Introduction
Late diagnosis, frequent regional lymph node
metastases and the high rate of local and regional recurrence that
is caused mainly by the persistence of malignant cells, are the
major causes for the poor prognosis of oral squamous cell carcinoma
(OSCC) (1,2). Recurrence leads to a significantly
decreased life expectancy of the affected patients (3). Early detection of recurrence improves
patient survival (4–7).
TNM classification, histological grade and the depth
of tumor invasion are the best known prognostic factors for
recurrence, even if the real prognostic value of these clinical and
pathological features is still controversial (2,8).
Additionally, the state of the margins after tumor resection has
been investigated in order to predict recurrence. However, even
following wide tumor excision, a local recurrence of OSCC occurs in
up to 30% of OSCC patients due to residual malignant cells that
could not be microscopically detected in tumor margins at the time
of surgery (9).
To solve these shortcomings of the clinical
parameters, there is an urgent need to determine highly sensitive
and tumor-specific biomarkers for the stratification of patients
with high risk of disease recurrence and for long-term surveillance
of OSCC as well as for restaging procedures to exclude or confirm
tumor recurrence at stages that allow a successful therapeutic
intervention (10). Unfortunately,
intensive research spanning several decades has failed to identify
new prognostic molecular markers in oral cancer. To date, relevant
prognostic indicators with sufficient sensitivity and specificity
are nonexistent (11). Therefore,
the importance of developing useful diagnostic and monitoring tools
is emphasized in order to improve the clinical outcome of patients
suffering from OSCC.
MicroRNAs (miRNAs) are predicted to control ~50% of
all gene expression (12). Thus,
altered miRNA expression has been associated with several diseases
including the development of different types of cancers (13,14).
In addition, miRNAs are released from cells and enter the
circulation, where they are highly stable. They have been found in
several human body fluids, including plasma, serum, whole blood
cells and saliva (15). The
alterations in cancer cells are thought to be reflected in the
extracellular space as affected cells release upregulated miRNAs,
fail to release apparently downregulated species or export
selectively the type of miRNAs which suppress tumorigenesis.
Therefore, the detection of altered patterns of
circulating miRNAs in the blood has been proposed as cancer
biomarkers used in minimally invasive assays that may aid in risk
assessment, diagnosis, prognosis, detection of recurrence,
monitoring of the clinical course of the disease, and in predicting
or monitoring treatment response as well as in therapy decision
making in various types of cancer (11,16–24)
including OSCC (25–27). Ultimately, it has been proven that
alterations in the patterns of miRNAs predict recurrence risk and
are associated with therapeutic response (28–30).
Recently, we demonstrated that the miRNAs, miR-494, miR-3651 and
miR-186, are differentially expressed in whole blood of OSCC
patients when compared to that of healthy volunteers. The altered
expression rates were significantly correlated to diagnosis.
Additionally, overexpression of miR-3651 was associated to lymph
node metastases, clinical stage and to more dedifferentiated
tumors. Hence, these miRNAs may be useful in diagnostic and
prognostic applications (31,32).
Nevertheless, little is known concerning altered miRNA expression
in whole blood of recurrent OSCC patients (33–36).
The present study aimed to evaluate the difference
in circulating miRNA expression in whole blood samples of OSCC
patients suffering from recurrence compared to patients who were
disease-free for at least 1 year after treatment in order to
clarify their impact in the early detection of recurrent
disease.
Materials and methods
Patients and sample collection
The present study was approved by the Ethics
Committee of the University of Erlangen-Nuremberg (Erlangen,
Germany) (approval no. 3962) and patient written informed consent
was obtained.
Patients who suffered from a primary OSCC in the
past were included. They were clinically and histopathologically
considered to be free of tumor cells after surgical removal of the
tumor. The patients were followed up for 1 year at time-points of
1, 3, 6, 9 and 12 months after primary treatment in order to detect
recurrence of disease. Based on these observations 2 groups were
established. The R-OC group included 21 OSCC patients who suffered
from recurrence within 1 year. Blood samples were immediately
obtained after clinical observation of the occurrence of recurrent
disease. The 21 patients of the NR-OC group were free of disease
for at least 1 year after primary treatment. Fifty-four patients
with an initial diagnosis of OSCC and 33 healthy volunteers served
as control groups. Demographic information concerning the tumor,
control, R-OC and NR-OC cohorts is presented in Table I.
 | Table I.Demographic characteristics of the
test (OSCC patients), control (healthy volunteers) and follow-up
groups. |
Table I.
Demographic characteristics of the
test (OSCC patients), control (healthy volunteers) and follow-up
groups.
| Group | Test group | Control group | R-OC group | NR-OC group |
|---|
| Number of
cases | 54 | 33 | 21 | 21 |
| Mean age ± SD
(years) | 65.04±10.97 | 60.5±20.68 | 60.43±8.25 | 62.1±12.96 |
| Age range
(years) | 35–93 | 15–88 | 51–83 | 33–81 |
| Gender, n (%) |
|
Male | 37 (68.5) | 23 (69.7) | 16 (76.2) | 14 (66.7) |
|
Female | 17 (31.5) | 10 (30.3) | 5 (23.8) | 7 (33.3) |
Sampling of whole blood and miRNA
isolation
Two aliquots of whole blood (2.5 ml) for each
subject (tumor, R-OC, NR-OC and control group) were collected in a
PAXgene Blood RNA Tube (PreAnalyticX, Hombrechtikon, Switzerland)
and stored at −80°C until RNA isolation.
Whole RNA was extracted using the PAXgene Blood
miRNA kit as recommended by the manufacturer. The RNA concentration
was assessed using NanoDrop spectrometer (PeqLab, Erlangen,
Germany), and sufficient quality of the samples for RT-qPCR
analysis was confirmed using A260/A280 ratios. Subsequently the RNA
samples were stored at −80°C.
Real-time quantitative reverse
transcription-PCR (RT-qPCR)
The values of the miRNAs, miR-186, miR-494 and
miR-3651, were determined by RT-qPCR. These analyses were carried
out on 500 ng of total RNA. In the first step, miRNA was reverse
transcribed using the miScript II RT kit according to the
manufacturer's recommendations (Qiagen, Hilden, Germany). Detection
of amplification was carried out on 2.5 ng of cDNA using the
miScript SYBR-Green PCR kit and miRNA-specific quantitative RT-PCR
primer sets for the miRNA of interest (Qiagen) were determined on
an ABI-7300 Sequence Detection System (Applied Biosystems, Foster
City, USA). The features of the miRNAs are summarized in Table II.
 | Table II.List of miRNAs, snRNAs (endogenous
controls) and miScript Primer Assay used in the RT-qPCR
analysis. |
Table II.
List of miRNAs, snRNAs (endogenous
controls) and miScript Primer Assay used in the RT-qPCR
analysis.
| Sanger ID | Sanger accession
Mature miRNA | Sequence | Ref. no.
(Qiagen) |
|---|
| miRNAs |
|
hsa-miR-186-5p | MIMAT0000456 |
CAAAGAAUUCUCCUUUUGGGCU | MS 00008883 |
|
hsa-miR-3651 | MIMAT0018071 |
CAUAGCCCGGUCGCUGGUACAUGA | MS 00023121 |
|
hsa-miR-494-5pa | MIMAT0026607 |
AGGUUGUCCGUGUUGUCUUCUC | MSC 0002535 |
| Endogenous
controls | Ref. Seq |
|
|
|
RNU6-2 | NR_002752 |
| MS 00033740 |
|
SNORD44 | NR_002750 |
| MS 00007518 |
The values from the RT-qPCR analyses were normalized
by the ΔCT method using the primer sets RNU6-2 (U6 snRNA, RNA U6
small nuclear 2) and SNORD44 (small nucleolar RNA, C/D box 44) as
internal controls (Qiagen; Table
II). The mean value of both controls was applied as the
normalization value.
Relative quantification of differences in expression
(RQ) between the 2 groups was carried out by the ΔΔCT method using
the formula (RQ = 2−ΔΔCT) (37). A 2-fold change in miRNA expression
rate (0.5 ≤ RQ ≤2) between the 2 groups was defined as
relevant.
Statistical analysis
For statistical evaluation, IBM® SPSS
Statistics 21 (SPSS, Inc., Chicago, IL, USA) was used. The mean
value of duplicate ΔCT values for each sample was used for the data
results. Expression data were controlled for normal distribution
using the Shapiro-Wilk test. Graphical diagrams are plotted as
Box-Whisker plots which represent the median (ME), the
interquartile range (IQR) and the standard deviation (SD) as well
as the minimum and maximum values of the ΔCT values. Statistical
relevance of the apparent expression between the 2 groups was
analyzed using Mann-Whitney U test. P-values ≤0.05 were considered
as statistically significant.
Furthermore, the expression profile of each
differentially expressed miRNA was used for creation of receiver
operating characteristics (ROC) curves, and for estimation of the
area under the curve (AUC). This method displays the discriminatory
accuracy of the marker for distinguishing between the 2 groups of
blood donors.
Using ROC curves the highest Youden index (Y) was
calculated. The Youden index is associated with the critical
expression point or the optimal threshold value respectively [named
cut off-point (COP)] for the biological marker. The COP indicates
which value of increased or decreased expression is relevant for
the discrimination between malignancy and normal samples and allows
assigniment of a particular sample to a certain group.
Based on these COPs the 2 groups were divided into 2
subgroups which exhibited an expression rate over or under the COP.
Afterwards, association between altered miRNA expression and
malignancy, clinical features and histopathological parameters were
calculated using the Chi-square test.
Results
Demographic characteristics of the
study participants
Whole blood samples were collected from 54 OSCC
patients (tumor-group), 33 healthy volunteers (control group), 21
patients who relapsed after primary treatment within 1 year, and
from 21 patients who did not suffer from a recurrence for at least
1 year after primary treatment. The demographical characteristics
of all study participants are summarized in Table I. None of the healthy volunteers had
marked oral mucosal pathologies, such as inflammation, hyperplasia
or dysplasia. All groups matched in regards to gender and age.
There were no statistically relevant differences determined by the
Mann-Whitney U test. The R-OC and the NR-OC groups matched in
regards to gender and age (P=0.5; P=0.359), respectively.
RT-qPCR screening for miRNA expression
differences between all groups
A significantly different expression of all assessed
miRNAs could be ascertained between the OSCC and control group
(pmiR-186=0.012; pmiR-3651=0.001;
pmiR-494=0.007) and the NR-OC group
(pmiR-186=0.001; pmiR-3651=0.0001;
pmiR-494=0.039), respectively. In contrast there was no
significant difference in the expression values between the OSCC
and the R-OC group (pmiR-186=0.94;
pmiR-3651=0.89; pmiR-494=0.21). The
comparison of the expression values between the control and the
NR-OC group revealed no statistical relevant discrepancy
(pmiR-186=0.07; pmiR-3651=0.41;
pmiR-494=0.45), while the expression of the miRNAs
significantly varied when the control group was compared to the
patients who developed recurrence (R-OC group)
(pmiR-186=0.03; pmiR-3651=0.0001;
pmiR-494=0.003). A statistically significant difference
in expression rate could also be assigned when the recurrence group
(R-OC) was compared to the patients who did not develop a
recurrence (NR-OC group) within 1 year (pmiR-186=0.03;
pmiR-3651=0.001; pmiR-494=0.003). The results
of the statistical evaluation are summarized in Table III.
 | Table III.Statistical evaluation of the
comparison between all different [test group (OSCC patients),
control (healthy volunteers) and follow-up] groups. |
Table III.
Statistical evaluation of the
comparison between all different [test group (OSCC patients),
control (healthy volunteers) and follow-up] groups.
|
|
P-valuesa |
|---|
|
|
|
|---|
| Compared
groups | hsa-miR-186-5p | hsa-miR-3651 | hsa-miR-494-5p |
|---|
| Test group vs.
control group |
0.012 |
0.001 |
0.007 |
| Test group vs. R-OC
group | 0.94 | 0.89 | 0.21 |
| Test group vs.
NR-OC group |
0.001 |
0.0001 |
0.039 |
| Control group vs.
R-OC group | 0.03 |
0.0001 |
0.003 |
| Control group vs.
NR-OC group | 0.07 | 0.41 |
0.451 |
| R-OC group vs.
NR-OC group |
0.003 |
0.001 |
0.003 |
RT-qPCR screening for miRNA expression
between the R-OC and the NR-OC group
The impact of the differences in the expression
levels of the miRNAs for the discrimination between the NR-OC and
the R-OC groups was more accurately evaluated statistically in
order to assess their usefulness for the detection of recurrence
and the clinical monitoring of the disease. A significantly
different expression could be determined between the NR-OC and the
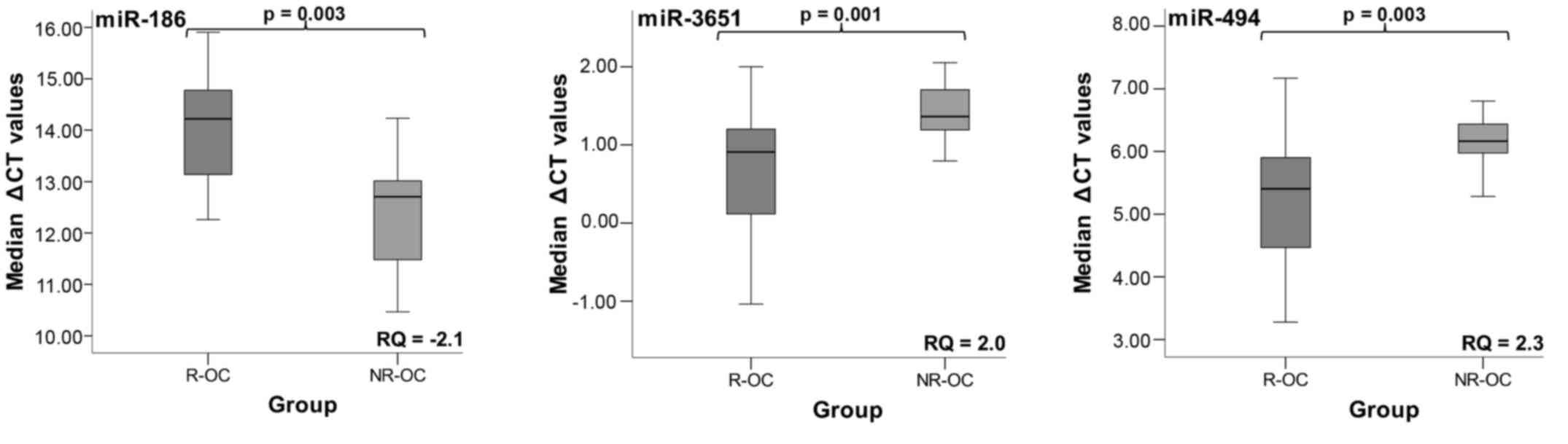
R-OC group for miR-186, miR-3651, and miR-494 (Table IV; Fig.
1). The results are graphically plotted as Box-Whisker plots.
Higher ΔCT values signify lower miRNA expression levels. RQ values
indicate the changes in the expression levels (Table IV; Fig.
1).
 | Table IV.Statistical evaluation of the
comparison of the R-OC (tumor patients suffering from recurrence
within 1 year) and the NR-OC group (recurrence-free patients) based
on the determined ΔCT values. |
Table IV.
Statistical evaluation of the
comparison of the R-OC (tumor patients suffering from recurrence
within 1 year) and the NR-OC group (recurrence-free patients) based
on the determined ΔCT values.
| miRNA | FC | AUC | Y | COP (ΔCT) |
P-valuea | Sensitivity
(%) | Specificity
(%) |
|---|
| hsa-miR-186-5p | −2.1 | 0.76 | 0.524 | 13.39 | 0.003 |
71,4 | 81 |
| hsa-miR-3651 |
2.0 | 0.80 | 0.525 |
1.16 | 0.001 | 81 |
71,4 |
| hsa-miR-494-5p |
2.3 | 0.78 | 0.476 |
5.87 | 0.003 |
71,4 |
76,2 |
miR-186 was significantly decreased in whole blood
when recurrence occurred (P=0.003). It was downregulated 2.1-fold.
The expression of the two other miRNAs was significantly increased.
The fold-change for miR-3651 was 2-fold whereas this value amounted
to 2.3 for miR-494. The increased expression was significantly
associated to recurrence (pmiR-3651=0.001;
pmiR-494=0.003). The results are presented in Fig. 1 and Table IV.
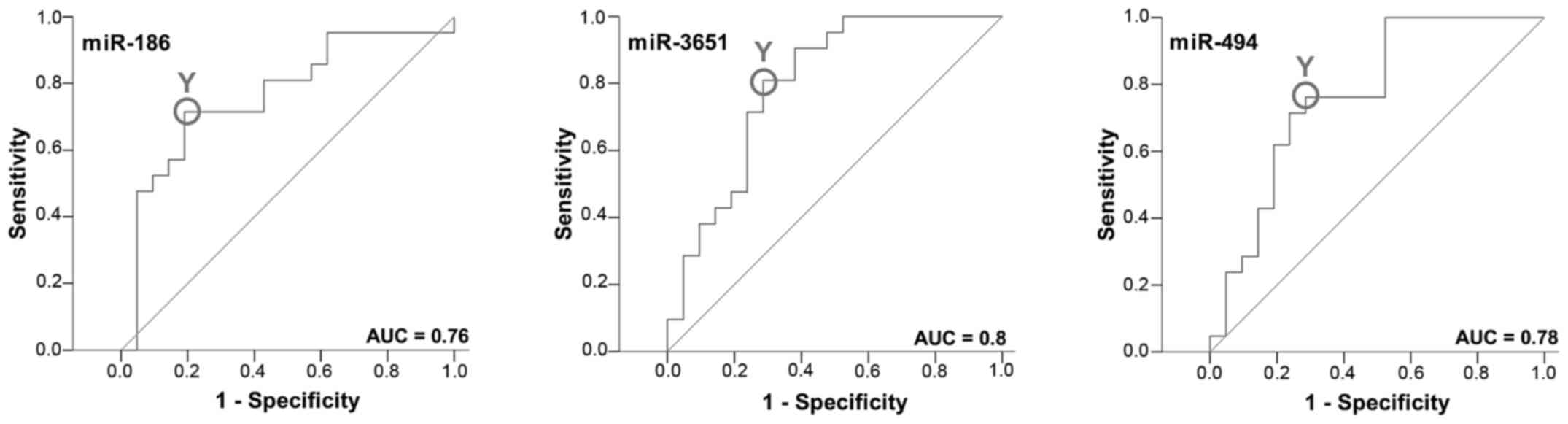
Between the R-OC and the NR-OC group an ROC curve
was established and the AUC was determined. All markers yielded a
significant AUC value. The upregulated miRNAs, miR-3651 and miR-494
yielded an AUC of 0.80 and 0.78, respectively. The AUC value of the
decreased miR-186 amounted to 0.76 (Fig. 2; Table
IV).
The highest Youden indices were 0.476 for miR-494,
0.525 for miR-3651 and 0.524 for miR-186 (Table IV). The optimal threshold
values/cut-off points (COP) expressed as a ΔCT value were 5.87 for
miR-494, 1.16 for miR-3651 and 13.39 for miR-186. For the miRNAs
miR-494 and miR-3651 a ΔCT under the COP (signifying upregulation)
was considered to be positive for the recurrence corresponding to
an increased abundance of the marker in whole blood. For miR-186 a
ΔCT value over the COP (signifying downregulation) was positive for
recurrence. Using the determined COPs the 2 groups were divided
into positive and negative specimens. The changes in the expression
rates of the miRNAs were statistically relevant and associated to
recurrence. The results are diagrammed in Fig. 3 and summarized in Table V.
 | Table V.Association between altered
expression rates of each individual miRNA and recurrence. |
Table V.
Association between altered
expression rates of each individual miRNA and recurrence.
|
| Positive n/% |
|
|---|
|
|
|
|
|---|
| miRNA | R-OC | NR-OC | P-value | Sensitivity | Specificity | Predictive value
(Positive/negative) |
|---|
| 186-5p | 15/71.4 |
3/14.3 | 0.001 |
0.714 |
0.857 |
0.83/0.75 |
| 3651 | 15/71.4 |
4/19.0 | 0.001 |
0.714 | 0.81 |
0.79/0.74 |
| 494-5p | 16/76.2 |
5/23.8 | 0.002 |
0.762 |
0.762 |
0.762/0.76 |
|
186/494a |
21/100 |
9/42.9 |
0.0001 | 1 |
0.571 | 0.7/1 |
|
186/3651a |
21/100 |
8/38.1 |
0.0001 | 1 |
0.619 |
0.81/1 |
|
494/3651a | 18/85.7 |
7/33.3 | 0.001 |
0.857 |
0.667 |
0.72/0.82 |
| Totalb |
21/100 | 11/52.4 |
0.0001 | 1 |
0.476 |
0.656/1 |
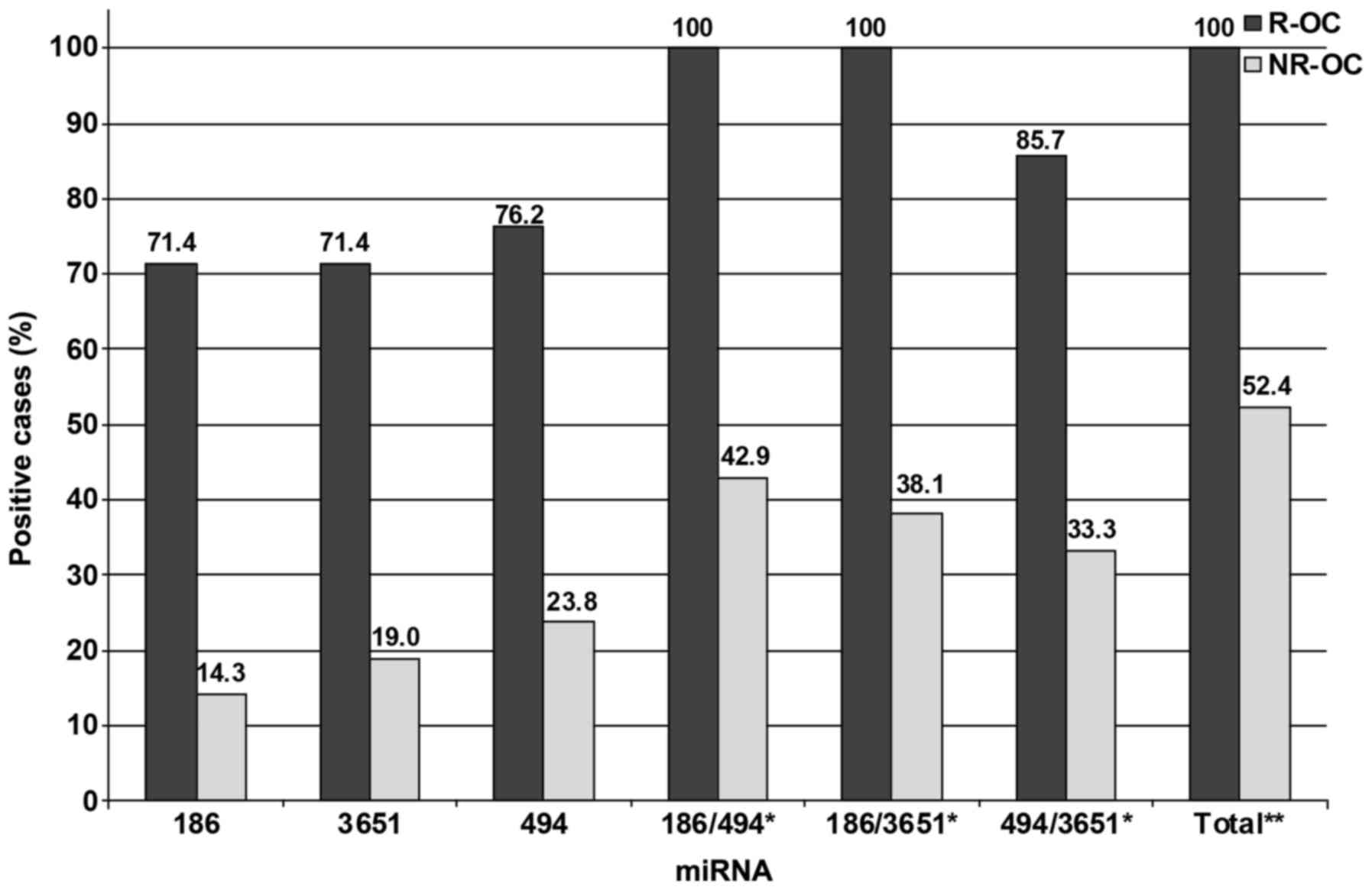
Out of the patients with recurrence 71.4% (15/21)
exhibited decreased levels of miR-186 and 71.4% (15/21) and 76.2%
(16/21) exhibited increased values of miR-3651 and miR-494,
respectively. Only 14.3% (3/21) of the whole blood samples from the
NR-OC group exhibited decreased levels of miR-186. Increased levels
in blood of patients suffering from NR-OC were revealed in 19.0%
(4/21) of the cases for miR-3651 and in 23.8% (5/21) for miR-494,
respectively. In order to ascertain the utility of the combination
of 2 miRNAs as prognostic tools 3 additional arrangements of the
investigated miRNAs were made. When a combination of
miR-186/miR-494 and miR-186/miR-3651 was assessed 100% of the R-OC
specimens exhibited positivity for altered expression, whereas only
42.9 and 38.1% of the NR-OC samples showed positivity. When, using
the combination miR-494/miR-3651 85.7% of the patients with
recurrence were positive, whereas only 33.3% of the patients with
no recurrence exhibited specific altered expression of 1 of these 2
markers. The correlation between recurrence and the detection of
altered expression rates was significant for all the investigated
miRNAs and their various combinations (P<0.01). Moreover, 100%
of the blood samples of patients suffering from recurrence
exhibited altered expression of at least 1 of the examined miRNAs
whereas in only 52.4% of the disease-free patients such an altered
abundance was evident. Thus, the association to recurrence was
statistically relevant (P=0.0001).
The sensitivity of miRNAs, miR-186, miR-3651 and
miR-494 amounted to 0.714, 0.714 and 0.762, respectively. The
values of the specificity of miR-186, miR-3651 and miR-494 were
0.857, 0.81 and 0.762. The analyzed combinations reached different
values for sensitivity: miR-186/miR-494, 1; miR-186/miR-3651, 1 and
miR-494/miR-3651, 0.857; and specificity: miR-186/miR-494, 0.571;
miR-186/miR-3651, 0.619 and miR-494/miR-3651, 0.667.
Discussion
Approximately 50% of patients with OSCC present with
metastatic disease or local recurrence at the time of initial
diagnosis or in the aftercare, leading to an unfavorable prognosis
(38). Therefore, early detection
of primary and/or recurrent OSCC by routine laboratory tests is
required (4,5,7).
Moreover, the monitoring of the response to successful therapy by
serial assessment of biomarkers, showing different pretreatment and
post-treatment levels is much-warranted (39). Tumor biomarkers in blood and saliva
may allow an earlier detection of the primary and/or recurrent
disease, and may serve as possible predictors of prognosis for
OSCC. Unfortunately, most of the identified markers lack the
required specificity and sensitivity. Therefore, the importance of
developing useful diagnostic and monitoring tools is emphasized in
order to increase initial recognition of patients with a higher
risk of recurrent disease and to improve long-term clinical
outcomes by allowing for a more aggressive treatment approach and
by carrying out a closer follow-up of these patients in the
aftercare (8).
miRNAs are important regulatory molecules and are
shown to be involved in disease pathogenesis. The perceived
opportunity for their use as clinical markers has loomed
particularly large in neoplastic disease, where alterations in
cancer cells are thought to be reflected in the extracellular space
as affected cells release upregulated miRNAs or fail to release
apparently downregulated species (18). Additionally, miRNAs are very stable
and can therefore be detected in several body fluids including
serum, plasma, saliva and whole blood. Several studies have
demonstrated the usefulness of circulating miRNAs as potential
biomarkers that may aid in risk assessment, diagnosis, prognosis,
and monitoring of disease and of treatment response for OSCC
(25–27,33,34,40–47).
In the present study the hypothesis that
differential expression of defined miRNAs in whole blood samples of
OSCC patients could be associated with recurrence of OSCC and can
therefore predict recurrent disease and provide clinically relevant
prognostic information for OSCC patients in a postoperative setting
was assessed.
In previous studies, we identified 3 differentially
expressed miRNAs in whole blood of OSCC patients when compared to
that of healthy volunteers (31,32).
The miR-494 and miR-3651 concentrations in OSCC patients were
increased, whereas miR-186 exhibited decreased abundance. The
altered expression rates of these miRNAs were significantly
associated to diagnosis. Additionally, overexpression of miR-3651
was significantly correlated to lymph node metastasis, clinical
stage and to more dedifferentiated tumors. Thus, it was postulated
that these miRNAs may be useful in prognostic applications and may
serve as the basis for establishing a minimally invasive method for
the detection and monitoring of OSCC (31,32).
In the present study, we confirmed the significant
changes in the expression of the investigated miRNAs in OSCC
patients compared to healthy controls. The diagnosis was
significantly correlated to the detection of altered expression
rates of all examined miRNAs. In addition, the changes in miRNA
concentrations were significantly different between healthy people
and patients with recurrence (R-OC), as well as in OSCC patients
compared to patients with no recurrence (NR-OC), respectively. In
contrast, these miRNAs were not differentially expressed in whole
blood of volunteers when compared to patients with no recurrence or
in whole blood of patients suffering from a primary OSCC compared
to test subjects who relapsed. These results strengthen the
hypothesis that the occurrence of primary as well as of recurrent
malignancies can be diagnosed by the expression analysis of these
miRNAs in whole blood.
It has already been demonstrated that increased
postsurgical concentrations of different miRNAs in plasma, serum
and saliva samples of OSCC patients were associated with recurrence
and predicted worse clinical outcome (46–49).
In the present study, we revealed a significantly different
expression of miR-186, miR-3651 and miR-494 in whole blood between
the group of patients with recurrence and those with no recurrence
that allows for the discrimination between the 2 groups. Moreover,
taking into account the COPs the presence of upregulated miR-494
and miR-3651 and downregulated miR-186 in individual pro-bands was
significantly correlated with the absence and presence of the
recurrence of the disease. Moreover, the markers showed high
sensitivity and specificity, and promising positive or negative
predictive values. Thus, it appears that these 3 miRNAs are
relevant biomarkers in the differentiation of patients with
recurrence to those with no recurrence. Thus, they could be useful
for the establishment of a minimally invasive method based on blood
assessment that may provide an early indication of the existence of
persistent or recurrent OSCC in an individual patient. Furthermore,
it was also revealed that all patients with recurrence were
positive for at least 1 differentially expressed miRNA. Thus the
sensitivity of this multimarker combination was excellent. However,
52.4% of the patients with no recurrence were determined as
positive. This led to a low degree of specificity. Additionally,
the paired combination of the 2 markers resulted in a higher
sensitivity of the test, but in contrast in a concomitant loss of
specificity. Therefore, in the future it may be useful to explore
more than 1 biomarker in order to enhance the diagnostic accuracy
(8,50). However, by establishing combinations
of markers both the sensitivity and specificity of the test may be
investigated and the ratio between these 2 values may be taken into
consideration in order to achieve methods with optimal sensitivity
as well as the highest possible specificity.
Besides the detection of recurrent disease, many
issues could be addressed toward clinical application. For several
tumors including OSCC numerous potential miRNAs as valuable
biomarkers in predicting the behavior of individual cancers and
monitoring therapeutic responses could be displayed. Studies on
various tumors evaluating blood miRNAs as biomarkers for OSCC
management have been carried out (27,34,43–46,51,52).
Moreover, these identified markers may also be applied in the risk
assessment of precancerous lesions (53). These studies led to the formulation
of the phrase ‘liquid biopsy’ that may allow for the early warning
of oncogenesis, relapse and treatment failure (18). Consequently, our results may
encourage the expansion of studies on the evaluation of blood
miRNAs as biomarkers for OSCC management and monitoring of the
disease. However, further validation in a larger cohort is
warranted to fully assess the utility of particular miRNAs as OSCC
biomarkers. Additionally, further investigations are warranted for
the screening of other miRNAs for clinical use which have already
been shown to be valuable tools for these clinical applications.
Moreover, their impact in clinical monitoring and prognosis has to
be evaluated by particular prospective follow-up studies. Finally
in order to translate the promising results into clinical practice
these markers have to be validated in well-designed and
sufficiently powered multi-centre studies.
In conclusion, altered levels of miR-494, miR-3651
and miR-186 which are differentially expressed in primary OSCC
patients were also observed upon disease relapse. Thus, the present
study clearly demonstrated the usefulness of circulating miRNAs in
the screening of OSCC patients and for the early detection of
recurrent and persistent disease. This may provide us with the
possibility of setting up a minimally invasive method for OSCC
patient stratification according to the risk of recurrence,
clinical monitoring and therapeutic response in order to optimize
the treatment of patients and to improve tumor outcome.
Acknowledgements
The authors would like to thank Mrs. S. Schönherr
and Mrs. E. Diebel for their valuable technical support.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yanamoto S, Yamada S, Takahashi H,
Yoshitomi I, Kawasaki G, Ikeda H, Minamizato T, Shiraishi T, Fujita
S, Ikeda T, et al: Clinicopathological risk factors for local
recurrence in oral squamous cell carcinoma. Int J Oral Maxillofac
Surg. 41:1195–1200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang B, Zhang S, Yue K and Wang XD: The
recurrence and survival of oral squamous cell carcinoma: A report
of 275 cases. Chin J Cancer. 32:614–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Agra IM, Carvalho AL, Pinto CA, Martins
EP, Filho JG, Soares FA and Kowalski LP: Biological markers and
prognosis in recurrent oral cancer after salvage surgery. Arch
Otolaryngol Head Neck Surg. 134:743–749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalavrezos N and Bhandari R: Current
trends and future perspectives in the surgical management of oral
cancer. Oral Oncol. 46:429–432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ord RA, Kolokythas A and Reynolds MA:
Surgical salvage for local and regional recurrence in oral cancer.
J Oral Maxillofac Surg. 64:1409–1414. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schwartz GJ, Mehta RH, Wenig BL, Shaligram
C and Portugal LG: Salvage treatment for recurrent squamous cell
carcinoma of the oral cavity. Head Neck. 22:34–41. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Søland TM and Brusevold IJ: Prognostic
molecular markers in cancer - quo vadis? Histopathology.
63:297–308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang TY, Hsu LP, Wen YH, Huang TT, Chou
YF, Lee CF, Yang MC, Chang YK and Chen PR: Predictors of
locoregional recurrence in early stage oral cavity cancer with free
surgical margins. Oral Oncol. 46:49–55. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chauhan SS, Kaur J, Kumar M, Matta A,
Srivastava G, Alyass A, Assi J, Leong I, MacMillan C, Witterick I,
et al: Prediction of recurrence-free survival using a protein
expression-based risk classifier for head and neck cancer.
Oncogenesis. 4:e1472015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Komatsu S, Ichikawa D, Hirajima S,
Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H,
Shiozaki A, et al: Plasma microRNA profiles: Identification of
miR-25 as a novel diagnostic and monitoring biomarker in
oesophageal squamous cell carcinoma. Br J Cancer. 111:1614–1624.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cortez MA and Calin GA: MicroRNA
identification in plasma and serum: A new tool to diagnose and
monitor diseases. Expert Opin Biol Ther. 9:703–711. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Momen-Heravi F, Trachtenberg AJ, Kuo WP
and Cheng YS: Genomewide study of salivary microRNAs for detection
of oral cancer. J Dent Res. 93 Suppl 7:86S–93S. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA. 105:pp.
10513–10518. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Witwer KW: Circulating microRNA biomarker
studies: Pitfalls and potential solutions. Clin Chem. 61:56–63.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cappelletti V, Appierto V, Tiberio P, Fina
E, Callari M and Daidone MG: Circulating biomarkers for prediction
of treatment response. J Natl Cancer Inst Monogr. 2015:60–63. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tiberio P, Callari M, Angeloni V, Daidone
MG and Appierto V: Challenges in using circulating miRNAs as cancer
biomarkers. BioMed Res Int. 2015:7314792015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Allegra A, Alonci A, Campo S, Penna G,
Petrungaro A, Gerace D and Musolino C: Circulating microRNAs: New
biomarkers in diagnosis, prognosis and treatment of cancer
(Review). Int J Oncol. 41:1897–1912. 2012.PubMed/NCBI
|
|
23
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gomes CC, de Sousa SF and Gomez RS:
MicroRNAs: Small molecules with a potentially role in oral squamous
cell carcinoma. Curr Pharm Des. 19:1285–1291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu BH, Xiong XP, Jia J and Zhang WF:
MicroRNAs: New actors in the oral cancer scene. Oral Oncol.
47:314–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Soga D, Yoshiba S, Shiogama S, Miyazaki H,
Kondo S and Shintani S: microRNA expression profiles in oral
squamous cell carcinoma. Oncol Rep. 30:579–583. 2013.PubMed/NCBI
|
|
28
|
Zahran F, Ghalwash D, Shaker O, Al-Johani
K and Scully C: Salivary microRNAs in oral cancer. Oral Dis.
21:739–747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanash SM, Baik CS and Kallioniemi O:
Emerging molecular biomarkers - blood-based strategies to detect
and monitor cancer. Nat Rev Clin Oncol. 8:142–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu JY, Yi C, Chung HR, Wang DJ, Chang WC,
Lee SY, Lin CT, Yang YC and Yang WC: Potential biomarkers in saliva
for oral squamous cell carcinoma. Oral Oncol. 46:226–231. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ries J, Vairaktaris E, Agaimy A, Kintopp
R, Baran C, Neukam FW and Nkenke E: miR-186, miR-3651 and miR-494:
Potential biomarkers for oral squamous cell carcinoma extracted
from whole blood. Oncol Rep. 31:1429–1436. 2014.PubMed/NCBI
|
|
32
|
Ries J, Vairaktaris E, Kintopp R, Baran C,
Neukam FW and Nkenke E: Alterations in miRNA expression patterns in
whole blood of OSCC patients. In Vivo. 28:851–861. 2014.PubMed/NCBI
|
|
33
|
Masood Y, Kqueen CY and Rajadurai P: Role
of miRNA in head and neck squamous cell carcinoma. Expert Rev
Anticancer Ther. 15:183–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ganci F, Sacconi A, Manciocco V, Sperduti
I, Battaglia P, Covello R, Muti P, Strano S, Spriano G, Fontemaggi
G, et al: MicroRNA expression as predictor of local recurrence risk
in oral squamous cell carcinoma. Head Neck. 38 Suppl 1:E189–E197.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiao W, Bao ZX, Zhang CY, Zhang XY, Shi
LJ, Zhou ZT and Jiang WW: Upregulation of miR-31* is negatively
associated with recurrent/newly formed oral leukoplakia. PLoS One.
7:e386482012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gorenchtein M, Poh CF, Saini R and Garnis
C: MicroRNAs in an oral cancer context - from basic biology to
clinical utility. J Dent Res. 91:440–446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kowalski LP and Sanabria A: Elective neck
dissection in oral carcinoma: A critical review of the evidence.
Acta Otorhinolaryngol Ital. 27:113–117. 2007.PubMed/NCBI
|
|
39
|
Grimm M, Kraut W, Hoefert S, Krimmel M,
Biegner T, Teriete P, Cetindis M, Polligkeit J, Kluba S, Munz A, et
al: Evaluation of a biomarker based blood test for monitoring
surgical resection of oral squamous cell carcinomas. Clin Oral
Investig. 20:329–338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gomes CC and Gomez RS: MicroRNA and oral
cancer: Future perspectives. Oral Oncol. 44:910–914. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoshizawa JM and Wong DT: Salivary
microRNAs and oral cancer detection. Methods Mol Biol. 936:313–324.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zen K and Zhang CY: Circulating microRNAs:
A novel class of biomarkers to diagnose and monitor human cancers.
Med Res Rev. 32:326–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Summerer I, Niyazi M, Unger K, Pitea A,
Zangen V, Hess J, Atkinson MJ, Belka C, Moertl S and Zitzelsberger
H: Changes in circulating microRNAs after radiochemotherapy in head
and neck cancer patients. Radiat Oncol. 8:2962013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gombos K, Horváth R, Szele E, Juhász K,
Gocze K, Somlai K, Pajkos G, Ember I and Olasz L: miRNA expression
profiles of oral squamous cell carcinomas. Anticancer Res.
33:1511–1517. 2013.PubMed/NCBI
|
|
45
|
Komatsu S, Ichikawa D, Takeshita H,
Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H,
Shiozaki A, et al: Circulating microRNAs in plasma of patients with
oesophageal squamous cell carcinoma. Br J Cancer. 105:104–111.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW
and Lin SC: Increase of microRNA miR-31 level in plasma could be a
potential marker of oral cancer. Oral Dis. 16:360–364. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS
and Chang KW: miR-24 up-regulation in oral carcinoma: Positive
association from clinical and in vitro analysis. Oral Oncol.
46:204–208. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu CJ, Lin SC, Yang CC, Cheng HW and
Chang KW: Exploiting salivary miR-31 as a clinical biomarker of
oral squamous cell carcinoma. Head Neck. 34:219–224. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Grimm M, Schmitt S, Teriete P, Biegner T,
Stenzl A, Hennenlotter J, Muhs HJ, Munz A, Nadtotschi T, König K,
et al: A biomarker based detection and characterization of
carcinomas exploiting two fundamental biophysical mechanisms in
mammalian cells. BMC Cancer. 13:5692013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Friedman EB, Shang S, de Miera EV, Fog JU,
Teilum MW, Ma MW, Berman RS, Shapiro RL, Pavlick AC, Hernando E, et
al: Serum microRNAs as biomarkers for recurrence in melanoma. J
Transl Med. 10:1552012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Maclellan SA, Lawson J, Baik J, Guillaud
M, Poh CF and Garnis C: Differential expression of miRNAs in the
serum of patients with high-risk oral lesions. Cancer Med.
1:268–274. 2012. View Article : Google Scholar : PubMed/NCBI
|