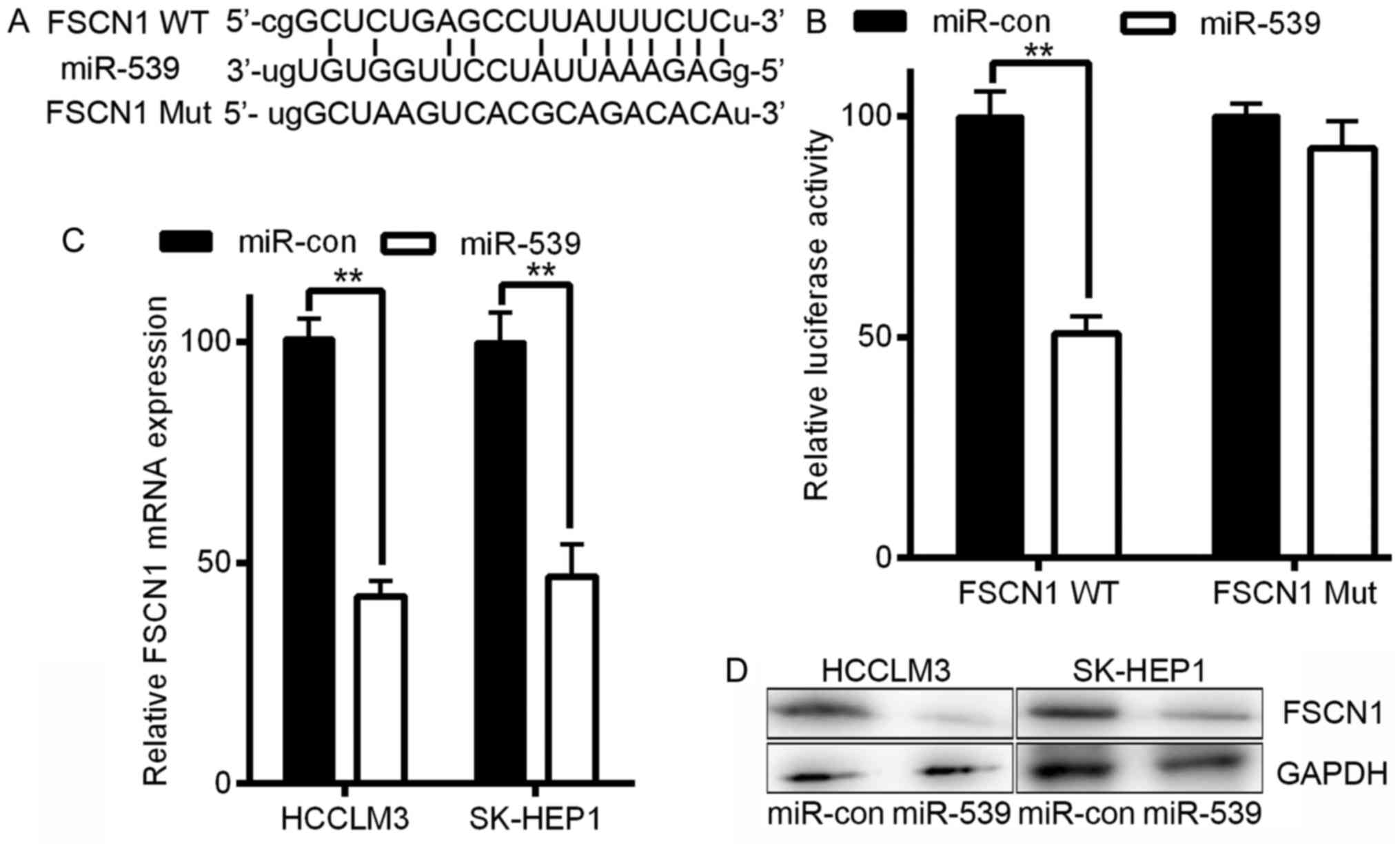
FSCN1 is a direct target of
miR-539
Next, we explored the underlying molecular mechanism
of the anti-tumorigenic effects of miR-539 in HCC cells. Because
miRNAs primarily mediate their biological functions in animal cells
by altering the expression of target genes, we performed a
bioinformatics search with publicly available databases
(TargetScan, RNAhybrid, and microRNA.org)
for putative mRNA targets of miRNAs that exhibited anti-tumor
properties. Fascin homologue 1 (FSCN1) was identified as a
potential target as it contains one potential miR-539 binding site
within its 3′-UTR (Fig. 4A). FSCN1
is a 55 kDa actin-binding protein that is an important regulator of
the maintenance and stability of filamentous actin and plays a
crucial role in the regulation of cell adhesion, migration, and
invasion (16,17).
Additionally, FSCN1 has been reported to regulate
the migration and invasion of HCC (18). To assess the potential regulation of
FSCN1 by miR-539 via this putative binding site, we constructed a
reporter vector consisting of the luciferase coding sequence
followed by the 3′-UTR of FSCN1. Either the wild-type
(pGL3-FSCN1-WT) or a mutated sequence (pGL3-FSCN1-Mut) with the
seed region sites was cloned into the modified pGL3 vector
(Fig. 4B). Co-transfection
experiments showed that miR-539 significantly decreased the
luciferase activity of the construct containing the wild-type
3′-UTR sequence in HCCLM3 cells (P<0.01, Fig. 4B), but this decreased activity was
not observed in the pGL3-FSCN1-Mut-transfected cells (Fig. 4B). Our data thus demonstrated that
FSCN1 is a direct target of miR-539. To further confirm that
miR-539 targets FSCN1, we transfected LV-miR-539 or LV-miR-con into
HCCLM3 and SK-HEP1 cells. Infection with miR-539 resulted in a
significant reduction in FSCN1 mRNA and protein expression as
measured by qRT-PCR and western blotting, respectively, in HCCLM3
and SK-HEP1 cells (P<0.01, Fig. 4C
and D). Taken together, these results indicate that in HCC
cells, FSCN1 is a direct target gene of miR-539, and this
regulation requires the miR-539 binding sequence in the 3′-UTR.
Restoring FSCN1 expression reverses
the effect of miR-539 on HCC cell migration and invasion
FSCN1 has been reported to play a critical role in
the migration and invasion of HCC (18). To confirm that miR-539 regulates
FSCN1, we co-transfected SK-HEP1 cells with LV-miR-539 and
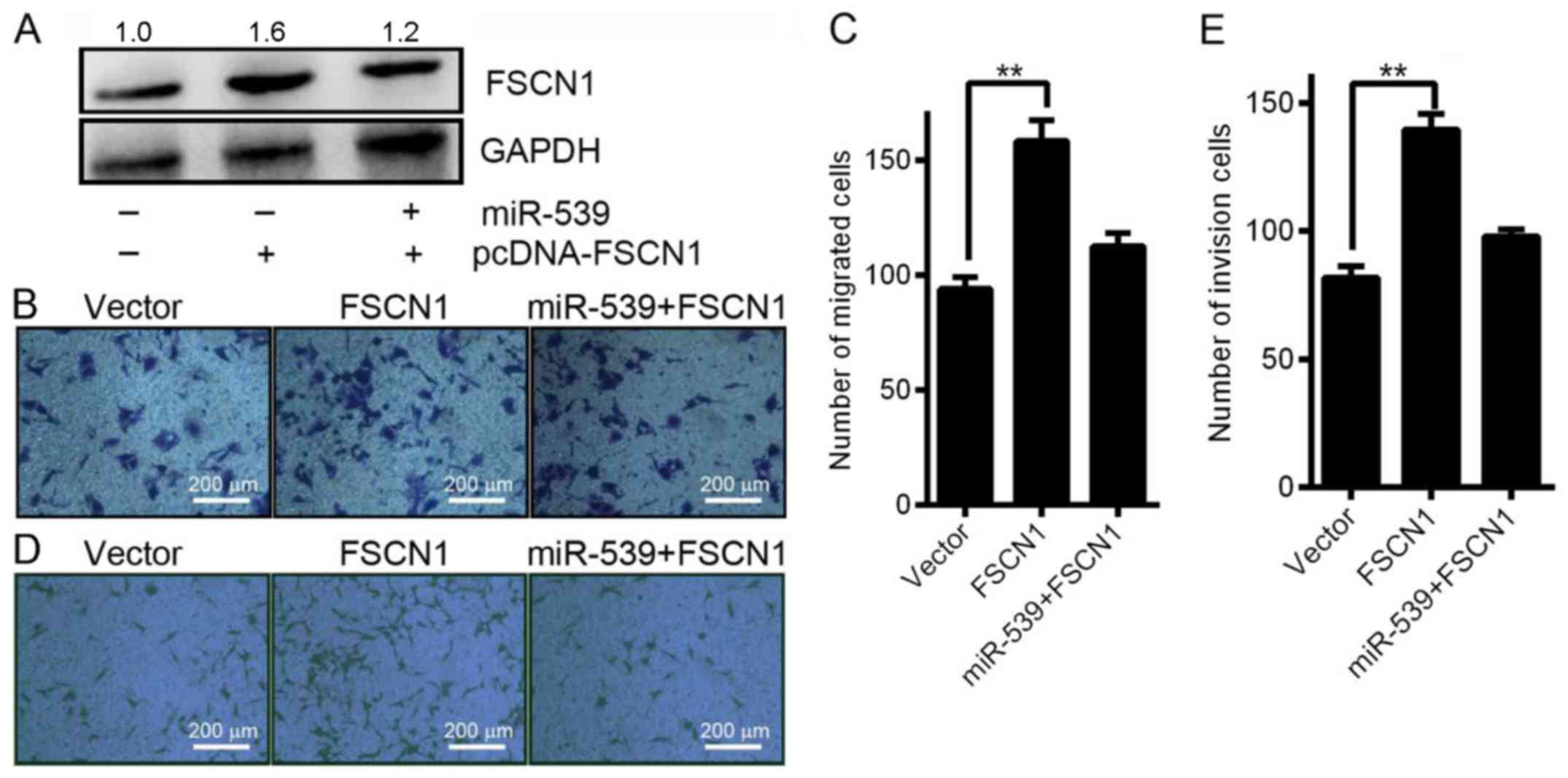
pcDNA-FSCN1 to perform a rescue experiment. As shown in Fig. 5A, western blotting analysis
indicated that simultaneous transfection of SK-HEP1 cells with
miR-539 and pcDNA-FSCN1 reduced the ability of miR-539 to
downregulate FSCN1 protein expression. Consistent with this result,
the Transwell migration assays revealed that co-transfection of
pcDNA-FSCN1 significantly reduced the ability of the miR-539 to
inhibit cell migration at 24 h after transfection (Fig. 5B and C). Additionally, the Transwell
invasion assays showed that co-transfection of pcDNA-FSCN1
abolished the increased cell invasion promoted by infection by
LV-miR-539 at 24 h (Fig. 5D and E).
Overall, the results indicate that miR-539 suppresses HCC cell
invasion and migration by regulating FSCN1.
miR-539 inhibits HCC xenograft tumor
growth in vivo
To further confirm the growth-attenuating effect of
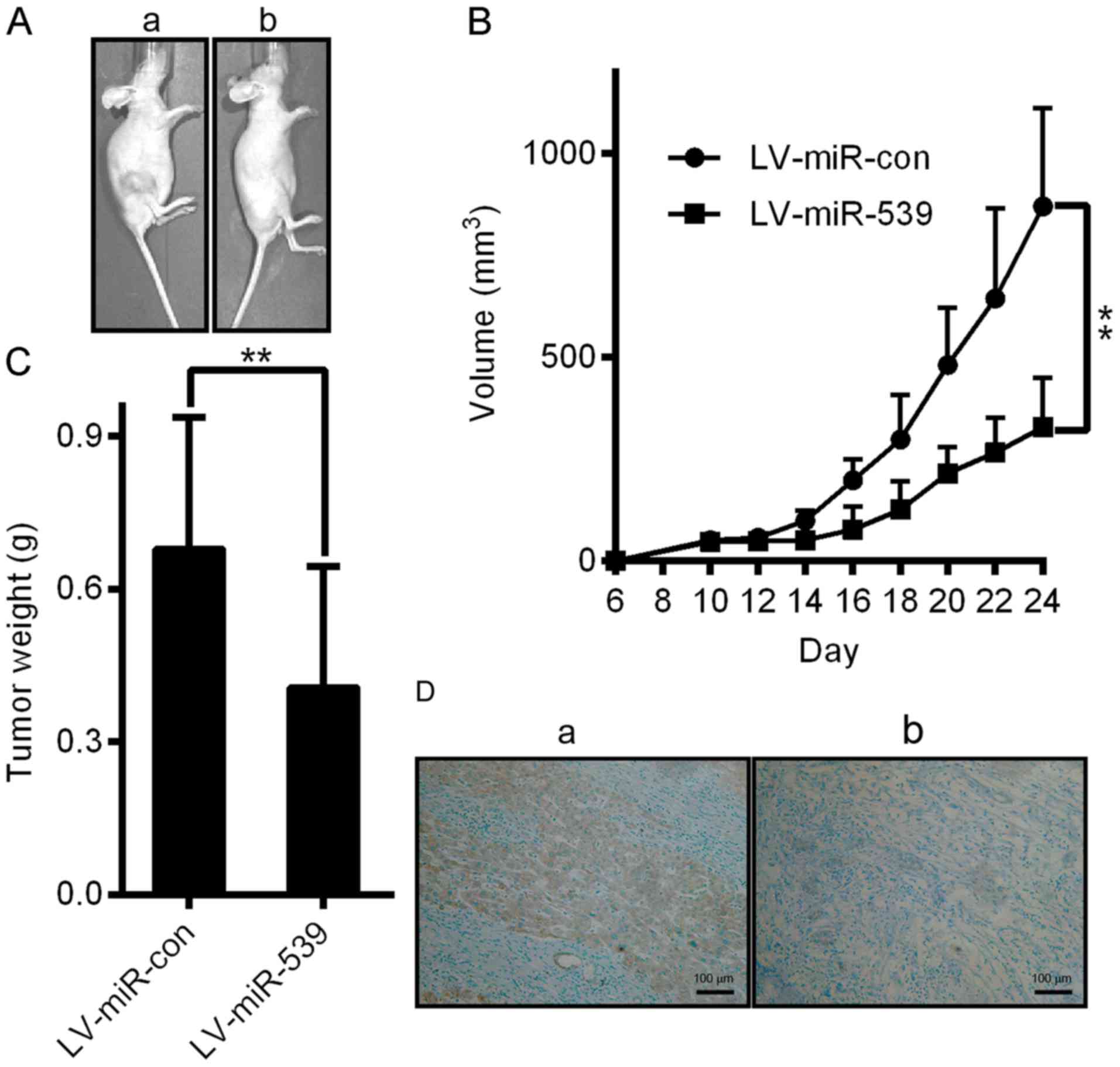
miR-539 on HCC, we performed a xenograft tumor growth assay.
Xenograft tumor growth models were established as described in the
methods section. As shown in Fig. 6A
and B, tumors grew at a slower rate and had smaller volumes in
the miR-539-overexpressing group than those of the miR-con group
(P<0.05). The average tumor weight in the miR-539-overexpressing
group was also significantly lower than that of the control group
(0.41±0.09 g vs. 0.68±0.10 g; Fig.
6C, P<0.05). We further performed IHC staining for Ki67 in
the tumors. Compared with the miR-con control, miR-539
overexpression suppressed proliferative activity, as indicated by
the percentage of cells positive for Ki67 staining (Fig. 6D). Our results revealed that miR-539
attenuated the proliferation of HCC cells in vivo. Thus,
this study demonstrates that miR-539 plays a tumor suppressive role
in HCC.
Discussion
There are several miRNAs with potential roles in
cancer development, and we selected miR-539 as the focus of this
study due to its tumor suppressor role in osteosarcoma cancer
(19) and its unclear roles in HCC.
miR-539 is present in the miRNA-rich intragenic region of mouse
chromosome 12 [NC_000078.6 (109728129 to 109728202)] between Rian
(RNA imprinted and accumulated in nucleus) and Mirg
(miRNA-containing gene). There are few verified targets of miR-539,
although miR-539 has been shown to inhibit migration and invasion
in several types of tumors, including thyroid cancer (20), prostate cancer (21), and osteosarcoma (19). Recently, Zhang et al
(21) showed that ectopic
overexpression of miR-539 drastically inhibited SPAG5 expression
and prostate cancer cell proliferation and metastasis in
vitro and in vivo. Additionally, Zhu et al
reported miR-539 was downregulated in HCC tissues and cells, and
suppresses tumor growth and tumorigenesis in vitro and in
vivo. miR-539 overcomes arsenic trioxide resistance in HCC
(22). There may be different
effects of miR-539 on different cellular processes involved in
tumor development, so the roles of miR-539 are still poorly
understood.
FSCN1, a 55 kDa actin-binding protein, regulates the
maintenance and stability of filamentous actin and plays a crucial
role in the regulation of cell adhesion, migration, and invasion
(16,17). The overexpression of FSCN1 was
associated with aggressive clinical course, poor prognosis, and
decreased survival in various cancers, including HCC (18,23),
indicating that FSCN1 may play a central role in tumorigenesis. The
tumorigenic function of FSCN1 may be conferred by its invasive
properties in cancer cells (24).
The knockdown of FSCN1 expression reduced the proliferation and
metastasis of GC cells, and higher expression of FSCN1 was
correlated with more advanced cancer stages and inversely
correlated with survival rates in gastric adenocarcinomas (25). A previous study identified FSCN1 as
a target of miR-145 in bladder cancer cells (26), but the clinical relevance of FSCN1
in HCC remains unknown.
HCC is characterized by rapid growth and relentless
invasion. Invasiveness and metastasis, the two leading causes of
cancer mortalities, are especially pronounced in HCC, which shows
strikingly high invasive and metastatic potential (27). We highlighted the therapeutic
potential of miR-539 in HCC by identifying miR-539 as a tumor
suppressor that attenuates the proliferation and invasion of HCC
cells both in vitro and in vivo and inhibited growth
and invasion of HCC cells in a nude mouse model. To identify novel
downstream targets of miR-539, we used the TargetScan, RNAhybrid,
and microRNA.org databases to perform screening
experiments. From several candidates, FSCN1 was selected for
further experiments due to the crucial roles of FSCN1 in tumor
migration and invasion. Several pieces of evidence suggest that
FSCN1 is a direct downstream target of miR-539. First, FSCN1
contains highly conserved sequences at the 3′-UTR that are
complementary to the miR-539 seed sequence. Second, we observed
that levels of miR-539 were substantially reduced in HCC samples
compared to adjacent tissues. The present study additionally shed
new light on the correlation between miR-539 and FSCN1 in HCC. Our
results suggest that miR-539 and FSCN1 both play important roles in
HCC invasion.
Our present study represents the first comprehensive
analysis of miR-539 in HCC. We selected miR-539 as a target
molecule based on miRNA profiling and then identified miR-539 as a
tumor suppressor that can attenuate the proliferation and invasion
of HCC in vitro and in vivo. Mechanistically, we
identified FSCN1 as a direct functional target of miR-539, which
expanding our understanding of the players and mechanisms
underlying HCC progression.
Although several studies have reported the
regulation of FSCN1 by miRNAs in many malignant tumors (26,28,29),
to the best of our knowledge, no study has identified FSCN1 as a
direct target of miR-539 in HCC. In this study, we confirmed that
miR-539 inhibits the expression of FSCN1 and directly targets the
3′-UTR of FSCN1 in HCC cells. Furthermore, the simultaneous
re-expression of FSCN1 compromised the miR-539 supression of cell
migration and invasion almost completely in miR-539-transfected
SK-HEP1 cells, which indicated that the repression of FSCN1 is
necessary for miR-539 to inhibit the migration and invasion of HCC
cells. Therefore, FSCN1 is an important downstream target of
miR-539 in the regulation of HCC migration and invasion.
In brief, our study showed that miR-539 is primarily
downregulated in HCC samples and cell lines and that miR-539
expression and FSCN1 expression are strongly negatively correlated
in HCC. In addition, we provided evidence that miR-539 inhibits
cell proliferation, migration, and invasion in HCC via targeting
FSCN1 directly. Therefore, restoring miR-539 expression should be
explored as a potential therapeutic strategy for treatment of
HCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81572734), the Scientific and
Technological Development Research Project Foundation by Shaanxi
Province (2016SF-121) and the Fundamental Research Funds for the
Central Universities in Xi'an Jiaotong University (no.
2013jdhz33).
References
|
1
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hao K, Luk JM, Lee NP, Mao M, Zhang C,
Ferguson MD, Lamb J, Dai H, Ng IO, Sham PC, et al: Predicting
prognosis in hepatocellular carcinoma after curative surgery with
common clinicopathologic parameters. BMC Cancer. 9:3892009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
White NM, Fatoohi E, Metias M, Jung K,
Stephan C and Yousef GM: Metastamirs: A stepping stone towards
improved cancer management. Nat Rev Clin Oncol. 8:75–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen R, Alvero AB, Silasi DA, Kelly MG,
Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T and Mor G:
Regulation of IKKbeta by miR-199a affects NF-kappaB activity in
ovarian cancer cells. Oncogene. 27:4712–4723. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li ZZ, Shen LF, Li YY, Chen P and Chen LZ:
Clinical utility of microRNA-378 as early diagnostic biomarker of
human cancers: A meta-analysis of diagnostic test. Oncotarget.
7:58569–58578. 2016.PubMed/NCBI
|
|
10
|
de Leeuw DC, Verhagen HJ, Denkers F,
Kavelaars FG, Valk PJ, Schuurhuis GJ, Ossenkoppele GJ and Smit L:
MicroRNA-551b is highly expressed in hematopoietic stem cells and a
biomarker for relapse and poor prognosis in acute myeloid leukemia.
Leukemia. 30:742–746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stewart SA, Dykxhoorn DM, Palliser D,
Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et
al: Lentivirus-delivered stable gene silencing by RNAi in primary
cells. RNA. 9:493–501. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan W, Chang Y, Li R, Xu Q, Lei J, Yin C,
Li T, Wu Y, Ma Q and Li X: Curcumin inhibits hypoxia inducible
factor-1α-induced epithelial-mesenchymal transition in HepG2
hepatocellular carcinoma cells. Mol Med Rep. 10:2505–2510.
2014.PubMed/NCBI
|
|
14
|
Lei J, Fan L, Wei G, Chen X, Duan W, Xu Q,
Sheng W, Wang K and Li X: Gli-1 is crucial for hypoxia-induced
epithelial-mesenchymal transition and invasion of breast cancer.
Tumour Biol. 36:3119–3126. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Ma Q, Xu Q, Liu H, Lei J, Duan W,
Bhat K, Wang F, Wu E and Wang Z: SDF-1/CXCR4 signaling induces
pancreatic cancer cell invasion and epithelial-mesenchymal
transition in vitro through non-canonical activation of Hedgehog
pathway. Cancer Lett. 322:169–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jayo A and Parsons M: Fascin: A key
regulator of cytoskeletal dynamics. Int J Biochem Cell Biol.
42:1614–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adams JC: Roles of fascin in cell adhesion
and motility. Curr Opin Cell Biol. 16:590–596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayashi Y, Osanai M and Lee GH: Fascin-1
expression correlates with repression of E-cadherin expression in
hepatocellular carcinoma cells and augments their invasiveness in
combination with matrix metalloproteinases. Cancer Sci.
102:1228–1235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mirghasemi A, Taheriazam A, Karbasy SH,
Torkaman A, Shakeri M, Yahaghi E and Mokarizadeh A: Down-regulation
of miR-133a and miR-539 are associated with unfavorable prognosis
in patients suffering from osteosarcoma. Cancer Cell Int.
15:862015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gu L and Sun W: MiR-539 inhibits thyroid
cancer cell migration and invasion by directly targeting CARMA1.
Biochem Biophys Res Commun. 464:1128–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang H, Li S, Yang X, Qiao B, Zhang Z and
Xu Y: miR-539 inhibits prostate cancer progression by directly
targeting SPAG5. J Exp Clin Cancer Res. 35:602016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu C, Zhou R, Zhou Q, Chang Y and Jiang
M: microRNA-539 suppresses tumor growth and tumorigenesis and
overcomes arsenic trioxide resistance in hepatocellular carcinoma.
Life Sci. 166:34–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iguchi T, Aishima S, Umeda K, Sanefuji K,
Fujita N, Sugimachi K, Gion T, Taketomi A, Maehara Y and Tsuneyoshi
M: Fascin expression in progression and prognosis of hepatocellular
carcinoma. J Surg Oncol. 100:575–579. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Machesky LM and Li A: Fascin: Invasive
filopodia promoting metastasis. Commun Integr Biol. 3:263–270.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsai WC, Jin JS, Chang WK, Chan DC, Yeh
MK, Cherng SC, Lin LF, Sheu LF and Chao YC: Association of
cortactin and fascin-1 expression in gastric adenocarcinoma:
Correlation with clinicopathological parameters. J Histochem
Cytochem. 55:955–962. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Inamoto T, Taniguchi K, Takahara K,
Iwatsuki A, Takai T, Komura K, Yoshikawa Y, Uchimoto T, Saito K,
Tanda N, et al: Intravesical administration of exogenous
microRNA-145 as a therapy for mouse orthotopic human bladder cancer
xenograft. Oncotarget. 6:21628–21635. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang ZC, Gao Q, Shi JY, Guo WJ, Yang LX,
Liu XY, Liu LZ, Ma LJ, Duan M, Zhao YJ, et al: Protein tyrosine
phosphatase receptor S acts as a metastatic suppressor in
hepatocellular carcinoma by control of epithermal growth factor
receptor-induced epithelial-mesenchymal transition. Hepatology.
62:1201–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xue M, Pang H, Li X, Li H, Pan J and Chen
W: Long non-coding RNA urothelial cancer-associated 1 promotes
bladder cancer cell migration and invasion by way of the
hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107:18–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen JJ, Cai WY, Liu XW, Luo QC, Chen G,
Huang WF, Li N and Cai JC: Reverse correlation between microRNA-145
and FSCN1 affecting gastric cancer migration and invasion. PLoS
One. 10:e01268902015. View Article : Google Scholar : PubMed/NCBI
|