Introduction
Dysregulation of metabolic pathways, including
regulation of glucose transporter, tricarboxylic acid cycle (TCA
cycle), pentose phosphate pathway and mitochondria respiratory
chain are observed in many types of cancers (1). In addition, amino acid glycine, serine
and glutamine metabolic pathways play important roles in cancers
(2,3). Recent evidence indicates that
significant different lipid metabolites and expression of lipid
metabolic enzymes are detected in cancer. These lipid metabolites
are associated with cell proliferation, cellular membrane synthesis
and signaling molecules (4–6). Some of the dysregulated metabolic
enzymes, such as the glucose transporter 1 (GLUT1), hexokinase 2,
lactate dehydrogenase A, glutaminase and fatty acid synthase have
been demonstrated to be novel therapeutic targets of cancers
(7). The metabolomics in serum or
plasma will be novel diagnostic approach in clinic (8). Therefore, investigating the
correlation between metabolites, metabolic enzymes and cancer is a
critical issue.
The latest statistics reveal that breast cancer is
still one of the most common cancer types and leading cause of
cancer death (9). The molecular
subtypes of breast cancer could be divided into four types: luminal
A, luminal B, triple-negative/basal-like and HER2 type. Luminal
type tends to express estrogen receptor (ER), HER2 type is HER2
(human epidermal growth factor receptor 2) positive and
progesterone receptor (PR), ER and HER2 expression is negative in
triple-negative/basal-like type (10,11).
Various studies have shown that the expression of metabolic enzymes
is associated with ER, PR and HER2. Triple-negative breast cancer
cells express the highest level of GLUT1 compared to other types of
breast cancer cells (12).
Immunohistochemistry assay shows that HER2 positive and
triple-negative breast cancer cells express relatively high level
of glutamate-metabolic enzymes (13). The evidence suggests that the
expression of ER, PR and HER2 is associated with various metabolic
enzymes in breast cancer.
Most breast cells acquire fatty acids from
circulation system. However, breast cancer cells synthesize fatty
acids for structured lipid synthesis (7). Fatty acid synthase (FAS) is an
important enzyme in lipid synthesis pathway. High FAS expression is
usually observed in HER2-positive breast cancer and the
HER2-FAS-related signaling pathway might promote proliferation,
metastasis and chemotherapy resistance (14–16).
Blockage of FAS induces apoptosis in breast cancer cells (17). Combination of trastuzumab
(monoclonal antibody against HER2) and FAS inhibitor results in
re-sensitization with trastuzumab in the trastuzumab-resistant
breast cancer cells (18).
Synergistic therapeutic effect is observed after combination of FAS
inhibitors and other chemotherapies (19,20).
Therefore, blockage of lipid metabolic enzymes might be a novel
strategy for breast cancer treatment.
In mammalian cells, the conversion between
free-fatty acid and fatty acid-CoA are catalyzed by a fatty
acyl-CoA synthase (ACS) which is classified by catalyzing
substrates. A free-fatty acid containing 14–20 carbons is the
substrate of long-chain acyl-CoA synthetases (ACSL) (21). The five isoforms of ACSL include
ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6. All enzymes have individual
functions in substrate preference and tissue specificity (22). Based on the sequence homology, the
five ACSL isoforms are divided into two groups: one is composed of
ACSL1, ACSL5, ACSL6 and the other ACSL3 and ACSL4 (23). ACSL family convert long-chain fatty
acid to fatty-acid-CoA which is essential component for β-oxidation
which was suggested to promote oncogenesis in breast cancer
(24,25). A study indicates that the ER
expression level is negatively associated with ACSL4 expression
through 10 published mRNA array datasets in breast cancer cell
lines (26). In addition, ACSL4 is
considered a biomarker for breast cancer and is associated with
aggressive breast cancer type (27). However, the association between
other ACSL isoforms and molecular subtypes of breast cancers is
poorly known. In the present study, we aimed to investigate this
issue in each breast cancer subtype from gene expression datasets
and in breast cancer cell lines.
Materials and methods
Bioinformatics analysis: mRNA
expression levels
The clinical data of breast cancer samples and mRNA
expression levels of ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6 was
downloaded as Z-Scores from the cBioPortal (http://www.cbioportal.org, Breast cancer, Metabric,
Nature 2012 & Nat Commun 2016, 2509 samples, Version 1.3.3)
(28,29). The expression levels of ACSL
isoforms were analyzed through Oncomine Research Edition which
includes Kao cohort (30), Hatzis
cohort (31), Minn cohort (32), Miyake cohort (33), van de Vijver cohort (34), and Wang cohort (35) (Thermo Fisher Scientific; http://www.oncomine.org, v4.5) and the Cancer Cell
Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle/home)
(36). The heatmap was draw by
GENE-E software.
Assessment of the patient survival
rate
The survival analysis in breast cancer patients with
different expression levels of ACSL isoforms were performed through
the KM-Plotter database (37). The
prognostic value of each gene was analyzed by splitting patient
samples into two groups by median, after the subtype of breast
cancer was restricted to different ER status. The relapse-free
survival rate was analyzed (2016.10.13 update, the breast cancer
database includes 5,143 samples).
Cell culture
Normal breast epithelial cell line H184B5F5/M10 were
obtained from the Bioresource Collection and Research Center
(Hsinchu, Taiwan). Human breast cancer cell lines MCF-7 and
MDA-MB-231 were kindly provided by Professor Ming-Derg Lai in
National Cheng-Kung University (38). Cells were maintained in recommended
media (H184B5F5/M10 was in alpha-Minimum Essential Medium (MEM).
MCF-7 and MDA-MB-231 cells were in defined MEM (Lonza,
Walkersville, MD, USA). Both media were supplemented with 10% fetal
bovine serum (FBS) and penicillin/streptomycin (100 U/0.1 mg/ml)
(Life Technologies, Inc., Grand Island, NY, USA).
Quantitative PCR
Total RNA of MCF-7, MDA-MB-231 and H184B5F5/M10 was
extracted using TRIzol (Invitrogen, Carlsbad, CA, USA).
Complementary DNA was produced from 500 ng total RNA was using a
PrimeScript RT reagent kit (Clontech Laboratories, Inc., Kusatsu,
Japan). The levels of ACSL isoforms were determined on a Real-Time
PCR system (StepOne Plus Real-Time PCT System; Applied Biosystems,
Foster City, CA, USA) using Fast SYBR-Green Master Mix (Applied
Biosystems). The primers of ACSL isoforms were obtained from a
previous report (39) and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were
5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-TTGATTTTGGAGGGATCTCG-3′. Relative
mRNA expression levels of ACSL isoforms were normalized to the
expression level of GAPDH and calculated by 2−ΔΔCt
method.
Western blot analysis
Cells were lysed in RIPA lysis buffer (Millipore,
Billerica, MA, USA) and protein concentration was quantitated by
BCA protein assay kit (Millipore). Each protein was detected by
using primary antibody (anti-ACSL antibody, #4047; Cell Signalling
Technology, Danvers, MA, USA), anti-ACSL4 (ab155282; Abcam,
Cambridge, UK), anti-ACSL5 (ab57210; Abcam) and GAPDH (MAB374;
Millipore). The results were analyzed on an imaging capture system
(Alpha Innovation).
Evaluation of proliferation rate
For cell proliferation measurement, WST-1 (Clontech
Laboratories) was used and then 2×103 MCF-7, MDA-MB-231
and H184B5F5/M10 cells were seeded in 96-well plates with different
concentration of ACSL inhibitors including triacsin C (Abcam),
rosiglitazone (Sigma-Aldrich) and 2-fluoropalmitic acid (Cayman
Chemical, Ann Arbor, MI, USA) in 0.8% dimethyl sulfoxide (DMSO).
The proliferation rate was determined at wavelength 450 nm on a
microplate spectrophotometer (PowerWave X340; BioTek Instruments,
Inc., Winooski, VT, USA) after 48 h of treatments.
Statistical analysis
All graphs were generated by GraphPad Prism 7
(GraphPad Software, Inc., San Diego, CA, USA). Students t-test or
one-way ANOVA was used for analysis of difference between two
groups and three groups, respectively. P<0.05 was considered to
indicate a statistically significant difference.
Results
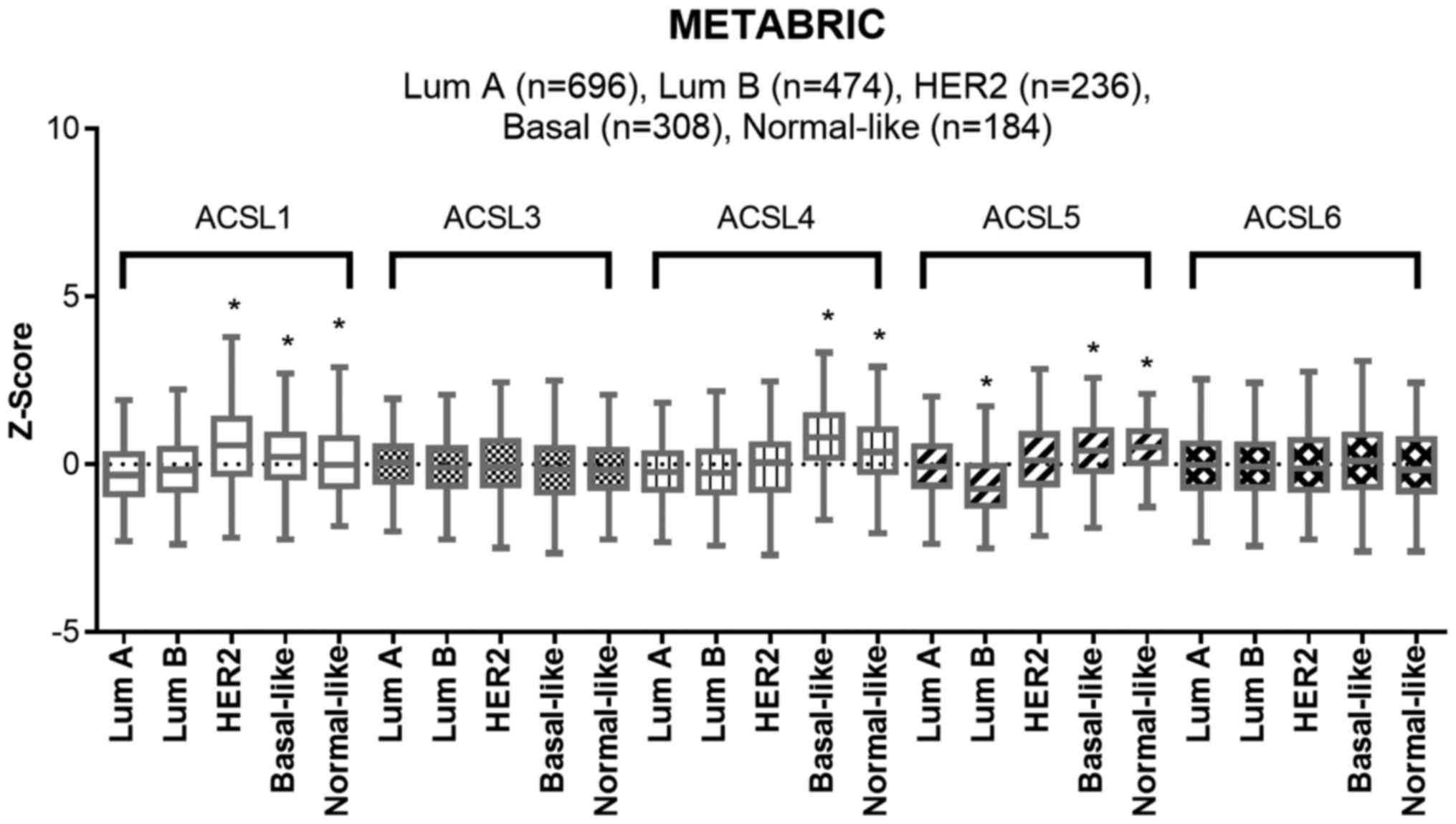
Low expression levels of ACSL1 and
ACSL5 is observed in luminal A subtype in the METABRIC dataset
We analyzed the expression levels of five ACSL
isoforms among five subtypes of breast cancer in METABRIC dataset
(Fig. 1). No significant difference
was detected in the expression levels of ACSL3 and ACSL6 among each
subtype of breast cancer. Compared to luminal A subtype, higher
mRNA levels of ACSL1, ACSL4 and ACSL5 were shown in basal-like and
normal-like subtypes. In addition, relatively high mRNA level of
ACSL1 was observed in HER2 subtype. Since high ER/PR expression and
low ER/PR expression is, respectively, a characteristic of luminal
A subtype and basal-like subtype, the results may imply that high
expression levels of ACSL1, ACSL4 and ACSL5 are associated with low
ER/PR expression. In addition, ACSL1 expression is associated with
HER2 expression.
The expression levels of ACSL1 and
ACSL5 is associated with ER and PR expression in breast cancer cell
lines
A previous report indicates that ACSL4 expression is
negatively associated with sex steroid hormone receptor in breast
cancer (26). To further
investigate the relationship between ACSL1, ACSL5 and ER/PR and
HER2 status, we analyzed it in human breast cancer cell lines. In
Fig. 2A, the mRNA expression of
ACSL1, ACSL4 and ACSL5 was analyzed through different probes in
several breast cancer cell lines within the Cancer Cell Line
Encyclopedia (CCLE) database. The status of ER and HER2 is based on
a previous report (40). The result
revealed that the lowest expression of ACSL1, ACSL4 and ACSL5 was
in MCF-7 cells (luminal A) compared to other cell lines. However,
the expression pattern of ACSL1, ACSL4 and ACSL5 is not associated
with the HER2 and basal-like subtypes. It suggests that all three
ACSL isoforms are associated with ER/PR. We further determine the
mRNA and protein expression levels of ACSL isoforms in MCF-7,
MDA-MB-231, and H184B5F5/M10 which is a normal breast epithelial
cell line. In Fig. 2B and C,
relatively high expression of ACSL isoforms was observed in
MDA-MB-231 cells. It might suggest that ACSL1, ACSL4 and ACSL5
expression is associated with ER/PR expression in breast cancer.
Notably, similar expression level of ACSL1 and ACSL4 was detected
between H184B5F5/M10 and MDA-MB-231 (Fig. 3C). The mRNA expression of ACSL5 in
H184B5F5/M10 was higher than MCF-7 (Fig. 3B).
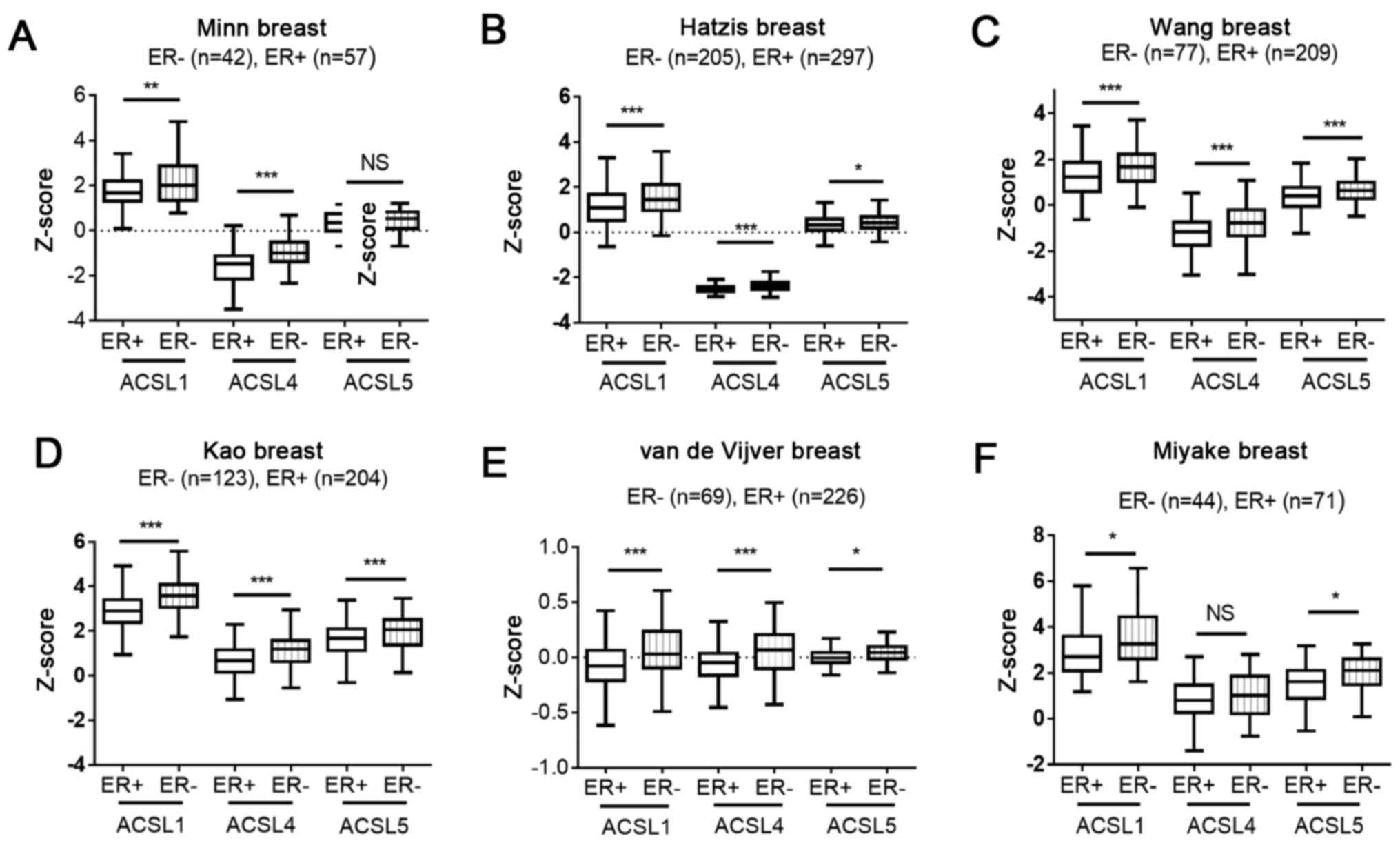
Investigating ACSL1 and ACSL5
expression in patients with different ER status within public
microarray datasets
To further determine whether ACSL1 and ACSL5
expression is associated with ER expression, we analyzed it within
six microarray datasets including Kao cohort (30), Hatzis cohort (31), Minn cohort (32), Miyake cohort (33), van de Vijver cohort (34) and Wang cohort (35). In Fig.
3A-F, ACSL1 levels in ER-negative group was higher than that in
ER-positive group. In addition, higher levels of ACSL4 and ACSL5
was respectively observed in Fig.
3A-E and 3B-F. The evidence
suggests the ER status is an important factor to regulate ACSL1,
ACSL4 and ACSL5 expression.
Investigation of ACSL1, ACSL4 and
ACSL5 as therapeutic targets of breast cancer
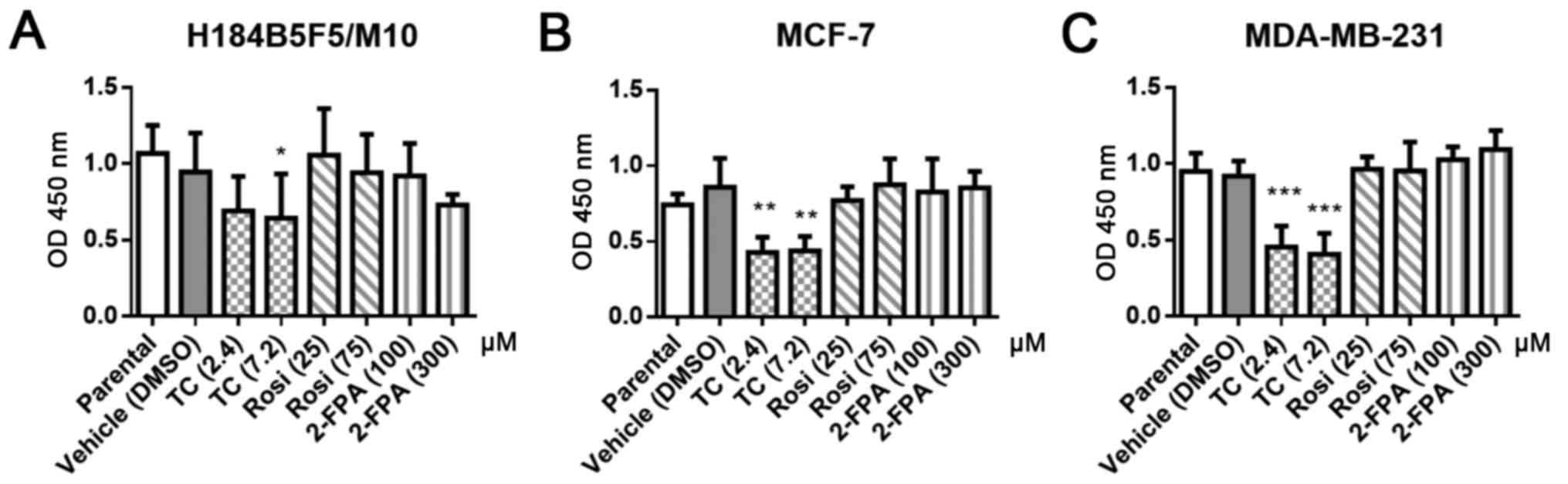
Previous reports indicate that inhibition of FAS is
a strategy to treat breast cancer (17–19).
We therefore investigated whether ACSL inhibitors resulted in
growth inhibition in the ER-positive cell line MCF-7, ER-negative
cell line MDA-MB-231 and the normal breast epithelial cell line
H184B5F5/M10. Three ACSL inhibitors were chosen. Triacsin C is an
analog of a polyunsaturated fatty acid and competitively inhibits
enzyme ACSL 1, 3, 4 and 5 (41,42).
2-Fluoropalmitic acid is an analog of palmitic acid and a
competitive inhibitor of ACSL (43). Rosiglitazone is an agonist of
peroxisome proliferator-activated receptor gamma (PPAR-γ) and
selectively suppresses ACSL4 activity over other ACSL isoforms
(44). Our results revealed that
2-fluoropalmitic acid and rosiglitazone did not affect cell growth
(Fig. 4). In contrast, the growth
of all three cell lines, including H184B5F5/M10, was inhibited by
triacsin C treatment. It might imply blockage of ACSL activity at
an appropriate concentration that may be a strategy to inhibit
breast cell growth.
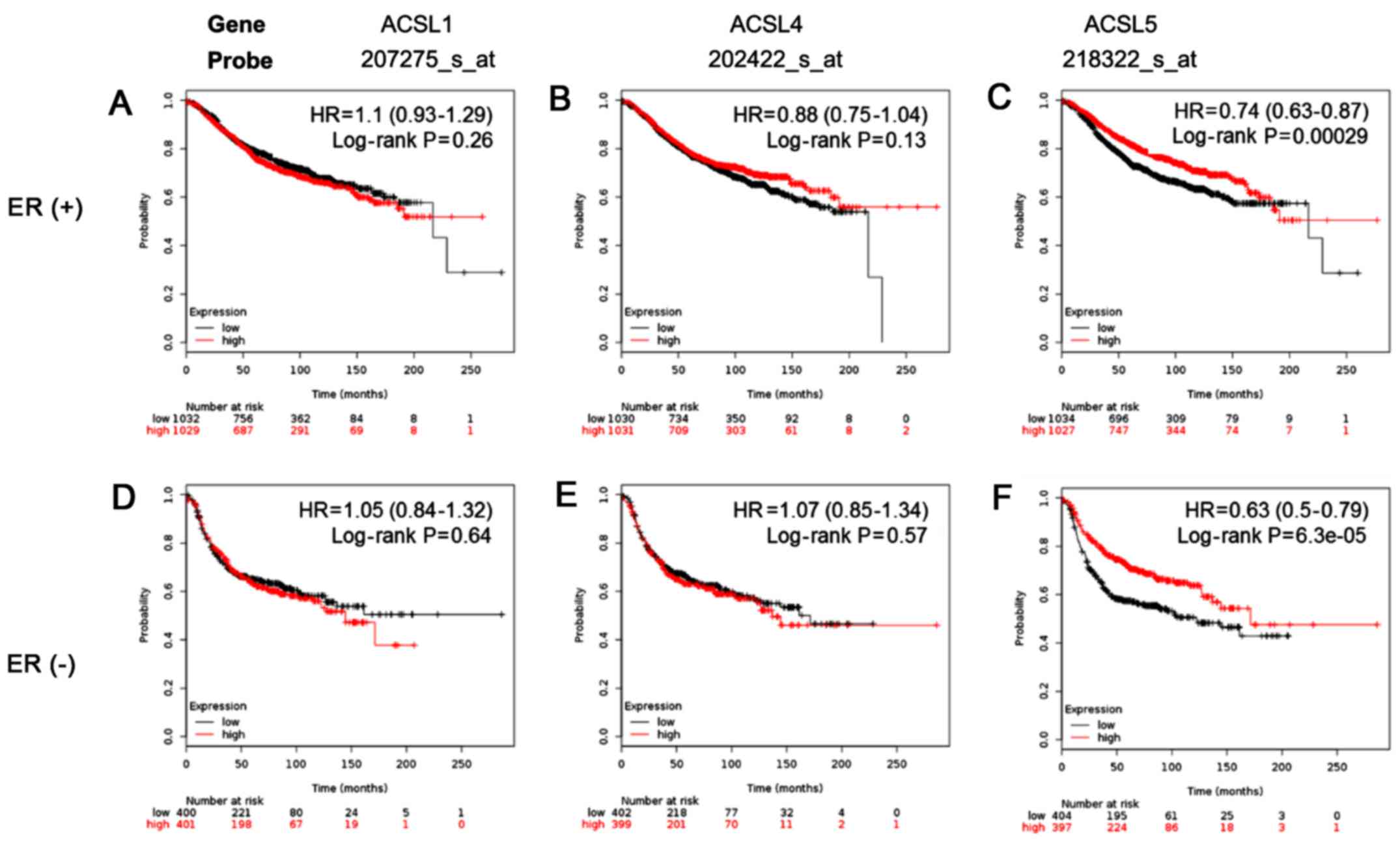
Investigation of ACSL1, ACSL4 and
ACSL5 could serve as markers for predicting the survival of
patients with breast cancer
Since high expression levels of ACSL1, ACSL4 and
ACSL5 was observed in ER-negative breast cancer patients, we
further investigated whether expression levels of ACSL1, ACSL4 and
ACSL5 were associated with survival of patients with different ER
status in breast cancer. Our results show that the expression of
ACSL1 and ACSL4 was not significantly associated with survival
rate. In contrast, ACSL5 was significantly associated with good
survival in all the patients (Fig.
5). It suggests ACSL5 is a potential novel biomarker for
predicting prognosis of breast cancer patients.
Discussion
Recently, a study which use a systematic analysis
through public microarray datasets indicated that different types
of ACSL isoforms reveal distinct function in different types of
cancers, including breast cancer (45). The present study shows that high
ACSL1 expression is correlated with poor overall survival rate and
high expression levels of ACSL4 and ACSL5 are correlated to good
overall survival rate in breast cancer (45). However, the function of ACSL
isoforms were not investigated in different subtypes of breast
cancer and the survival analysis was performed in a relatively
small cohort. In this study, we found that the ACSL1, ACSL4 and
ACSL5 expression was negatively associated with ER expression in
breast cancer patients in large cohorts. Similar expression pattern
was detected in breast cancer cell lines. Treatment of ACSL
inhibitor triacsin C inhibited cell proliferation in H184B5F5/M10,
MCF-7 and MDA-MB-231 cells. In addition, only ACSL5 could be a
potential marker for good survival of breast cancer patients.
The role of ACSL4 has been investigated in several
studies. A report indicates that overexpression of ACSL4 in MCF-7
which expresses low endogenous ACSL4 enhances the ability of cell
growth, invasion, anchorage-independent growth in vitro and
tumor growth in nude mice (27).
However, another study demonstrated that silencing ACSL4 expression
in MDA-MB-231 did not affect growth rate, but MDA-MB-231 cells
sensitize triacsin C treatment (26). In addition, low dose (<100 mM) of
ACSL4 inhibitor rosiglitazone did not significantly decrease cell
viability in MDA-MB-231 and in MDA-MB-231 xenograft model (46,47).
The growth inhibitory effect of high-dose rosiglitazone might be
through PPAR-γ but not ACSL4 pathway (44,47).
Similar results were observed in the present study. The evidence
suggests that ACSL4 is not a critical enzyme to increase cell
growth and viability in ER-negative breast cancer. On the other
hand, a recent study demonstrates that ACSL4-silencing breast
cancer cells resist ferroptosis and the w6 fatty acid acids are
enriched in cellular membrane under ferroptosis stimulation
(48). The mammalian target of
rapamycin (mTOR) signaling pathway is regulated by ACSL4 in breast
cancer cells (46). ACSL4 involves
in deiminase isoform 2-mediated oncogenic pathway in an ER-positive
MCF-7 breast cancer cell line (49). The physiological role and regulatory
mechanism of ACSL4 needs to be investigated in the future.
The role of ACSL1 and ACSL5 is little-known in
breast cancer. The ACSL1 and ACSL5 are in the same group and ACSL4
is another group based on their sequence homology (23). Previous studies have shown that
substrate preference of ACSL1 is unsaturated fatty acids oleate (18
carbons) and linoeate (18 carbons) and ACSL5 is palmitic acid (16
carbons), palmitoleic acid (16 carbons), oleic acid (18 carbons)
and linoleic acid (18 carbons). Besides, both enzymes are detected
in nucleus and mitochondria (50,51).
Although ACSL1 and ACSL5 have similar substrate preference and
subcellular location, only ACSL5 is associated with survival of
patients with ER-positive and ER-negative breast cancer. In the
present study, mRNA and protein expression of ACSL1, ACSL4 and
ACSL5 in H184B5F5/M10 was higher than that in MCF-7 cells (Fig. 2B and C). We therefore, suppose that
the ACSL5 function could be compensated in high ER expression
breast cancer, such as Luminal A subtype. Knockdown of ACSL5 in
hepatocytes decrease triglyceride synthesis (52). In addition, overexpression of ACSL5
induces neosynthesis of ceramide which is a signaling molecule in
the apoptosis pathway (53). The
correlation between ER signaling pathways and lipid metabolites
should be investigated in breast cancer in further studies.
Targeting fatty acid synthesis is a strategy for
cancer treatment. Blockage of FAS enzyme activity-mediated de
novo fatty acid synthesis shows antitumor potential in multiple
types of cancer (17–20). However, potential side-effects of
FAS inhibition is still a concern (54). In our results (Fig. 4) and a previous study (26), triacsin C is a relatively potent
inhibitor to induce apoptosis in breast cancer cells in comparison
with other inhibitors. Triacsin C but not 2-fluoropalmitic acid and
rosiglitazone inhibits de novo synthesis of triacylglycerol,
diacylglycerol and cholesterol esters and synthesis of phospholipid
(55). These results imply that
ACSL metabolic products within de novo synthesis pathway are
important for proliferation of breast cancer. Although ACSL
isoforms might serve as alternative cancer therapeutic targets in
process of de novo fatty acid synthesis, high-dose triacsin
C (7.2 mM) inhibits growth in normal breast cells (Fig. 4). Low-dose triacsin C might be
suitable for breast cancer treatment. However, current evidence
could not provide a specific ACSL isoform as the best target for
breast cancer treatment.
Estrogen affects reactive oxygen species production
in mitochondria in breast cancer (56). In addition, ERα and ERβ are found in
mitochondria and ERβ interacts with a mitochondrial protein HADHB
which is required for β-oxidation in breast cancer (57). β-oxidation is reported to promote
oncogenesis in breast cancer (24,25).
In addition, ACSL1, ACSL4 and ACSL5 are observed in mitochondria
and cytosol and the metabolic of ACSL family is essential for
β-oxidation. We suppose that the interaction of ACSL1, ACSL4, ACSL5
and ER in mitochondria might play an important role in development
of breast cancer.
In summary, our results have shown that the high
expression of ACSL1, ACSL4 and ACSL5 is associated with ER-negative
breast cancer. Inhibition of ACSL activity through low-dose
triacsin C might be a strategy to suppress growth in breast cancer
cell. Furthermore, our results suggest that high ACSL5 expression
is associated with good prognosis in patients with breast cancer
(Fig. 6).
Acknowledgements
The present study was supported by grants from the
Ministry of Science and Technology (MOST 105-2314-B-037-037-MY3;
MOST 104-2314-B-037-053-MY4; MOST 103-2320-B-037-006-MY3), the
‘KMU-KMUH Co-Project of Key Research’ (grant no. KMU-DK 105002 from
Kaohsiung Medical University) and the Kaohsiung Medical University
Hospital Research Foundation (KMUH104-4M24; KMUH104-4M56).
References
|
1
|
Chen JQ and Russo J: Dysregulation of
glucose transport, glycolysis, TCA cycle and glutaminolysis by
oncogenes and tumor suppressors in cancer cells. Biochim Biophys
Acta. 1826:370–384. 2012.PubMed/NCBI
|
|
2
|
Locasale JW: Serine, glycine and
one-carbon units: Cancer metabolism in full circle. Nat Rev Cancer.
13:572–583. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hensley CT, Wasti AT and DeBerardinis RJ:
Glutamine and cancer: Cell biology, physiology, and clinical
opportunities. J Clin Invest. 123:3678–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferro M, Terracciano D, Buonerba C,
Lucarelli G, Bottero D, Perdonà S, Autorino R, Serino A, Cantiello
F, Damiano R, et al: The emerging role of obesity, diet and lipid
metabolism in prostate cancer. Future Oncol. 13:285–293. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hilvo M, Denkert C, Lehtinen L, Müller B,
Brockmöller S, Seppänen-Laakso T, Budczies J, Bucher E, Yetukuri L,
Castillo S, et al: Novel theranostic opportunities offered by
characterization of altered membrane lipid metabolism in breast
cancer progression. Cancer Res. 71:3236–3245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Long JP, Li XN and Zhang F: Targeting
metabolism in breast cancer: How far we can go? World J Clin Oncol.
7:122–130. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Trezzi JP, Vlassis N and Hiller K: The
role of metabolomics in the study of cancer biomarkers and in the
development of diagnostic tools. Adv Exp Med Biol. 867:41–57. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: A population-based study from the California cancer
Registry. Cancer. 109:1721–1728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, et al: Immunohistochemical and clinical characterization of the
basal-like subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choi J, Jung WH and Koo JS:
Metabolism-related proteins are differentially expressed according
to the molecular subtype of invasive breast cancer defined by
surrogate immunohistochemistry. Pathobiology. 80:41–52. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim S, Kim DH, Jung WH and Koo JS:
Expression of glutamine metabolism-related proteins according to
molecular subtype of breast cancer. Endocr Relat Cancer.
20:339–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim S, Lee Y and Koo JS: Differential
expression of lipid metabolism-related proteins in different breast
cancer subtypes. PLoS One. 10:e01194732015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang YY, Kuhajda FP, Li J, Finch TT, Cheng
P, Koh C, Li T, Sokoll LJ and Chan DW: Fatty acid synthase as a
tumor marker: Its extracellular expression in human breast cancer.
J Exp Ther Oncol. 4:101–110. 2004.PubMed/NCBI
|
|
16
|
Vazquez-Martin A, Ortega-Delgado FJ,
Fernandez-Real JM and Menendez JA: The tyrosine kinase receptor
HER2 (erbB-2): From oncogenesis to adipogenesis. J Cell Biochem.
105:1147–1152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou W, Simpson PJ, McFadden JM, Townsend
CA, Medghalchi SM, Vadlamudi A, Pinn ML, Ronnett GV and Kuhajda FP:
Fatty acid synthase inhibition triggers apoptosis during S phase in
human cancer cells. Cancer Res. 63:7330–7337. 2003.PubMed/NCBI
|
|
18
|
Menendez JA, Vellon L, Mehmi I, Oza BP,
Ropero S, Colomer R and Lupu R: Inhibition of fatty acid synthase
(FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in
cancer cells. Proc Natl Acad Sci USA. 101:pp. 10715–10720. 2004;
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Menendez JA, Vellon L, Colomer R and Lupu
R: Pharmacological and small interference RNA-mediated inhibition
of breast cancer-associated fatty acid synthase (oncogenic
antigen-519) synergistically enhances Taxol (paclitaxel)-induced
cytotoxicity. Int J Cancer. 115:19–35. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vazquez-Martin A, Ropero S, Brunet J,
Colomer R and Menendez JA: Inhibition of fatty acid synthase (FASN)
synergistically enhances the efficacy of 5-fluorouracil in breast
carcinoma cells. Oncol Rep. 18:973–980. 2007.PubMed/NCBI
|
|
21
|
Soupene E and Kuypers FA: Mammalian
long-chain acyl-CoA synthetases. Exp Biol Med (Maywood).
233:507–521. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coleman RA and Lee DP: Enzymes of
triacylglycerol synthesis and their regulation. Prog Lipid Res.
43:134–176. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Soupene E, Fyrst H and Kuypers FA:
Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes.
Proc Natl Acad Sci USA. 105:pp. 88–93. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park JH, Vithayathil S, Kumar S, Sung PL,
Dobrolecki LE, Putluri V, Bhat VB, Bhowmik SK, Gupta V, Arora K, et
al: Fatty acid oxidation-driven Src links mitochondrial energy
reprogramming and oncogenic properties in triple-negative breast
cancer. Cell Rep. 14:2154–2165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pucci S, Zonetti MJ, Fisco T, Polidoro C,
Bocchinfuso G, Palleschi A, Novelli G, Spagnoli LG and Mazzarelli
P: Carnitine palmitoyl transferase-1A (CPT1A): A new tumor specific
target in human breast cancer. Oncotarget. 7:19982–19996.
2016.PubMed/NCBI
|
|
26
|
Monaco ME, Creighton CJ, Lee P, Zou X,
Topham MK and Stafforini DM: Expression of long-chain fatty
acyl-CoA synthetase 4 in Breast and prostate cancers is associated
with sex steroid hormone receptor negativity. Transl Oncol.
3:91–98. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu X, Li Y, Wang J, Wen X, Marcus MT,
Daniels G, Zhang DY, Ye F, Wang LH, Du X, et al: Long chain fatty
Acyl-CoA synthetase 4 is a biomarker for and mediator of hormone
resistance in human breast cancer. PLoS One. 8:e770602013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kao KJ, Chang KM, Hsu HC and Huang AT:
Correlation of microarray-based breast cancer molecular subtypes
and clinical outcomes: Implications for treatment optimization. BMC
Cancer. 11:1432011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Itoh M, Iwamoto T, Matsuoka J, Nogami T,
Motoki T, Shien T, Taira N, Niikura N, Hayashi N, Ohtani S, et al:
Estrogen receptor (ER) mRNA expression and molecular subtype
distribution in ER-negative/progesterone receptor-positive breast
cancers. Breast Cancer Res Treat. 143:403–409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu
W, Giri DD, Viale A, Olshen AB, Gerald WL and Massagué J: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miyake T, Nakayama T, Naoi Y, Yamamoto N,
Otani Y, Kim SJ, Shimazu K, Shimomura A, Maruyama N, Tamaki Y, et
al: GSTP1 expression predicts poor pathological complete response
to neoadjuvant chemotherapy in ER-negative breast cancer. Cancer
Sci. 103:913–920. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van t Veer LJ, Dai H, van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM,
Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME and
Yu J: Gene-expression profiles to predict distant metastasis of
lymph-node-negative primary breast cancer. Lancet. 365:671–679.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The Cancer Cell Line Encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cho CY, Lee KT, Chen WC, Wang CY, Chang
YS, Huang HL, Hsu HP, Yen MC, Lai MZ and Lai MD: MST3 promotes
proliferation and tumorigenicity through the VAV2/Rac1 signal axis
in breast cancer. Oncotarget. 7:14586–14604. 2016.PubMed/NCBI
|
|
39
|
Golej DL, Askari B, Kramer F, Barnhart S,
Vivekanandan-Giri A, Pennathur S and Bornfeldt KE, Barnhart S,
Vivekanandan-Giri A, Pennathur S and Bornfeldt KE: Long-chain
acyl-CoA synthetase 4 modulates prostaglandin E2 release from human
arterial smooth muscle cells. J Lipid Res. 52:782–793. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, et
al: The expression patterns of ER PR, HER2, CK5/6, EGFR, Ki-67 and
AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer (Auckl). 4:35–41. 2010.PubMed/NCBI
|
|
41
|
Kaemmerer E, Peuscher A, Reinartz A,
Liedtke C, Weiskirchen R, Kopitz J and Gassler N: Human intestinal
acyl-CoA synthetase 5 is sensitive to the inhibitor triacsin C.
World J Gastroenterol. 17:4883–4889. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vessey DA, Kelley M and Warren RS:
Characterization of triacsin C inhibition of short-, medium-, and
long-chain fatty acid: CoA ligases of human liver. J Biochem Mol
Toxicol. 18:100–106. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Soltysiak RM, Matsuura F, Bloomer D and
Sweeley CC: D,L-alpha-Fluoropalmitic acid inhibits sphingosine base
formation and accumulates in membrane lipids of cultured mammalian
cells. Biochim Biophys Acta. 792:214–226. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Askari B, Kanter JE, Sherrid AM, Golej DL,
Bender AT, Liu J, Hsueh WA, Beavo JA, Coleman RA and Bornfeldt KE:
Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid
partitioning to diacylglycerol and triacylglycerol via a peroxisome
proliferator-activated receptor-gamma-independent mechanism in
human arterial smooth muscle cells and macrophages. Diabetes.
56:1143–1152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen WC, Wang CY, Hung YH, Weng TY, Yen MC
and Lai MD: Systematic analysis of gene expression alterations and
clinical outcomes for long-chain acyl-coenzyme A synthetase family
in cancer. PLoS One. 11:e01556602016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Orlando UD, Castillo AF, Dattilo MA,
Solano AR, Maloberti PM and Podesta EJ: Acyl-CoA synthetase-4, a
new regulator of mTOR and a potential therapeutic target for
enhanced estrogen receptor function in receptor-positive and
-negative breast cancer. Oncotarget. 6:42632–42650. 2015.PubMed/NCBI
|
|
47
|
Mody M, Dharker N, Bloomston M, Wang PS,
Chou FS, Glickman TS, McCaffrey T, Yang Z, Pumfery A, Lee D, et al:
Rosiglitazone sensitizes MDA-MB-231 breast cancer cells to
anti-tumour effects of tumour necrosis factor-alpha, CH11 and
CYC202. Endocr Relat Cancer. 14:305–315. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Doll S, Proneth B, Tyurina YY, Panzilius
E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A,
et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular
lipid composition. Nat Chem Biol. 13:91–98. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Khurana P, Gokhale RS and Mohanty D:
Genome scale prediction of substrate specificity for acyl adenylate
superfamily of enzymes based on active site residue profiles. BMC
Bioinformatics. 11:572010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kanter JE, Tang C, Oram JF and Bornfeldt
KE: Acyl-CoA synthetase 1 is required for oleate and linoleate
mediated inhibition of cholesterol efflux through ATP-binding
cassette transporter A1 in macrophages. Biochim Biophys Acta.
1821:358–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lopes-Marques M, Cunha I, Reis-Henriques
MA, Santos MM and Castro LF: Diversity and history of the
long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates.
BMC Evol Biol. 13:2712013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bu SY and Mashek DG: Hepatic long-chain
acyl-CoA synthetase 5 mediates fatty acid channeling between
anabolic and catabolic pathways. J Lipid Res. 51:3270–3280. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gassler N, Roth W, Funke B, Schneider A,
Herzog F, Tischendorf JJ, Grund K, Penzel R, Bravo IG, Mariadason
J, et al: Regulation of enterocyte apoptosis by acyl-CoA synthetase
5 splicing. Gastroenterology. 133:587–598. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Röhrig F and Schulze A: The multifaceted
roles of fatty acid synthesis in cancer. Nat Rev Cancer.
16:732–749. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Igal RA, Wang P and Coleman RA: Triacsin C
blocks de novo synthesis of glycerolipids and cholesterol esters
but not recycling of fatty acid into phospholipid: Evidence for
functionally separate pools of acyl-CoA. Biochem J. 324:529–534.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sastre-Serra J, Nadal-Serrano M, Pons DG,
Valle A, Oliver J and Roca P: The effects of 17β-estradiol on
mitochondrial biogenesis and function in breast cancer cell lines
are dependent on the ERα/ERβ ratio. Cell Physiol Biochem.
29:261–268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhou Z, Zhou J and Du Y: Estrogen receptor
beta interacts and colocalizes with HADHB in mitochondria. Biochem
Biophys Res Commun. 427:305–308. 2012. View Article : Google Scholar : PubMed/NCBI
|