Introduction
Laryngeal carcinoma has been reported as the second
most common head and neck squamous carcinoma, in which more than
95% of cases are laryngeal squamous cell carcinoma (LSCC) (1). This disease is a serious threat to
patients health and quality of life, especially for males, with a
global incidence rate of 5.1/100,000 in 2008 (2,3).
Advanced laryngeal carcinoma usually indicates poor treatment
efficacy and a higher recurrence rate. Patients with invasion and
metastasis generally have a much worse prognosis and have a 5-year
survival rate of ~60% (4).
Therefore, more in-depth research of the molecular mechanisms may
aid us in finding new diagnostic and/or therapeutic approaches to
LSCC to improve the prognosis of LSCC patients (5).
Long non-coding RNAs (lncRNAs) usually range from
200 nt to over 100 kb in length and are defined as endogenous
cellular RNAs. At first, lncRNAs were discovered as
‘transcriptional noise’. Currently, lncRNAs are considered to be a
primary element of the human transcriptome. However, there is
little knowledge regarding most of these lncRNAs, which require
functional explanation (6). More
new evidence has shown that in tumorigenesis and in cancer
progression, lncRNAs play an important regulatory role (7). More lncRNAs have been discovered and
identified as oncogenic or anti-oncogenic in head and neck cancer,
including TUG1 (8), HOTAIR
(9), ANRIL (10), CCAT2 (11), MEG3 (12), LOC285194 (13) and 91H (14). However, little is known about the
role of long non-coding RNA in predicting metastasis and patient
prognosis of LSCC. Furthermore, the underlying mechanisms of lncRNA
in regulating LSCC metastasis remain unclear.
Microarray analysis of LSCC tissues showed abnormal
expression of the lncRNA RP11-169D4.1. It was demonstrated that
RP11-169D4.1 levels are significantly decreased in LSCC tissues,
and decreased expression of RP11-169D4.1 indicates a poor prognosis
and increased lymph node metastasis in patients with LSCC (15). However, the role of the lncRNA
RP11-169D4.1 in LSCC remains unknown. In the present study, we
further explored the role of the lncRNA RP11-169D4.1 and its
potential underlying mechanism in LSCC.
Materials and methods
Clinical specimens
A total of 51 patients with laryngeal squamous cell
carcinoma were analyzed in the present study at the First
Affiliated Hospital of Sun Yat-sen University (Guangzhou, China)
between February 2012 and March 2014. All LSCC patients signed
informed consent. The diagnosis of LSCC was histopathologically
confirmed. Tumor and corresponding adjacent normal tissues were
selected from each patient. Normal human laryngeal tissues were
obtained at a minimum of >10 mm from the edge of the cancerous
area. Tissue samples were resected and immediately frozen in liquid
nitrogen. They were stored at −80°C until RNA extraction. The
following clinicopathological data were collected: age, sex, tumor
origin, TNM stage, lymph node metastasis, clinical stage and
histological differentiation.
Cell culture
Human LSCC cell lines (SNU899 and SNU46) were
obtained from Hong Kong University. All LSCC cells were maintained
in Dulbeccos modified Eagles medium (DMEM) with RPMI-1640, which
was supplemented with 10% fetal bovine serum (FBS). All LSCC cells
were incubated in a humidified incubator with 5% CO2 at
37°C.
RNA extraction
Total RNA was extracted using TRIzol®
(Invitrogen, Carlsbad, CA, USA). A NanoDrop 1000 spectrophotometer
(Thermo Fisher Scientific, Waltham, MA, USA) was used to confirm
the RNA quality. The criterion of acceptable purity is an
OD260/280 ratio of ~1.8. Reverse transcription was
achieved with a First Strand cDNA Synthesis kit (Qiagen, Hilden,
Germany) according to the manufacturers protocol.
Transfection assays
The RP11-169D4.1 sequence (Gene: LINC01537,
ENSG00000227467) was synthesized according to the full length
RP11-169D4.1 sequence (based on the RP11-169D4.1 sequence) and then
cloned into a pLVX vector (Invitrogen; Thermo Fisher Scientific).
Then, either RP11-169D4.1-pLVX or empty vector was transfected into
LSCC cells using Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific). The empty pLVX vector was used as the
control.
Real-time quantitative RT-PCR
Real-time PCR was performed using the FastStart
Universal Probe Master (Roche Applied Science, Indianapolis, IN,
USA) on a LightCycler® 480 (Roche Applied Science).
Primers for real-time PCR were purchased from Integrated DNA
Technologies (Coralville, IA, USA). The detection probe was
obtained from Roche Applied Science. The reaction was incubated at
95°C for 10 min followed by 55 cycles of 95°C for 15 sec and 60°C
for 1 min. The mRNA was normalized to GAPDH levels using the
2−ΔΔCt method. The micro-RNA was normalized to U6 levels
using the 2−ΔΔCt method.
Proliferation assay
LSCC cells (1×104) treated with either
RP11-169D4.1-pLVX or empty vector were seeded into 96-well plates
and cultured for 24, 48 and 72 h. Before the indicated time-point,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
(0.5 mg/ml, pH 4.7; Sigma-Aldrich, St. Louis, MO, USA) was added
for 4 h. At the indicated times, the supernatant was removed and
150 µl of dimethyl sulfoxide (DMSO) was added to the plate before
shaking for 15 min at room temperature. A microplate reader (Thermo
Fisher Scientific) was used to measure the absorbance at 490 nm.
The cell growth value was calculated from the mean values of 6
identical wells.
Flow cytometric analysis
LSCC cells (2–5×105) treated with either
RP11-169D4.1-pLVX or negative control (NC) were seeded into 6-well
plates. The cells were harvested by trypsinization after incubation
for 48 h. The cells were double stained with Annexin V and 7-AAD
(Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) in the dark for
30 min at 37°C. The cells were collected and analyzed on a flow
cytometer (FACScan; BD Biosciences, Franklin Lakes, NJ, USA) to
determine the apoptosis levels.
Wound healing assay
Transfected cells (2–5×104/well) were
cultured in 6-well plates and serum starved for 24 h, after which
the medium was replaced with medium containing serum (10% FBS). A
100-µl pipette tip was used to scratch the cell monolayer, which
was imaged at 0 and 24 h after the wounding.
Cell invasion assays
Cells (2×104/well) in 200 µl RPMI-1640
were seeded into the upper chamber of a Transwell apparatus with
Matrigel (BD Biosciences) after transfection for 24 h. The lower
chambers were filled with media containing 15% FBS. The LSCC cells
were incubated for 24 h. After the cells invaded the membrane, they
were fixed with 95% ethanol for 30 min. Cells on the lower surface
were stained with 0.1% crystal violet, photographed in three
independent fields and counted.
Western blot analysis
Total protein was isolated from LSCC cells. A BCA
protein quantification kit was used to determine the protein
concentration. Proteins were separated on a 10% sodium dodecyl
sulfate-polyacrylamide gel using SDS-PAGE and transferred
electrophoretically onto polyvinylidene difluoride membranes
(Whatman, Maidstone, UK). Then, the membranes were blocked for 1 h
in 5% skim milk and washed three times with Tris-buffered saline
containing 20% Tween-20 (TBST) at room temperature. The membranes
were incubated with the primary antibodies overnight at 4°C, washed
the following day with TBST and incubated with secondary antibody
for 1 h at room temperature. Finally, the immunoreactivity was
visualized by enhanced chemiluminescence.
Mouse monoclonal anti-GAPDH (cat. no. KM9002;
1:5,000; Tianjin Sungene Biotech, Co., Ltd,. Tianjin, China),
rabbit monoclonal anti-vimentin (cat. no. 5741S; 1:1,000; Cell
Signaling Technology, Danvers, MA, USA), rabbit monoclonal
E-cadherin (cat. no. A0965; 1:1,000; ABclonad, Inc., Seoul, Korea),
rabbit monoclonal SNAIL2 (cat. no. A0572; 1:500; ABclonad), rabbit
monoclonal AKT (cat. no. 4691S; 1:1,000; Cell Signaling Technology)
and rabbit monoclonal p-AKT (cat. no. 4060S; 1:2,000; Cell
Signaling Technology) primary antibodies were used. Goat anti-mouse
IgG-HRP (cat. no. BA1050; 1:5,000; Wuhan Boster Bio-engineering,
Co., Ltd., Wuhan, China) and goat anti-rabbit IgG-HRP (cat. no.
BA1055; 1:5,000; Wuhan Boster Bio-engineering) were used as
secondary antibodies.
Argonaute 2 (AGO2) protein
immunoprecipitation
LSCC cells were transfected with either miR-205-5p
inhibitor or negative control (Qiagen) using the Lipofectamine 2000
reagent. Subsequently, a human Argonaute 2 (Ago2) miRNA isolation
kit (Wako Pure Chemical Industries, Osaka, Japan) was used to
isolate the Ago2 complex. After transfection, the cells were lysed,
and anti-Ago2 monoclonal antibody-immobilized beads (Wako Pure
Chemical Industries) were added to the cell lysate. After
incubation for 2 h at 4°C, the beads were washed with wash buffer,
and the Ago2 complex was eluted from the beads. The levels of
miR-205-5p and RP11-169D4.1 were measured from the eluted Ago2
complex. The PCR primer-probe pairs for RP11-169D4.1 quantification
were as follows: forward primer, 5-CCGGAATTCCCCAGACACAG
GGCAGCCTTCC-3 and reverse primer, 5-ATAAGAATG
CGGCCGCTTTTATATAATATTTTGAAT-3; probe, #15 from the Universal Probe
Library (Roche Applied Science). The PCR primer-probe pair for
miR-205-5p quantification was as follows:
5-TCCTTCATTCCACCGGAGTCTG-3; probe #61. The expression level of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
quantified using the Universal Probe Library Human GAPD Gene Assay
(Roche Applied Science).
Statistical analysis
Data were reported as the means ± standard deviation
from at least three independent experiments. All of the statistical
analyses were performed using the SPSS 20.0 statistical software
(IBM, New York, NY, USA) with either Students t-test (two tailed)
or one-way analysis of variance (ANOVA) for multiple groups.
Differences were considered statically significant at the
probability of P<0.05.
Results
RP11-169D4.1 expression is
downregulated in LSCC tissues and cell lines
The expression levels of lncRNA RP11-169D4.1 were
evaluated by qRT-PCR in 51 paired LSCC tissues and adjacent normal
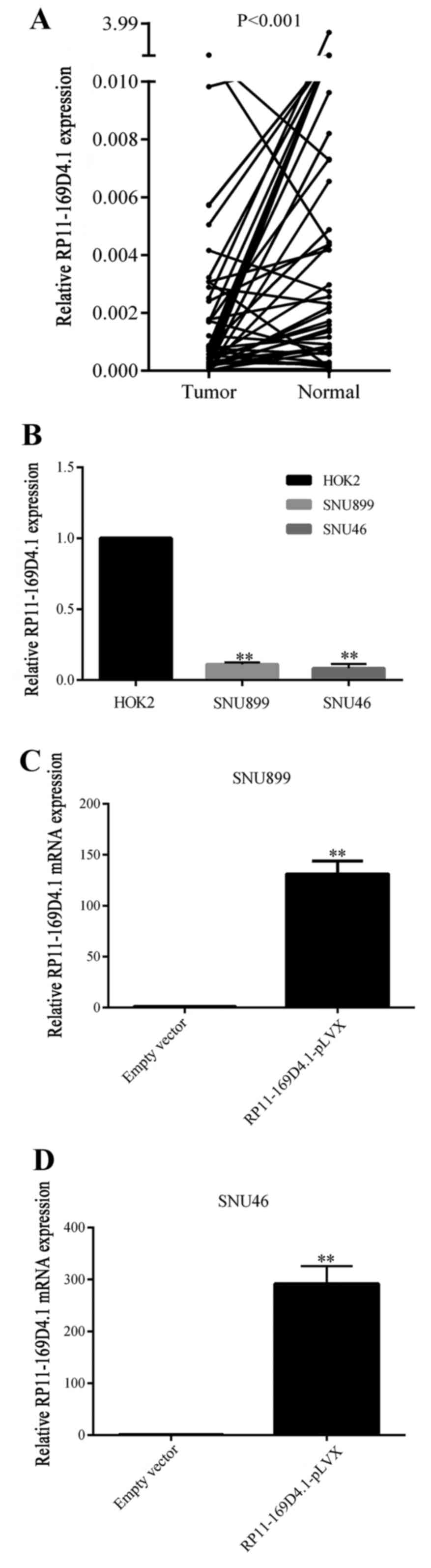
tissues. As shown in Fig. 1A,
RP11-169D4.1 expression was much lower in LSCC tissues than that in
normal tissues (P<0.001). The clinicopathological
characteristics of the 51 patients, including age, sex, tumor
origin, TNM stage, lymph node metastasis, clinical stage and
histological differentiation are presented in Table I. LncRNA RP11-169D4.1 expression in
cancer tissues was associated with lymph node metastasis (P=0.029).
The expression of RP11-169D4.1 was significantly downregulated in
LSCC cell lines compared with normal throat epithelial cells as
shown in Fig. 1B.
 | Table I.Relationship between RP11-169D4.1
expression and tumor clinicopathological features in LSCC. |
Table I.
Relationship between RP11-169D4.1
expression and tumor clinicopathological features in LSCC.
|
|
| RP11-169D4.1
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases | Low (26) | High (25) |
P-valuea |
|---|
| Sex |
|
Male | 50 | 25 | 25 | 0.322 |
|
Female | 1 | 1 | 0 |
|
| Age (years) |
|
<60 | 29 | 11 | 18 | 0.032a |
|
≥60 | 22 | 15 | 7 |
|
| T stage |
|
T1-2 | 20 | 8 | 12 | 0.208 |
|
T3-4 | 31 | 18 | 13 |
|
| Lymph node |
|
Positive | 20 | 14 | 6 | 0.029a |
|
Negative | 31 | 12 | 19 |
|
| Clinical stage |
|
Early | 16 | 6 | 10 | 0.193 |
|
Advance | 35 | 20 | 15 |
|
| Histological
differentiation |
| Well
and moderately differentiated | 29 | 13 | 16 | 0.313 |
| Poor
and undifferentiated | 22 | 13 | 9 |
|
| CDH1
expression |
|
Low | 26 | 17 | 9 | 0.036a |
|
High | 25 | 9 | 16 |
|
| The correlation
between RP11-169D4.1 and CDH1 |
| Pearson
correlation | 0.744 |
|
|
|
| P-value |
<0.001b |
|
|
|
RP11-169D4.1 suppresses proliferation
and promotes apoptosis in LSCC cells
To investigate the role of RP11-169D4.1 in the
regulation of cell proliferation and apoptosis, SNU899 and SNU46
cells were transfected with RP11-169D4.1-pLVX. qRT-PCR was used to
measure the expression of RP11-169D4.1, which was greatly increased
(Fig. 1C and D).
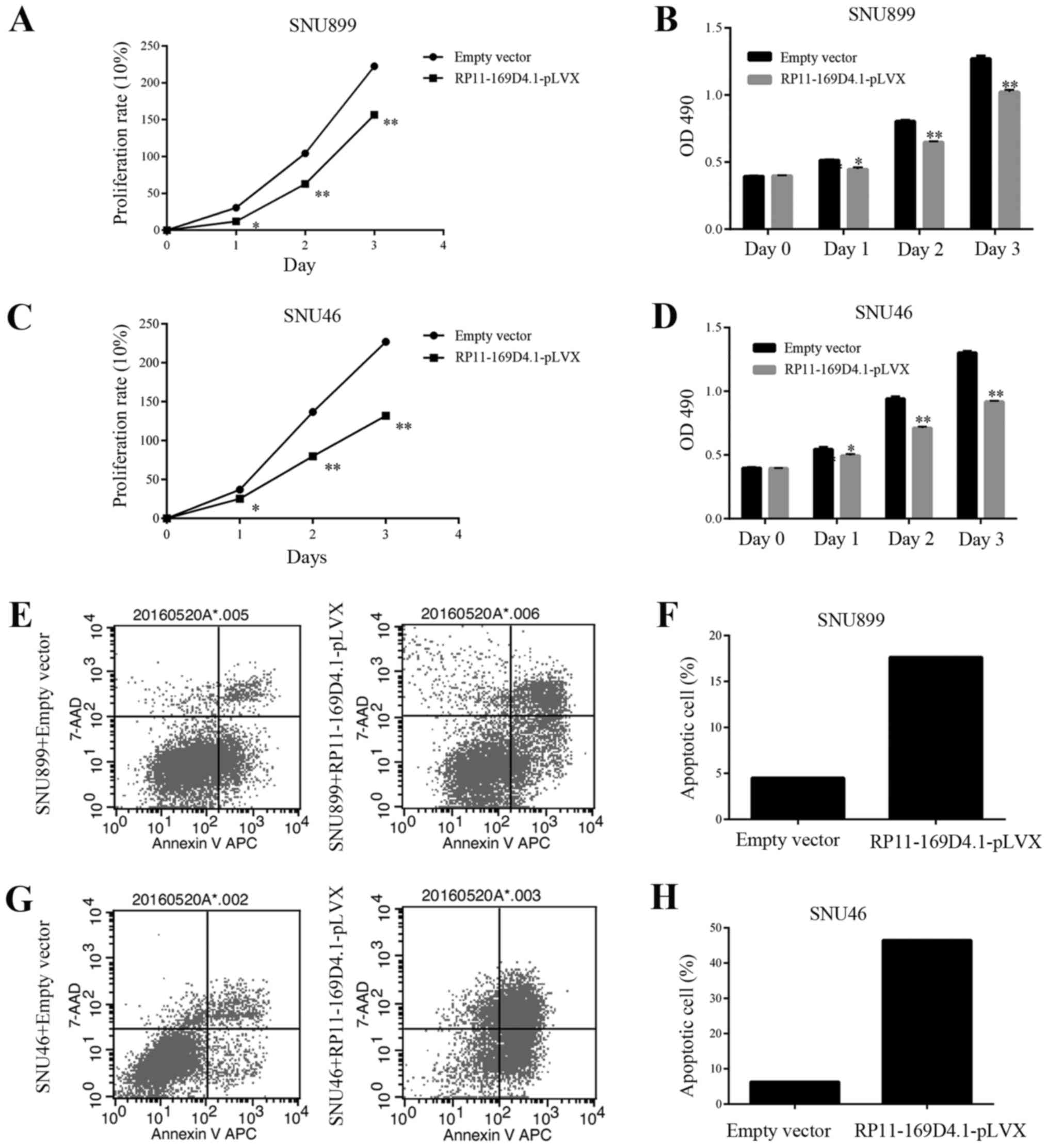
The growth curves determined by the MTT assay showed
that overexpression of RP11-169D4.1 could suppress the
proliferation of SNU899 and SNU46 cells at 24, 48 and 72 h after
transfection (Fig. 2A-D). The
apoptosis assay showed that the percentage of apoptotic cells was
significantly increased in response to RP11-169D4.1 overexpression
compared with NC overexpression in SNU899 and SNU46 cells (Fig. 2E-H). These results indicated the
anti-proliferative and pro-apoptotic role of RP11-169D4.1 in LSCC
cells.
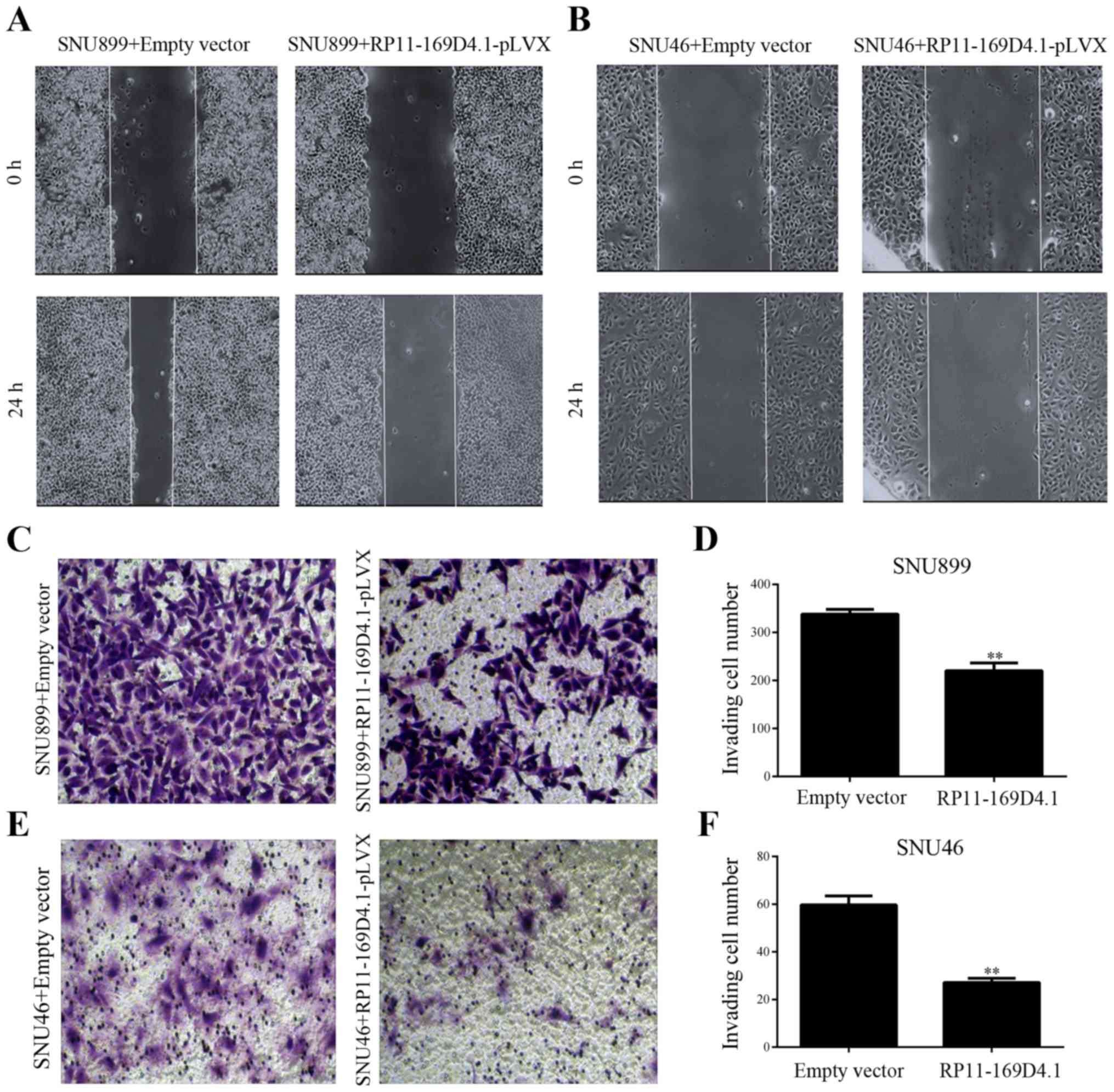
RP11-169D4.1 inhibits migration and
invasion in LSCC cells
To examine the effect of RP11-169D4.1 on migration
and invasion of LSCC cells, wound healing and Transwell invasion
assays were conducted. We found that LSCC cells transfected with
RP11-169D4.1-pLVX showed less wound closure than cells transfected
with empty pLVX vector (Fig. 3A and
B). The Transwell assays showed that overexpression of
RP11-169D4.1 inhibited the invasion of LSCC cells transfected with
RP11-169D4.1-pLVX (Fig. 3C-F).
These results suggest that RP11-169D4.1 contributes to the
inhibition of the migratory and invasive capacity of LSCC
cells.
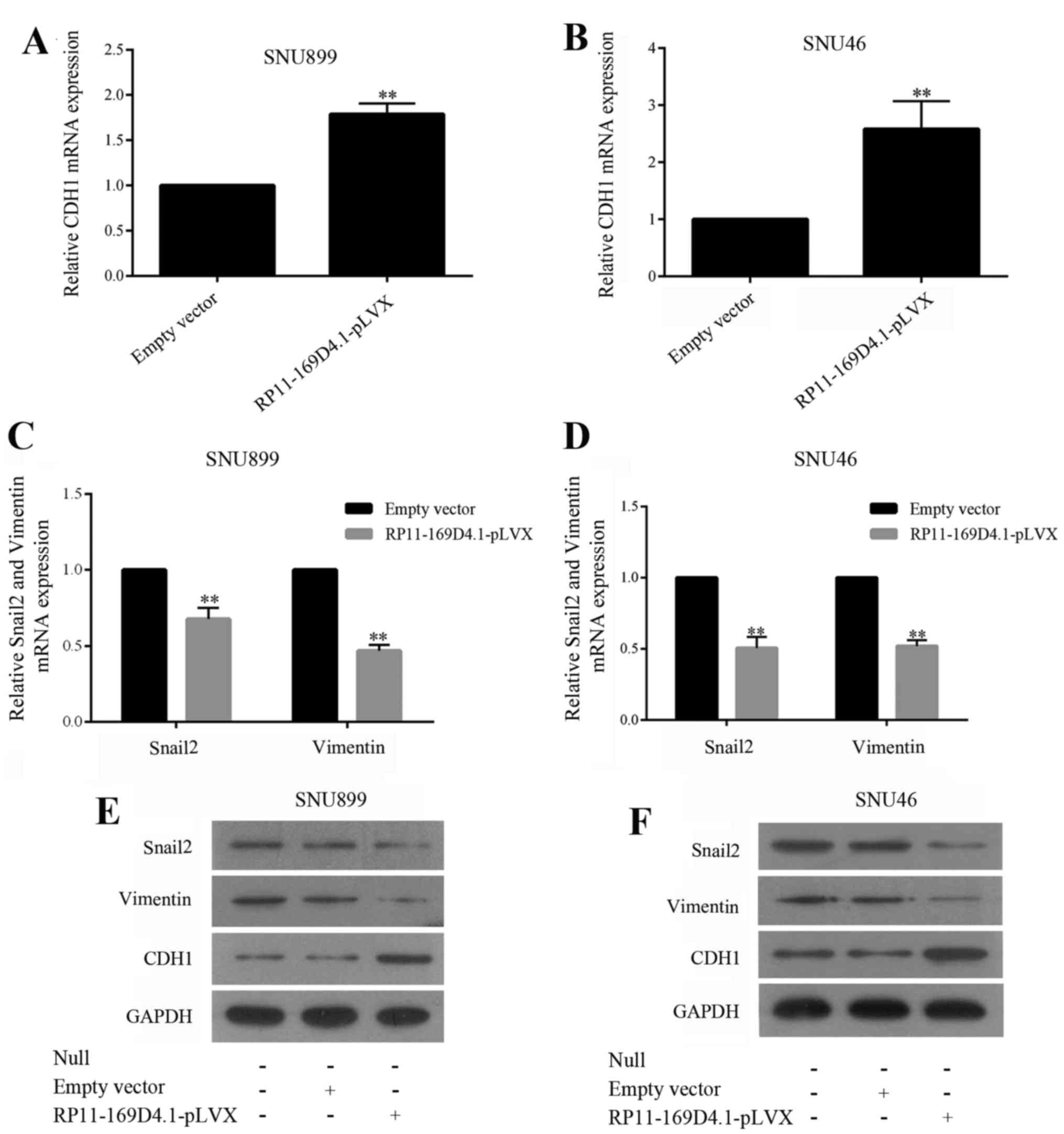
RP11-169D4.1 inhibits EMT in LSCC
cells
To determine whether overexpression of RP11-169D4.1
inhibits epithelial-mesenchymal transitions (EMT) in LSCC cells, we
enhanced RP11-169D4.1 expression in SNU899 and SNU46 cells and
examined the mRNA expression of EMT markers by RT-PCR and the
protein levels by western blot assay. As illustrated in Fig. 4, the level of CDH1 was improved, and
the levels of Snail2 and vimentin were reduced in cells transfected
with RP11-169D4.1-pLVX. These results showed that RP11-169D4.1 was
able to inhibit EMT in LSCC cells.
RP11-169D4.1 was targeted and
inhibited by miR-205-5p
Given the observation that RP11-169D4.1 played an
important role in regulating the biological properties of LSCC
cells, we next investigated the potential mechanisms of
RP11-169D4.1 using the bioinformatics tool RNA22 (16). RNA22 predicted that RP11-169D4.1 was
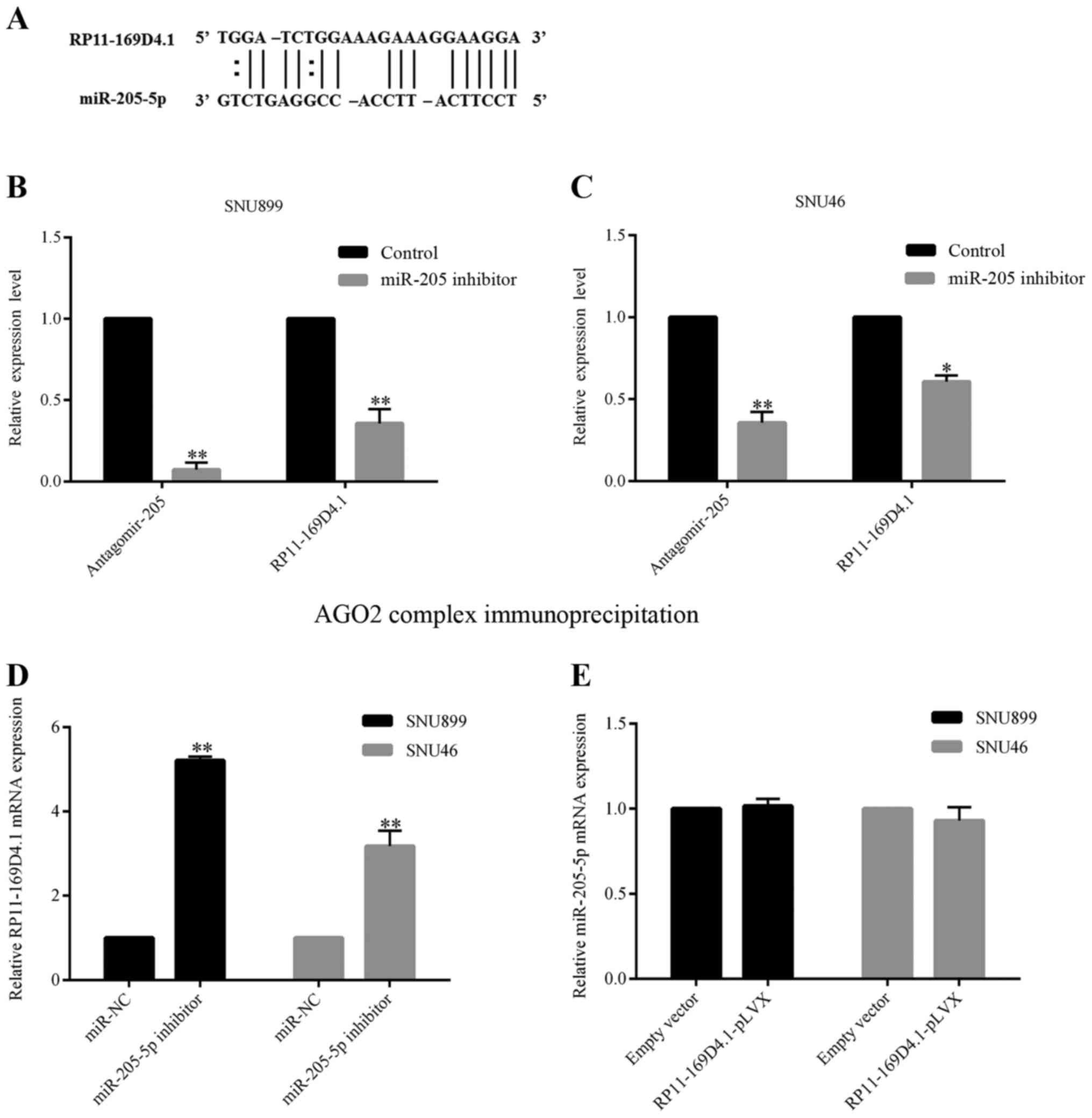
a target of miR-205-5p (Fig. 5A).
We transfected LSCC cells with either a miR-205-5p inhibitor or
miR-NC and confirmed the transfection efficiency using RT-PCR.
SNU899 and SNU46 cells with low expression of miR-205-5p showed
higher levels of RP11-169D4.1 compared with the negative control
cells (Fig. 5D). However, the
overexpression of RP11-169D4.1 did not affect the expression of
miR-205-5p in LSCC cells (Fig. 5E),
which indicates that miR-205-5p might inhibit the expression of
RP11-169D4.1.
We performed immunoprecipitation of endogenous
protein-mRNA complexes in SNU899 and SNU46 cells. Both miR-205-5p
and RP11-169D4.1 were enriched in the immunopurified AGO2 complex,
suggesting that RP11-169D4.1 is an AGO2-selected transcript in the
LSCC cell lines. In the cell lines transfected with the miR-205-5p
inhibitor, the transcript level of mature miR-205-5p and
RP11-169D4.1 dropped significantly compared with the cell lines
transfected with miR-NC (Fig. 5B and
C). The results indicated that mature miR-205-5p could possibly
bind to the RP11-169D4.1 transcript and hinder RP11-169D4.1
expression by 3-UTR-mediated mRNA degradation.
CDH1 expression is downregulated and
correlated with RP11-169D4.1 in LSCC tissues
CDH1, also known as E-cadherin, is a
well-established tumor suppressor (17–19).
Loss of CDH1 can trigger EMT and also has a strong association with
the invasive metastasis of various tumors (20,21).
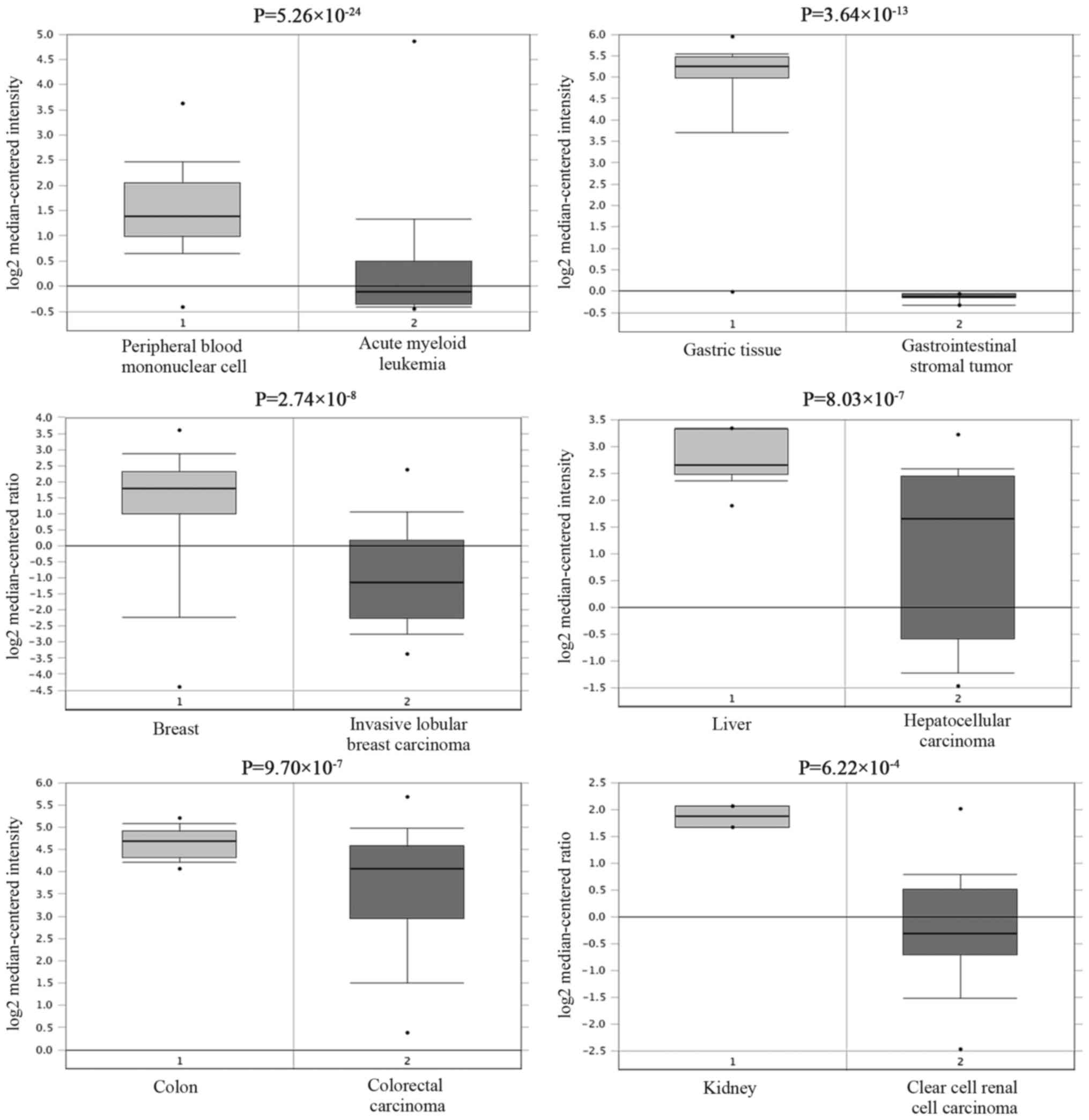
By using the Oncomine StarBase, large sets of data that show
reduced CDH1 mRNA levels in various cancerous tissues compared to
normal tissues can be searched (Fig.
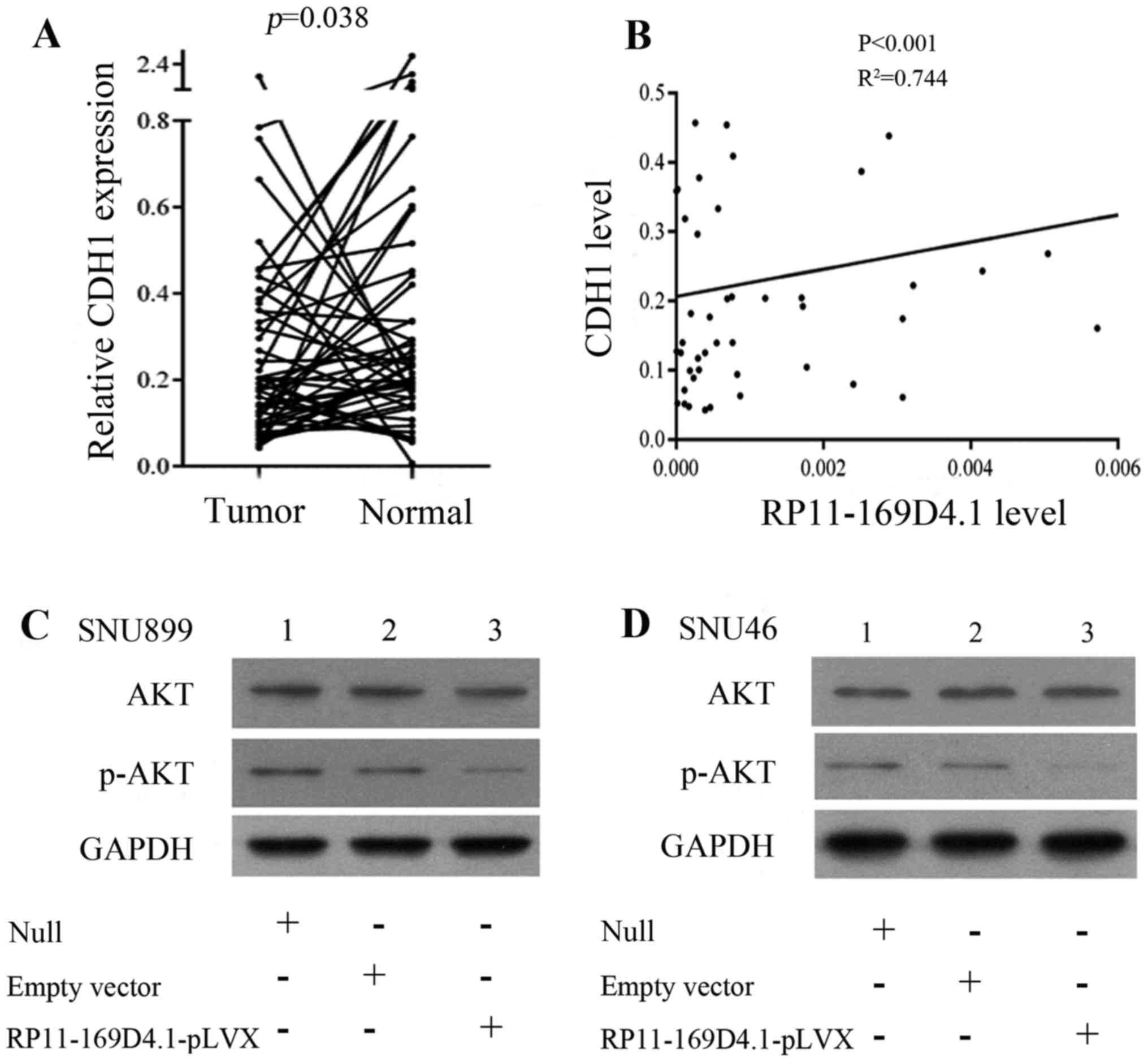
6). To determine the correlation between RP11-169D4.1 and CDH1,
we further analyzed the expression of CDH1 in 51 paired LSCC
tissues and adjacent normal tissues. CDH1 was identified as having
lower expression in LSCC tissues than in normal tissues (P=0.038;
Fig. 7A). In addition, the
correlation analysis revealed that there was a positive correlation
between the expression of RP11-169D4.1 and CDH1 (P<0.001;
R2=0.744) (Table I;
Fig. 7B).
As shown in Fig. 4, the upregulation of RP11-169D4.1
could enhance the level of CDH1 protein
Furthermore, we found that the expression of CDH1
mRNA was significantly improved in the cells transfected with
RP11-169D4.1-pLVX compared to cells transfected with empty vector.
These results indicated that RP11-169D4.1 could modulate the
expression of CDH1.
RP11-169D4.1 exerts its function via the AKT
signaling pathway. According to studies that showed that loss of
CDH1 could activate AKT signaling, we proposed that RP11-169D4.1
could regulate the AKT signaling pathway through CDH1. We detected
the levels of AKT and phospho-AKT in LSCC cell lines. As expected,
the level of p-AKT was reduced after cells were transfected with
RP11-169D4.1-pLVX and total AKT levels were constant (Fig. 7C and D). These results suggest that
RP11-169D4.1 may be a major player in the AKT signaling
pathway.
Discussion
With the fast development of human genome and
transcriptome sequencing technologies, much attention has been
focused on lncRNA. lncRNAs could serve as potential diagnostic
biomarkers and effective therapeutic targets. In the near future,
in-depth research of lncRNAs is an attractive avenue to discover
novel biomarkers or targets (22).
To date, many studies have explored the various functions of
lncRNAs in head and neck neoplasms (23). For example, the overexpression of
UCA1 could promote the metastasis of TSCC cells (24). HOTAIR participated in PTEN
methylation in Hep-2 cells (25)
and had close correlation with miR-21 in LSCC (26). MALAT1 was a novel target of miR-217
and miR-101 and could stimulate the invasion and metastasis of ESCC
cells (27).
It was observed that RP11-169D4.1 expression was
downregulated in LSCC tissues and metastatic neck lymph nodes, and
lower expression of RP11-169D4.1 predicted poor prognosis. In
another study, the downregulation of RP11-169D4.1 in LSCC tissues
was also observed (28). However,
these studies were limited to its abnormal expression in LSCC.
However, the specific function of RP11-169D4.1 and its potential
mechanisms in LSCC still remain unknown.
In the present study, we first clarified the
function of RP11-169D4.1, which was thought to play a
tumor-suppressive role in LSCC. Moreover, RP11-169D4.1 expression
was significantly correlated with LSCC metastasis to the neck lymph
nodes. These data suggested that RP11-169D4.1 might participate in
the metastasis of LSCC. The molecular mechanisms that control the
expression of RP11-169D4.1 are now being elucidated. Through the
bioinformatics tool RNA22, we know that RP11-169D4.1 might be a
target of miR-205-5p. As an oncogenic microRNA, miR-205-5p has been
well characterized for its role in LSCC (29). Some studies have indicated that
miR-205-5p promotes the proliferation, migration and invasion of
LSCC (30). Our findings highlight
the interaction between miR-205-5p and the lncRNA RP11-169D4.1
during tumorigenesis and the progression of LSCC cells.
Next, we explored the molecular mechanisms
underlying RP11-169D4.1 inhibition of EMT. EMT has been identified
as a paramount event in the early periods of metastatic
dissemination in various tumor cells. During these periods, tumor
cells become more active and have a stronger invasive ability
(31). The CDH1 gene encodes
E-cadherin, which is a transmembrane glycoprotein and a
prototypical member of the classical cadherin family.
E-cadherin/CDH1 plays a critical role in preserving cell polarity
as well as maintaining epithelial integrity. It was reported that
CDH1 expression was correlated with the metastasis to neck lymph
nodes and the TNM stages in LSCC (32). As CDH1 is the most commonly
expressed gene during EMT, we investigated whether RP11-169D4.1
could regulate the expression of CDH1.
To further explore the mechanisms of RP11-169D4.1,
we tried to find the potential signaling pathway of RP11-169D4.1.
Previous studies have shown that CDH1 expression is regulated
through the AKT pathway (33), and
miR-205-5p promotes tumor metastasis by activating the AKT
signaling (34), which indicates
that RP11-169D4.1 might regulate the AKT signaling pathway by
modulating CDH1. The AKT pathway is considered to be closely
related to laryngeal carcinoma (35,36).
These results suggest that the miR-205-5p/RP11-169D4.1/CDH1/AKT
signaling pathway may play an important role in the development of
LSCC.
EMT is a process that results in the migration,
invasion and metastasis of cancer cells. At the same time, loss of
CDH1 (E-cadherin) is considered as a fundamental event in EMT.
Several studies indicate that patients with lymph node metastasis
tend to have higher recurrence rate and poor prognosis and
RP11-169D4.1 can be considered as a predictor of lymph node
metastasis in patients. However, the specific mechanism of
regulating CDH1 for RP11-169D4.1 still needs further exploration.
Continued study of these molecules and an improved understanding of
the lncRNA RP11-169D4.1 will facilitate the development of more
effective therapies against human laryngeal carcinoma.
In conclusion, the lncRNA RP11-169D4.1 is
downregulated in LSCC and is associated with lymph node metastasis.
Overexpression of RP11-169D4.1 in LSCC cells decreased cell
migration and invasion in vitro. miR-205-5p acted as the
upstream molecule of RP11-169D4.1 activity. Furthermore,
RP11-169D4.1 could suppress the process of EMT by modulating CDH1
expression. The miR-205-5p/RP11-169D4.1/CDH1/AKT signaling pathway
is an important part of the molecular mechanisms of EMT in LSCC.
RP11-169D4.1 may be a novel and valuable therapeutic target in
predicting outcomes of patients with LSCC.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang Z, Meng Q, Luo J, Lu Q, Li X, Li G
and Wan C: Development and validation of the simplified Chinese
version of EORTC QLQ-H&N35 for patients with head and neck
cancer. Support Care Cancer. 20:1555–1564. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hunter KD, Parkinson EK and Harrison PR:
Profiling early head and neck cancer. Nat Rev Cancer. 5:127–135.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marioni G, Marchese-Ragona R, Cartei G,
Marchese F and Staffieri A: Current opinion in diagnosis and
treatment of laryngeal carcinoma. Cancer Treat Rev. 32:504–515.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Denaro N, Merlano MC, Russi EG and Lo
Nigro C: Non coding RNAs in head and neck squamous cell carcinoma
(HNSCC): A clinical perspective. Anticancer Res. 34:6887–6896.
2014.PubMed/NCBI
|
|
6
|
Rühle F and Stoll M: Long non-coding RNA
databases in cardiovascular research. Genomics Proteomics
Bioinformatics. 14:191–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu Y, Wang J, Qiu M and Xu L, Li M, Jiang
F, Yin R and Xu L: Upregulation of the long noncoding RNA TUG1
promotes proliferation and migration of esophageal squamous cell
carcinoma. Tumour Biol. 36:1643–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu
ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al: Upregulation of the long
non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma
metastasis and poor prognosis. Mol Carcinog. 52:908–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen D, Zhang Z, Mao C, Zhou Y, Yu L, Yin
Y, Wu S, Mou X and Zhu Y: ANRIL inhibits p15INK4b through the TGFβ1
signaling pathway in human esophageal squamous cell carcinoma. Cell
Immunol. 289:91–96. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q,
Yin R and Xu L: Long noncoding RNA CCAT2 correlates with smoking in
esophageal squamous cell carcinoma. Tumour Biol. 36:5523–5528.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia LF, Wei SB, Gan YH, Guo Y, Gong K,
Mitchelson K, Cheng J and Yu GY: Expression, regulation and roles
of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J
Cancer. 135:2282–2293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong YS, Zhou XL, Wang XW, Wu QQ, Yang TX,
Lv J, Yang JS, Zhu B and Cao XF: Association of decreased
expression of long non-coding RNA LOC285194 with chemoradiotherapy
resistance and poor prognosis in esophageal squamous cell
carcinoma. J Transl Med. 12:2332014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao T, He B, Pan Y, Xu Y, Li R, Deng Q,
Sun H and Wang S: Long non-coding RNA 91H contributes to the
occurrence and progression of esophageal squamous cell carcinoma by
inhibiting IGF2 expression. Mol Carcinog. 54:359–367. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen Z, Li Q, Deng H, Lu D, Song H and Guo
J: Long non-coding RNA profiling in laryngeal squamous cell
carcinoma and its clinical significance: Potential biomarkers for
LSCC. PLoS One. 9:e1082372014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Loher P and Rigoutsos I: Interactive
exploration of RNA22 microRNA target predictions. Bioinformatics.
28:3322–3323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiao F, Hu H, Han T, Zhuo M, Yuan C, Yang
H and Wang L and Wang L: Aberrant expression of nuclear HDAC3 and
cytoplasmic CDH1 predict a poor prognosis for patients with
pancreatic cancer. Oncotarget. 7:16505–16516. 2016.PubMed/NCBI
|
|
18
|
Wang Q, Wang B, Zhang YM and Wang W: The
association between CDH1 promoter methylation and patients with
ovarian cancer: A systematic meta-analysis. J Ovarian Res.
9:232016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berx G, Cleton-Jansen AM, Nollet F, De
Leeuw WJ, van de Vijver M, Cornelisse C and van Roy F: E-cadherin
is a tumour/invasion suppressor gene mutated in human lobular
breast cancers. EMBO J. 14:6107–6115. 1995.PubMed/NCBI
|
|
20
|
Zhou J, Tao D, Xu Q, Gao Z and Tang D:
Expression of E-cadherin and vimentin in oral squamous cell
carcinoma. Int J Clin Exp Pathol. 8:3150–3154. 2015.PubMed/NCBI
|
|
21
|
Kowalski PJ, Rubin MA and Kleer CG:
E-cadherin expression in primary carcinomas of the breast and its
distant metastases. Breast Cancer Res. 5:R217–R222. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cellular and molecular life sciences. Cell Mol Life Sci.
73:2491–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen H, Xin Y, Zhou L, Huang JM, Tao L,
Cheng L and Tian J: Cisplatin and paclitaxel target significant
long noncoding RNAs in laryngeal squamous cell carcinoma. Med
Oncol. 31:2462014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang Z, Wu L, Wang L, Yang Y, Meng Y and
Yang H: Increased expression of the long non-coding RNA UCA1 in
tongue squamous cell carcinomas: A possible correlation with cancer
metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 117:89–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J
and Liu M: Long intergenic noncoding RNA HOTAIR is overexpressed
and regulates PTEN methylation in laryngeal squamous cell
carcinoma. Am J Pathol. 182:64–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M
and Tian L: Combined detection of serum exosomal miR-21 and HOTAIR
as diagnostic and prognostic biomarkers for laryngeal squamous cell
carcinoma. Med Oncol. 31:1482014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zou AE, Ku J, Honda TK, Yu V, Kuo SZ,
Zheng H, Xuan Y, Saad MA, Hinton A, Brumund KT, et al:
Transcriptome sequencing uncovers novel long noncoding and small
nucleolar RNAs dysregulated in head and neck squamous cell
carcinoma. RNA. 21:1122–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cao P, Zhou L, Zhang J, Zheng F, Wang H,
Ma D and Tian J: Comprehensive expression profiling of microRNAs in
laryngeal squamous cell carcinoma. Head Neck. 35:720–728. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tian L, Zhang J, Ge J, Xiao H, Lu J, Fu S,
Liu M and Sun Y: MicroRNA-205 suppresses proliferation and promotes
apoptosis in laryngeal squamous cell carcinoma. Med Oncol.
31:7852014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ahmed RA, Shawky AA and Hamed RH:
Prognostic significance of cyclin D1 and E-cadherin expression in
laryngeal squamous cell carcinoma. Pathol Oncol Res. 20:625–633.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Su L and Liu X: Loss of CDH1
up-regulates epidermal growth factor receptor via phosphorylation
of YBX1 in non-small cell lung cancer cells. FEBS Lett.
587:3995–4000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mao Y, Wu S, Zhao R and Deng Q: MiR-205
promotes proliferation, migration and invasion of nasopharyngeal
carcinoma cells by activation of AKT signalling. J Int Med Res.
44:231–240. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu B, Di W, Wang H, Ma H, Li J and Zhang
Q: Tumor suppressor TSLC1 is implicated in cell proliferation,
invasion and apoptosis in laryngeal squamous cell carcinoma by
regulating Akt signaling pathway. Tumour Biol. 33:2007–2017. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang N, Hui L, Wang Y, Yang H and Jiang X:
SOX2 promotes the migration and invasion of laryngeal cancer cells
by induction of MMP-2 via the PI3K/Akt/mTOR pathway. Oncol Rep.
31:2651–2659. 2014.PubMed/NCBI
|