Introduction
Multiple myeloma (MM), characterized by a
heterogeneous clonal proliferation of plasma cells, is the
second-most common hematological malignancy (1). MM is responsible for 15–20% of deaths
from hematological malignancy (1).
Targeted therapies such as the proteasome inhibitor (PI) bortezomib
(Bz), the immunomodulatory drugs (IMiDs) lenalidomide and
thalidomide (Th) have vastly improved the prognosis of MM patients
(2–4). Unfortunately, MM is a heterogeneous
disease with various disease courses, responses to therapy and
survival outcome, and many patients still have to face challenges
from disease progression (DP) and eventually succumb to MM
(5). Thus, research to identify
novel therapeutic agents is urgently needed. One promising area of
recent investigation is the development of immunotherapies that
target and eliminate myeloma cells more selectively (6–11). The
premise of these therapies is the identification of cancer-specific
and/or cancer-associated antigens (CAA) through the immune system.
Moreover, finding an effective serological molecular marker which
can forecast the prognoses and therapeutic responses of MM patients
will be a valuable tool for MM therapy.
MM-associated antigen-8 (MMSA-8) (GenBank no.
AY952889.1) is a new MM-associated antigen (MAA) that we previously
identified. We serologically assessed tumor antigens using a
recombinant cDNA expression library (SEREX) of MM to identify
MM-associated antigens (MMSAs). MMSA-8 was confirmed as a new
antigen that specifically interacted with the serum from patients
with allogeneic myeloma but did not interact with normal donors
(12). Differences in the MMSA-8
sequence with the novel available working version of the human
genome suggests that MMSA-8 is a new RPS27A-related transcription
variant that is specifically associated with MM. The corresponding
gene is human RPS27A, which is located on chromosome 2p16. RPS27A
transcript variant 1, RPS27A transcript variant 2 and RPS27A
transcript variant 3 are distinct transcripts of RPS27A, but they
encode the same protein (13,14).
Further exploration of the correlation between MMSA-8 and RPS27A
and of MMSA-8 expression patterns in MM thus became necessary.
Therefore, the full-length cDNA sequence of MMSA-8 was cloned and
its expression pattern was investigated in MM patients.
Materials and methods
Cell line culture
U266 cells were purchased from the Institute for
Cancer Research of the Medical School of Xi'an Jiaotong University
(China). The cells were cultured with RPMI-1640 medium (Gibco-BRL,
Life Technologies Ltd., Paisley, Scotland) that was supplemented
with 15% fetal bovine serum (FBS), 0.1 mg/ml streptomycin and 100
U/ml penicillin G and incubated at 37°C in a humidified atmosphere
with 5% CO2. Cells in a log phase of growth were used
for all experiments.
Patients
Fifteen healthy hematopoietic stem cell transplant
donors were obtained as controls. The donors and 85 MM patients
were enrolled in the study at the Department of Clinical Hematology
of the Second Affiliated Hospital, Medical School of Xi'an Jiaotong
University, between March 2011 and December 2014. Diagnoses were
based on the 2008 World Health Organization criteria. The
distribution of the clinical parameters and clinicopathological
features of the patients are displayed in Table I. Protocols and response definitions
were based on the 2011 MM Clinical Practice Guidelines of the
National Comprehensive Cancer Network (NCCN) Clinical Practice
Guidelines in Oncology. Patients received drugs, including Bz, Th,
dexamethasone (D), vincristine (V), adriamycin (A) and prednisolone
(P), in varying combinations. Thirty five patients used VAD (V + A
+ D) regimens, 30 patients used PD (Bz + D) regimens, 16 patients
used a Th-Bz combination and 4 patients used PAD (Bz + A + D)
regimens combination as induction therapy. Th (100–200 mg) was
taken orally each day during chemotherapy intervals. Most patients
received a Th-P combination as maintenance therapy. Therapeutic
responses were defined as complete remission (CR), partial
remission (PR), progression or relapse. Follow-ups were performed
on 68 MM patients, and the 17 MM patients were assessed at the
first time of diagnosis and relapse. The duration of the median
follow-up was 25.5 months, with a range of 5 to 45 months. All
collected samples had the written informed consent of the subjects.
The Ethics Committee of the School of Medicine of Xi'an Jiaotong
University approved the study and all procedures were conducted in
compliance with the Declaration of Helsinki.
 | Table I.The expression rates and expression
level of MMSA-8 with respect to the clinical characteristics of the
MM cases. |
Table I.
The expression rates and expression
level of MMSA-8 with respect to the clinical characteristics of the
MM cases.
|
|
| Expression rates of
MMSA-8 |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
|
Characteristics | Total, N=85 | Positive n=60 | Negative n=25 | P-value | Expression level of
MMSA-8 | P-value |
|---|
| Sex,
female/male | 37/48 | 28/32 | 9/16 |
0.473 | 40.05/50.68 | 0.601 |
| Age (years),
≥65/<65 | 20/65 | 14/46 | 6/9 |
1.000 | 54.60/43.42 | 0.569 |
| ISS stage |
| I | 23 | 9 | 14 |
0.008 |
3.26 | <0.001 |
| II | 39 | 29 | 10 |
0.042 | 39.90 | <0.001 |
|
III | 23 | 22 | 1 | <0.001 | 99.28 | <0.001 |
| Isotype |
|
|
|
0.578 |
|
|
|
igG | 42 | 31 | 11 |
| 48.40 | 0.899 |
|
igA | 26 | 18 | 8 |
| 47.54 |
|
|
igM | 11 | 7 | 4 |
| 38.46 |
|
|
igE | 1 | 1 | 0 |
| 50.10 |
|
| Light
chain | 5 | 3 | 2 |
| 34.52 |
|
| Bone lesions |
| 0 | 24 | 16 | 8 |
0.383 | 44.18 | 0.294 |
|
1–3 | 36 | 28 | 8 |
0.261 | 58.13 | 0.092 |
|
>3 | 25 | 16 | 9 |
1.000 | 30.45 | 0.412 |
| Renal failure |
|
(<2.0/≥2.0 Scr mg/dl) | 74/11 | 50/4 | 24/7 |
0.089 | 48.24/31.32 | 0.323 |
| Hb (>85/≤85
g/l) | 68/17 | 44/16 | 24/1 |
0.018 | 44.84/50.91 | 0.229 |
| Calcium (mg/dl)
(<12/≥12) | 69/16 | 46/14 | 25/2 |
0.133 | 47.04/41.79 | 0.609 |
| CRP (≤10/>10
mg/l) | 41/44 | 24/36 | 17/8 |
0.031 | 17.83/72.35 | <0.001 |
| LDH (<250/≥250
IU/l) | 44/41 | 23/37 | 18/4 | <0.001 | 15.43/78.91 | <0.001 |
| Cytogenetics |
|
Absent | 28 | 10 | 18 |
|
8.63 |
|
|
Hyperdiploid | 6 | 4 | 2 | 0.202 (vs.
absent) |
24.46 | 0.231 |
|
t(11;14) | 8 | 7 | 1 | 0.016 (vs.
absent) |
44.99 | 0.013 |
|
t(4;14) | 3 | 3 | 0 | 0.064 (vs.
absent) |
60.56 | 0.035 |
|
t(14;16) | 3 | 3 | 0 | 0.064 (vs.
absent) |
56.11 | 0.027 |
| del
(17p) | 10 | 9 | 1 | 0.008 (vs.
absent) |
72.81 | <0.001 |
| del
(13q) | 15 | 15 | 0 | <0.001(vs.
absent) |
93.06 | <0.001 |
| Double
abnormalities | 6 | 6 | 0 | 0.006 (vs.
absent) | 104.00 | <0.001 |
| Not
examined | 6 |
|
|
|
|
|
| p53 deletion |
|
Absent | 71 | 49 | 22 |
0.098 | 40.60 | 0.001 |
|
Present | 8 | 8 | 0 |
| 120.44 |
|
| Not
examined | 6 |
|
|
|
|
|
| Myeloma cells
(<10/≥10%) | 34/51 | 18/42 | 16/9 |
0.007 | 8.35/71.19 | <0.001 |
Characterization of the MMSA-8
transcript using 3′- and 5′-RACE
To characterize the intact MMSA-8 transcript, 3′-
and 5′- rapid amplification of cDNA ends (RACE) was performed using
the SMARTer™ RACE cDNA Amplification kit (Clontech Laboratories,
Inc., Mountain View, CA, USA) according to the manufacturer's
instructions. The existing MMSA-8 sequences were used as a guide to
design primers using Primer Premier 5.0 (12). Briefly, total RNA was isolated using
TRIzol (Gibco-BRL, NY, USA). Total RNA (1 µg) was used to generate
3′- or 5′-RACE-ready cDNA using special primers, and associated
reagents provided in the Moloney Murine Leukemia Virus (MMLV)
reverse transcriptase kit. The nested PCR reactions for 5′- and
3′-RACE began with primary PCR, consisting of incubation at 95°C
for 3 min before 33 cycles of denaturation at 94°C for 30 sec,
annealing at 58°C for 30 sec, an extension at 72°C for 60 sec and a
reconditioning step at 72°C for 7 min. Primary PCR was followed by
secondary PCR, consisting of the same process as the primary PCR.
The 3′- and 5′-RACE products were electrophoretically separated in
a 1% agarose gel with 1X TAE buffer. The resulting bands were
eluted with the Agarose Gel DNA Purification kit (Takara, Dalian,
China) and were then cloned into the pGEM-T Easy vector (Promega,
MI, USA). Four clone products from the same band were then
sequenced. The DNA sequencing service was executed at Sangon
Biological Engineering Technology and Service Co., Ltd. (Shanghai,
China). Its corresponding primer pairs are listed in Table II.
 | Table II.Primers used in the study. |
Table II.
Primers used in the study.
| Primer | Technique | Sequence 5′-3′ |
|---|
| RT primer |
|
TCTTGTGCTTATTCTTCTTGGGAGTGGTG |
| GSP1 | 5′-RACE |
TTCCTTATCCTGGATCTTGGCCTTTACAT |
| GSP2 | 5′-RACE |
AGAAAGAAGGTTAAGCTGGCTGTCCTGAA |
| GSP3 | 3′-RACE |
CTTGGCCTTTACATTTTCTATCGTATCCG |
| GSP4 | 3′-RACE |
GTGCTGGGGTGTTTATGGCAAGTCA |
| MMSA-8F | DNA-cloning |
ATGTGGTGCTGGGGTGTTTAT |
| MMSA-8R | DNA-cloning |
TTACTTGTCTTCTGGTTGGTGAAC |
| β-actinF | qPCR |
TAGTTGCGTTACACCCTTTCTTG |
| β-actinR | qPCR |
TCACCTTCACCGTTCCAGTTT |
| MMSA-8F | qPCR |
ACCCTCGGATACGATAGAAAATG |
| MMSA-8R | qPCR |
CACCACCACGAAGTCTCAACA |
Isolation and sequencing of MMSA-8
transcript
We used the sequences of the 3′- and 5′-RACE
products of MMSA-8 as a guide for designing primers to isolate the
full length sequence of MMSA-8. RT-PCR was performed with the High
Fidelity Prime Script™ RT-PCR kit (Takara). The product was
electrophoretically separated in a 1% agarose gel with 1X TAE
buffer. The resulting bands were cloned and the four clone products
from each of the bands were then sequenced as previously described.
Its corresponding primer pairs are listed in Table II.
Bioinformatics
Our newly cloned full-length sequences were then
analyzed using Basic Local Alignment Search Tool (BLAST) search
with GenBank at the NCBI. The potential coding sequences were
validated with the open reading frame (ORF) finder of the NCBI.
Real-time quantitative PCR assay
Leukocyte cells were isolated from bone marrow
samples using density gradient centrifugation with Ficoll-Hypaque.
Total RNA was extracted from 1×106 leukocytes using
TRIzol (Gibco-BRL, Gaithersburg, MD, USA). RNA quality was visually
assessed by confirmation of intact 28 and 18S ribosomal bands
following agarose gel electrophoresis and ethidium bromide
staining. cDNA was synthesized using the RevertAid First Strand
cDNA synthase kit (Fermentas, Lithuania) according to the
manufacturer's instructions. Primers for qRT-PCR were designed
using Primer Premier 5.0 software. Human β-actin primers were used
as an internal control. qRT-PCR was performed using the SYBR-Green
PCR kit (Takara, Japan) according to the manufacturer's
instructions on an ABI PRISM 7500 real-time PCR system. The
following thermal cycling conditions were used: 95°C for 3 min,
followed by 40 cycles of 7 sec at 95°C and 10 sec at 57°C. The
qRT-PCR reactions were performed in total volumes of 20 µl that
contained 2 µl of sample cDNA and 0.4 µM of each primer. The
resulting qRT-PCR products were 151 and 176 bp, respectively. The
comparative CT method was used, and the data are presented as
2−ΔΔCT (15). Its
corresponding primer pairs are listed in Table II.
Western blotting assay
Total protein was isolated and extracted with RIPA
buffer (Pierce, Rockford, IL, USA) for 15 min on ice. Protein
concentrations were detected using the BCA assay (Pierce). Blotted
with specific anti-MMSA-8 primary Ab [anti-RPS27A antibody ab111598
(Abcam, Cambridge, MA, USA)]. Incubated in secondary antibody and
then detected with enhanced chemiluminescence technique (Amersham
Pharmacia Biotech). The protein values of MMSA-8 and β-actin were
quantified by Quantity One 4.2.2 Software (Bio-Rad, Hercules, CA,
USA).
Statistical analysis
Mann-Whitney U tests and Fisher's exact test were
used. Spearman correlation was used to detect the relationship
between the expression value and levels of MMSA-8 and the clinical
parameters. The Kaplan-Meier method was used to calculate the
progression-free survival (PFS) and overall survival (OS).
Differences between survival curves were analyzed using the
log-rank test. With the Cox proportional hazards model, a
multivariate analysis of survival time was performed to identify
the adjusted impact of MMSA-8 overexpression on PFS and OS. The
SPSS software package (SPSS17.0) was used for evaluation of
statistical analyses and P<0.05 was considered as statistically
significant.
Results
Sequence analysis
The MMSA-8 sequence from the cDNA libraries may not
represent full-length and/or mature transcripts. Therefore, we
detected the 3′- and 5′-ends of MMSA-8 using SMART-RACE with the
U266 cell line. We then performed RT-PCR to clone and confirm the
new, full-length MMSA-8 cDNA sequence by primers that were designed
using the newly obtained 3′- and 5′-RACE sequences from the U266
cell line. We did not detect other variants of MMSA-8 in the U266
cells. Therefore, we identified a full-length MMSA-8 mRNA of 1,063
bp in the U266 cells via assembly of all the RACE and RT-PCR
clones. We analyzed our sequence using BLAST search on GenBank of
the NCBI. The corresponding mRNA was assigned as Homo sapien
ribosomal protein S27a (RPS27A), transcript variant 1 (NCBI
reference sequence: NM_002954.5). The cDNA sequence of MMSA-8 was
100% identical to RPS27A, transcript variant 1. The corresponding
gene was assigned as RPS27A [Homo sapien (human)] and gene
ID: 6233. These results confirmed that MMSA-8 is RPS27A-related
transcript variant 1.
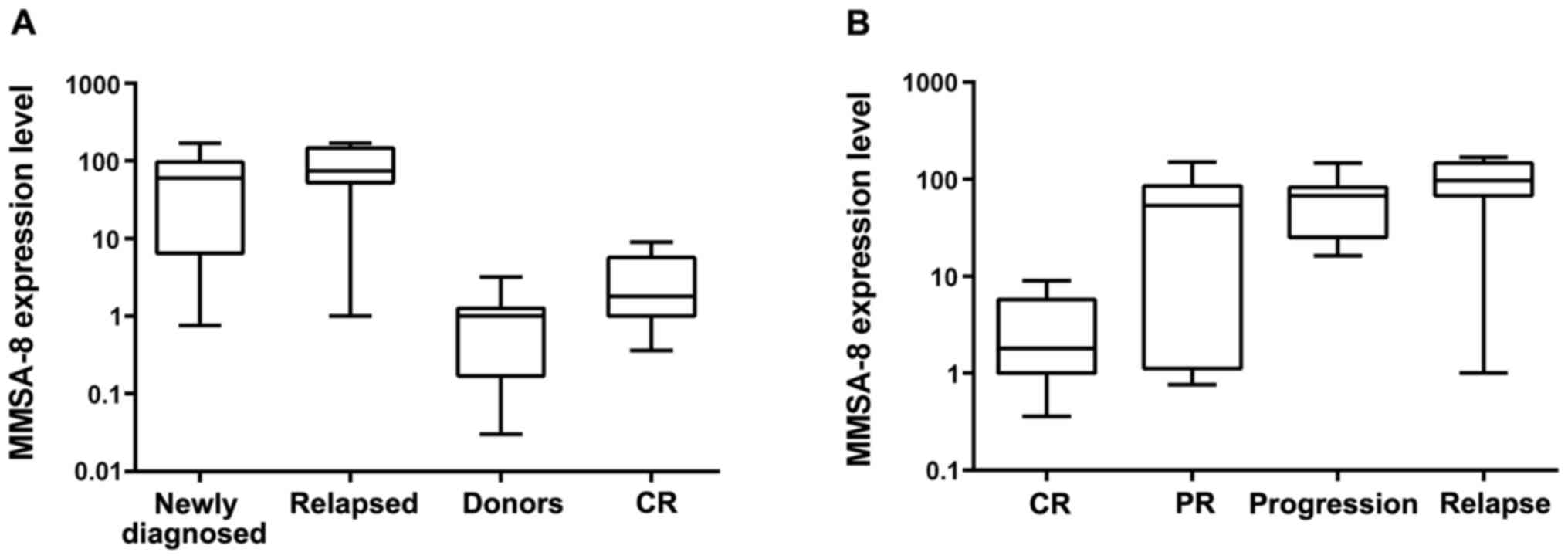
MMSA-8 expression in MM patients
MMSA-8 mRNA and protein levels were assessed in
healthy donors and in newly diagnosed, relapsed and CR groups using
qRT-PCR and western blotting. The mean relative value of MMSA-8
expressed in the control group was 9.10E-01. An MMSA-8 expression
value that was one log grade higher than the expression value in
the control group was defined as positive; lower values were
considered to be negative. MMSA-8 transcript levels revealed a
striking increase in patients with DP (i.e., newly diagnosed or
relapsed) relative to normal donors (P<0.001 for both groups;
Fig. 1A). Greater upregulation of
MMSA-8 mRNA expression was observed in relapsed patients relative
to the CR group (P<0.001, Fig.
1A). There was also a significant statistical difference
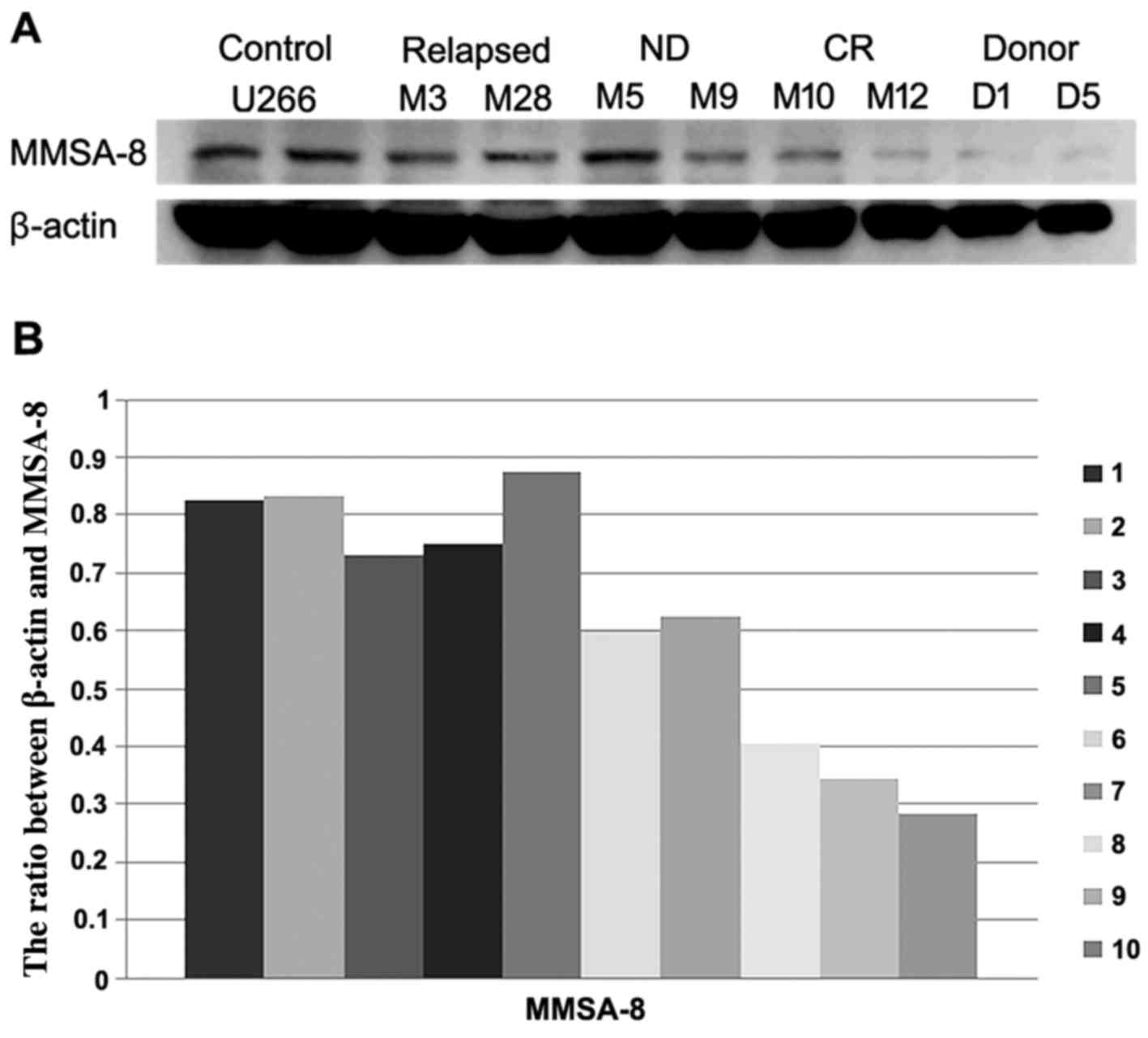
between healthy donors and the CR group (P=0.03; Fig. 1A). Western blot assay detected
MMSA-8 protein in most MM patient samples (Fig. 2). β-actin expression was used as a
loading control. A significant correlation was detected between
MMSA-8 protein levels and MMSA-8 mRNA expression levels (r=0.813,
P<0.001) (data not shown).
Association of MMSA-8 expression
levels with patient clinicopathological features
Table I presents the
clinicopathological characteristics of the MM patients at
diagnosis. The expression values of MMSA-8 were significantly
correlated with international staging system (ISS) stage, myeloma
cell burden, lactate dehydrogenase (LDH) and C-reactive protein
(CRP) levels (Table I). The
expression levels of MMSA-8 were higher in patients with
unfavorable cytogenetic abnormalities, including the presence of
t(11;14), t(4;14), t(14;16), del(17p), del(13q) and double
abnormalities; this correlation was statistically significant
(Table I). There were no
significant relationships between mRNA expression of MMSA-8 and
age, sex, immunoglobulin isotype, bone lesions, renal failure,
anemia, hypercalcemia or hyperdiploid abnormalities (Table I).
Association of MMSA-8 expression with
therapeutic response
The association of MMSA-8 expression with treatment
response was examined; treatment responses were classified as CR,
PR, progression or relapse. The mean MMSA-8 levels were 5.11E+01 in
patients with PR, 6.58E+01 in patients with DP and 9.93E+01 in
patients with relapse. The three groups exhibited much higher
MMSA-8 expression than the CR group (P=0.001, P<0.001 and
P<0.001, respectively; Fig. 1B).
A significant difference between the PR and relapse groups was also
found (P=0.004; Fig. 1B).
Association of MMSA-8 expression with
p53 deletion
Table I shows the
prevalence of the p53 deletion in MM patients, as assessed using
fluorescence in situ hybridization (FISH) at diagnosis. The
p53 deletion was detected in 8 of the 79 patients, and all 8 (100%)
of the p53-deletion patients were positive for MMSA-8 expression.
The positive MMSA-8 expression rates were lower in patients without
p53 deletion (69.01%), although this tendency was not statistically
significant (P=0.098). The level of MMSA-8 expression was higher in
groups with the p53 deletion than in groups lacking this genetic
aberration; this tendency was statistically significant (P=0.001;
Table I).
Correlation of MMSA-8 expression with
clinical outcome
The median duration of follow-up was 25.5 months,
with a range of 5 to 45 months. MM relapsed and progressed in 29
patients, and therapeutic mortality was observed in 25 patients.
The rates of OS and PFS were much lower in the MMSA-8-positive
group (25% OS and 0% PFS) than in the MMSA-8-negative group (60% OS
and 11.4% PFS, P<0.001 for both; Fig. 3). Patients in the MMSA-8-positive
group had a significantly shorter PFS (median 11.72 months) than
patients in the MMSA-8-negative group (median 28.53 months,
P<0.001; Fig. 3A). The OS was
significantly shorter (median 27.05 months) for patients in the
MMSA-8-positive group than for patients in the MMSA-8-negative
group (median 41.23 months, P<0.001; Fig. 3B). With univariate risk factor
analysis, including age, sex, ISS stage, immunoglobulin isotype,
bone lesions, renal failure, Hb, hypercalcemia, CRP, LDH, p53
deletion, myeloma cell burden, t(4;14), t(11;14), t(14;16),
del(17p), del(13q), double cytogenetic abnormalities and MMSA-8
expression level it was revealed that ISS stage, CRP, LDH, myeloma
cell burden, del(17P), del(13q), double cytogenetic abnormalities,
p53 deletion and MMSA-8 expression were significant (Table III). A multivariate analysis
confirmed myeloma cell burden, p53 deletion and MMSA-8 expression
level as risk factors for PFS and OS (Table IV).
 | Table III.Univariate analysis of PFS and OS in
MM patients. |
Table III.
Univariate analysis of PFS and OS in
MM patients.
|
|
P-valuea |
|---|
|
|
|
|---|
| Variable | PFS | OS |
|---|
| Sex |
|
Female/male |
0.735 |
0.957 |
| Age (years) |
| ≥65 vs.
<65 |
0.975 |
0.213 |
| ISS stage |
| I vs.
II and III | <0.001 | <0.001 |
| Immunoglobulin
isotype |
|
IgG/IgA/IgM/IgE/light chain
only |
0.665 |
0.905 |
| Bone lesions |
|
0/1–3/>3 |
0.579 |
0.584 |
| Renal failure (Scr
mg/dl) |
| <2.0
vs. ≥2.0 |
0.742 |
0.745 |
| Hb (g/l) |
| >85 vs. ≤85 |
0.499 |
0.253 |
| Hypercalcemia
calcium (mg/dl) |
| <12
vs. ≥12 |
0.494 |
0.864 |
| CRP (mg/l) |
| ≤10 vs.
>10 |
0.004 |
0.012 |
| LDH (IU/L) |
| <250
vs. ≥250 | <0.001 |
0.002 |
| Myeloma cell burden
(%) | <0.001 | <0.001 |
| t(11;14) |
| Absent
vs. present |
0.909 |
0.908 |
| t(14;16) |
| Absent
vs. present |
0.789 |
0.857 |
| t(4;14) |
| Absent
vs. present |
0.568 |
0.917 |
| Hyperdiploid |
| Absent
vs. present |
0.328 |
0.393 |
| Del(17p) |
| Absent
vs. present |
0.051 |
0.040 |
| Del(13q) |
| Absent
vs. present | <0.001 |
0.028 |
| Double
abnormalities |
| Absent
vs. present |
0.028 |
0.055 |
| p53 deletion |
| Absent
vs. present | <0.001 |
0.001 |
| MMSA-8 expression
level |
|
Negative vs. positive | <0.001 | <0.001 |
 | Table IV.Multivariate analysis of PFS and OS
in MM patients. |
Table IV.
Multivariate analysis of PFS and OS
in MM patients.
|
| PFS | OS |
|---|
|
|
|
|
|---|
| Variables | P-value | RR (95% CI) | P-value | RR (95% CI) |
|---|
| ISS stage | 0.051 |
| 0.054 |
|
| I vs.
II and III |
| CRP
(mg/l) |
| ≤10 vs.
>10 | 0.354 | 1.385
(0.696–2.758) | 0.274 | 0.479
(0.128–1.792) |
| LDH (IU/l) |
| <250
vs. ≥250 | 0.785 | 1.109
(0.529–2.323) | 0.357 | 0.546
(0.151–1.978) |
| Myeloma cell burden
(%) | 0.014 | 5.908
(1.437–24.291) | 0.007 | 31.102
(2.598–372.374) |
| Del(17p) |
| Absent
vs. present | 0.107 | 1.919
(0.869–4.241) | 0.200 | 2.288
(0.645–8.113) |
| Del(13q) |
| Absent
vs. present | 0.965 | 1.018
(0.452–2.294) | 0.115 | 0.275
(0.055–1.372) |
| Double
abnormalities |
| Absent
vs. present | 0.729 | 1.190
(0.445–3.180) | 0.056 | 5.761
(0.955–34.747) |
| P53 deletion |
| Absent
vs. present | 0.008 | 3.775
(1.410–10.102) | 0.001 | 12.851
(2.948–56.017) |
| MMSA-8 expression
level |
|
Negative vs. positive | <0.001 | 7.093
(2.887–17.427) | 0.033 | 8.406
(1.191–59.314) |
Discussion
MMSA-8 is an MM-associated antigen that was
identified by SEREX. The present study first determined the 3′- and
5′-ends of the MMSA-8 cDNA sequence in MM using SMART-RACE. The
MMSA-8 cDNA sequence was fully concordant with RPS27A transcript
variant 1(13,14). The RPS27A gene encodes a fusion
protein comprised of ubiquitin (Ub) at the N terminus and ribosomal
protein (RP) S27a at the C terminus. RPS27a is a ribosomal protein
with a Ub C-terminal extension. The Ub-RPS27a precursor protein is
rapidly processed by hydrolysis into an individual Ub monomer
(16–18). RPS27a may perform extra-ribosomal
functions in addition to its role in ribosome biogenesis and in
post-translational protein modification. RPS27a is overexpressed in
mouse liver cancer and in some human tumors (19–21).
The exact extra-ribosomal function of RPS27a is not clear.
Expression of RPS27a was increased in HMy2 cells, PRMI8226 cells
and U266 cells in the present study. RNA interference (RNAi)
technology demonstrated that MMSA-8 downregulation inhibited U266
cell growth, increased cell apoptosis and the number of cells in
the S phase. Our results suggest that MMSA-8 exerts a significant
role in the generation and progression of MM.
This study investigated MMSA-8 mRNA and protein
expression in MM patients and normal donors, and its relationship
with certain clinicopathological characteristics in prognosis. We
detected that the expression levels of MMSA-8 were significantly
upregulated in MM patients compared with those in normal donors and
that relapsed patients exhibited a greater upregulation of MMSA-8
expression compared to the CR group. These results confirmed our
previous hypothesis that MMSA-8 expression is closely related to
DP. The difference in the expression of MMSA-8 in various
characteristic groups indicated its unfavorable associated
clinicopathological characteristics. Age, ISS stage, bone lesions
and creatinine, LDH, CRP, Hb and calcium levels are independent
prognostic factors of MM, and the present study indicated that
patients in ISS stage III with higher LDH or CRP levels also had
higher MMSA-8 expression levels (22–25).
The MMSA-8 value was also higher in patients with unfavorable
cytogenetic abnormalities, including the presence of t(11;14),
t(4;14), t(14;16), del(17p), del(13q) and double abnormalities, and
this association was statistically significant (26–30).
However, there was no statistically significant association between
MMSA-8 expression and age, anemia or hypercalcemia, thereby
contradicting some previously published studies (31,32).
This discrepancy may be the result of the multiplicity and
heterogeneity of myeloma cells in our studies (33). Positive MMSA-8 expression was
associated with unfavorable clinical features at diagnosis. Strong
evidence confirms that MMSA-8 is an unfavorable prognostic factor
in MM. We also observed the lowest MMSA-8 mRNA expression level in
the CR group, compared to patients with PR and DP. Relapsed
patients exhibited the highest MMSA-8 expression, further
supporting the hypothesis that MMSA-8 is a strong prognostic factor
of response to therapy in MM patients.
Deletion of p53 was detected in 8 of the 79 patients
in our study and was associated with increased MMSA-8 mRNA levels,
suggesting that MMSA-8 is related to p53 deletion. p53 is a
tumor-suppressor gene that has been involved in the control of cell
proliferation, differentiation, invasion and apoptosis (34). p53 gene deletions in MM are
associated with poor patient survival (28,35–37).
As a direct transcriptional target of p53, the RPS27A gene is often
overexpressed in response to DNA damage (38). RPS27a also interacts with MDM2 to
suppress MDM2-associated p53 ubiquitination and then leading to the
activation of p53 and cell cycle arrest (39). RPS27a may perform extra-ribosomal
functions in addition to its role in ribosome biogenesis and
post-translational protein modification (40,41).
Previously published research support our observations and
conclusion, but the intrinsic mechanism involved in p53-RPS27a
interactions requires further investigation (39).
We also analyzed the correlation between MMSA-8
expression and clinical outcome. The results demonstrated that the
patients in the MMSA-8-positive group had lower rates of PFS and OS
than the MMSA-8-negative patients. Patients with positive MMSA-8
expression also had significantly shorter PFS and OS. Univariate
risk factor analysis found that ISS stage, CRP, LDH, myeloma cell
burden, del(17P), del(13q), double cytogenetic abnormalities, p53
deletion and MMSA-8 expression were significantly significant.
Multivariate analysis revealed that only the expression level of
MMSA-8, p53 deletion and myeloma cell burden were independent risk
factors in MM patients. These findings are consistent with
previously published studies (42–44).
However, we are the first to observe the relevance of the
expression level of MMSA-8 to patient survival.
In conclusion, the full-length cDNA sequence of
MMSA-8 was cloned in MM and it was hypothesized that MMSA-8 is
MM-associated RPS27A transcript variant 1. We firstly demonstrated
the specific expression of MMSA-8 in MM patients and the
association of its dysregulation with unfavorable clinical features
at diagnosis and poorer therapeutic outcome in MM. Univariate and
multivariate analyses revealed that MMSA-8 may be an independent
prognostic factor in MM. The present study supports further
investigation to ascertain MMSA-8 as a promising diagnostic marker
and therapeutic target in MM. Future studies should focus on its
pathophysiological relevance in myeloma cells, especially with
regard to its interaction with p53.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81172257).
References
|
1
|
Group IMW; International Myeloma Working
Group, : Criteria for the classification of monoclonal
gammopathies, multiple myeloma and related disorders: A report of
the International Myeloma Working Group. Br J Haematol.
121:749–757. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar SK, Rajkumar SV, Dispenzieri A, Lacy
MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust
JA, et al: Improved survival in multiple myeloma and the impact of
novel therapies. Blood. 111:2516–2520. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pulte D, Gondos A and Brenner H:
Improvement in survival of older adults with multiple myeloma:
Results of an updated period analysis of SEER data. Oncologist.
16:1600–1603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brenner H, Gondos A and Pulte D: Expected
long-term survival of patients diagnosed with multiple myeloma in
2006–2010. Haematologica. 94:270–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kumar SK, Lee JH, Lahuerta JJ, Morgan G,
Richardson PG, Crowley J, Haessler J, Feather J, Hoering A, Moreau
P, et al International Myeloma Working Group, : Risk of progression
and survival in multiple myeloma relapsing after therapy with IMiDs
and bortezomib: A multicenter international myeloma working group
study. Leukemia. 26:149–157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tai YT, Li X, Tong X, Santos D, Otsuki T,
Catley L, Tournilhac O, Podar K, Hideshima T, Schlossman R, et al:
Human anti-CD40 antagonist antibody triggers significant antitumor
activity against human multiple myeloma. Cancer Res. 65:5898–5906.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bae J, Prabhala R, Voskertchian A, Brown
A, Maguire C, Richardson P, Dranoff G, Anderson KC and Munshi NC: A
multiepitope of XBP1, CD138 and CS1 peptides induces
myeloma-specific cytotoxic T lymphocytes in T cells of smoldering
myeloma patients. Leukemia. 29:218–229. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosenblatt J, Avivi I, Vasir B, Uhl L,
Munshi NC, Katz T, Dey BR, Somaiya P, Mills H, Campigotto F, et al:
Vaccination with dendritic cell/tumor fusions following autologous
stem cell transplant induces immunologic and clinical responses in
multiple myeloma patients. Clin Cancer Res. 19:3640–3648. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong S, Li H, Qian J, Yang J, Lu Y and Yi
Q: Optimizing dendritic cell vaccine for immunotherapy in multiple
myeloma: Tumour lysates are more potent tumour antigens than
idiotype protein to promote anti-tumour immunity. Clin Exp Immunol.
170:167–177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Anderson LD Jr, Cook DR, Yamamoto TN,
Berger C, Maloney DG and Riddell SR: Identification of MAGE-C1
(CT-7) epitopes for T-cell therapy of multiple myeloma. Cancer
Immunol Immunother. 60:985–997. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Michalek J, Ocadlikova D, Matejkova E,
Foltankova V, Dudová S, Slaby O, Horvath R, Pour L and Hajek R:
Individual myeloma-specific T-cell clones eliminate tumour cells
and correlate with clinical outcomes in patients with multiple
myeloma. Br J Haematol. 148:859–867. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou FL, Zhang WG, Chen G, Zhao WH, Cao
XM, Chen YX, Tian W, Liu J and Liu SH: Serological identification
and bioinformatics analysis of immunogenic antigens in multiple
myeloma. Cancer Immunol Immunother. 55:910–917. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lamesch P, Li N, Milstein S, Fan C, Hao T,
Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al: hORFeome
v3.1: A resource of human open reading frames representing over
10,000 human genes. Genomics. 89:307–315. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gonzalez-Begne M, Lu B, Han X, Hagen FK,
Hand AR, Melvin JE and Yates JR: Proteomic analysis of human
parotid gland exosomes by multidimensional protein identification
technology (MudPIT). J Proteome Res. 8:1304–1314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Redman KL and Rechsteiner M:
Identification of the long ubiquitin extension as ribosomal protein
S27a. Nature. 338:438–440. 1989. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shabek N and Ciechanover A: Degradation of
ubiquitin: The fate of the cellular reaper. Cell Cycle. 9:523–530.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Komander D, Clague MJ and Urbé S: Breaking
the chains: Structure and function of the deubiquitinases. Nat Rev
Mol Cell Biol. 10:550–563. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adams SM, Sharp MG, Walker RA, Brammar WJ
and Varley JM: Differential expression of translation-associated
genes in benign and malignant human breast tumours. Br J Cancer.
65:65–71. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong JM, Mafune K, Yow H, Rivers EN,
Ravikumar TS, Steele GD Jr and Chen LB: Ubiquitin-ribosomal protein
S27a gene overexpressed in human colorectal carcinoma is an early
growth response gene. Cancer Res. 53:1916–1920. 1993.PubMed/NCBI
|
|
21
|
Kanayama H, Tanaka K, Aki M, Kagawa S,
Miyaji H, Satoh M, Okada F, Sato S, Shimbara N and Ichihara A:
Changes in expressions of proteasome and ubiquitin genes in human
renal cancer cells. Cancer Res. 51:6677–6685. 1991.PubMed/NCBI
|
|
22
|
Vincent Rajkumar S: Multiple myeloma: 2014
update on diagnosis, risk-stratification, and management. Am J
Hematol. 89:999–1009. 2014.PubMed/NCBI
|
|
23
|
Tarkun P, Atalay F, Atesoglu EB, Mehtap O,
Simsek M, Terzi E, Geduk A, Balli F, Batman A, Baydemir C, et al:
Treatment of patients with multiple myeloma over 65 yr: More
tolerability or better response? Eur J Haematol. 94:424–430. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chim CS, Sim J, Tam S, Tse E, Lie AK and
Kwong YL: LDH is an adverse prognostic factor independent of ISS in
transplant-eligible myeloma patients receiving bortezomib-based
induction regimens. Eur J Haematol. 94:330–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kiba T, Ito T, Nakashima T, Okikawa Y,
Kido M, Kimura A, Kameda K, Miyamae F, Tanaka S, Atsumi M, et al:
Bortezomib and dexamethasone for multiple myeloma: Higher AST and
LDH levels associated with a worse prognosis on overall survival.
BMC Cancer. 14:4622014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tricot G, Barlogie B, Jagannath S, Bracy
D, Mattox S, Vesole DH, Naucke S and Sawyer JR: Poor prognosis in
multiple myeloma is associated only with partial or complete
deletions of chromosome 13 or abnormalities involving 11q and not
with other karyotype abnormalities. Blood. 86:4250–4256.
1995.PubMed/NCBI
|
|
27
|
Fonseca R, Barlogie B, Bataille R, Bastard
C, Bergsagel PL, Chesi M, Davies FE, Drach J, Greipp PR, Kirsch IR,
et al: Genetics and cytogenetics of multiple myeloma: A workshop
report. Cancer Res. 64:1546–1558. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fonseca R, Blood E, Rue M, Harrington D,
Oken MM, Kyle RA, Dewald GW, van Ness B, van Wier SA, Henderson KJ,
et al: Clinical and biologic implications of recurrent genomic
aberrations in myeloma. Blood. 101:4569–4575. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Facon T, Avet-Loiseau H, Guillerm G,
Moreau P, Geneviève F, Zandecki M, Laï JL, Leleu X, Jouet JP,
Bauters F, et al: Intergroupe Francophone du Myélome: Chromosome 13
abnormalities identified by FISH analysis and serum
beta2-microglobulin produce a powerful myeloma staging system for
patients receiving high-dose therapy. Blood. 97:1566–1571. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oh S, Koo DH, Kwon MJ, Kim K, Suh C, Min
CK, Yoon SS, Shin HJ, Jo DY, Kwak JY, et al: Korean Multiple
Myeloma Working Party (KMMWP): Chromosome 13 deletion and
hypodiploidy on conventional cytogenetics are robust prognostic
factors in Korean multiple myeloma patients: Web-based multicenter
registry study. Ann Hematol. 93:1353–1361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kyrtsonis MC, Maltezas D, Tzenou T,
Koulieris E and Bradwell AR: Staging systems and prognostic factors
as a guide to therapeutic decisions in multiple myeloma. Semin
Hematol. 46:110–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Younes M, Hachfi H, Hammouda F, Younes K,
Ben Hammouda S, Jguirim M, Zrour S, Béjia I, Touzi M and Bergaoui
N: Survival prognosis factors in multiple myeloma. Tunis Med.
92:399–405. 2014.(In French). PubMed/NCBI
|
|
33
|
Lohr JG, Stojanov P, Carter SL,
Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B,
Gould J, Saksena G, et al: Multiple Myeloma Research Consortium:
Widespread genetic heterogeneity in multiple myeloma: Implications
for targeted therapy. Cancer Cell. 25:91–101. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Levine AJ: p53, the cellular gatekeeper
for growth and division. Cell. 88:323–331. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Drach J, Ackermann J, Fritz E, Krömer E,
Schuster R, Gisslinger H, DeSantis M, Zojer N, Fiegl M, Roka S, et
al: Presence of a p53 gene deletion in patients with multiple
myeloma predicts for short survival after conventional-dose
chemotherapy. Blood. 92:802–809. 1998.PubMed/NCBI
|
|
36
|
Königsberg R, Zojer N, Ackermann J, Krömer
E, Kittler H, Fritz E, Kaufmann H, Nösslinger T, Riedl L,
Gisslinger H, et al: Predictive role of interphase cytogenetics for
survival of patients with multiple myeloma. J Clin Oncol.
18:804–812. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chang H, Qi C, Yi QL, Reece D and Stewart
AK: p53 gene deletion detected by fluorescence in situ
hybridization is an adverse prognostic factor for patients with
multiple myeloma following autologous stem cell transplantation.
Blood. 105:358–360. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nosrati N, Kapoor NR and Kumar V: DNA
damage stress induces the expression of ribosomal protein S27a gene
in a p53-dependent manner. Gene. 559:44–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun XX, DeVine T, Challagundla KB and Dai
MS: Interplay between ribosomal protein S27a and MDM2 protein in
p53 activation in response to ribosomal stress. J Biol Chem.
286:22730–22741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Warner JR and McIntosh KB: How common are
extraribosomal functions of ribosomal proteins? Mol Cell. 34:3–11.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wool IG: Extraribosomal functions of
ribosomal proteins. Trends Biochem Sci. 21:164–165. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gertz MA, Lacy MQ, Dispenzieri A, Greipp
PR, Litzow MR, Henderson KJ, van Wier SA, Ahmann GJ and Fonseca R:
Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and
−17p13 in myeloma patients treated with high-dose therapy. Blood.
106:2837–2840. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
van de Donk NW and Sonneveld P: Diagnosis
and risk stratification in multiple myeloma. Hematol Oncol Clin
North Am. 28:791–813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mangiacavalli S, Pochintesta L, Cocito F,
Pompa A, Bernasconi P, Cazzola M and Corso A: Correlation between
burden of 17P13.1 alteration and rapid escape to plasma cell
leukaemia in multiple myeloma. Br J Haematol. 162:555–558. 2013.
View Article : Google Scholar : PubMed/NCBI
|