Introduction
Long non-coding RNAs (lncRNAs) are non-protein
coding transcripts with a base length of over 200 nucleotides;
lncRNAs often lack a functional open reading frame (1). lncRNAs modulate the expression of
protein-coding or non-protein-coding genes by adjusting
cis-acting elements and trans-acting factors
(2). lncRNAs also participate in
the regulation of biological behavior, such as cell
differentiation, development and carcinogenesis (3,4).
Therefore, lncRNAs are potential biomarkers and therapeutic targets
for malignant tumors, providing a comprehensive approach for
exploring cancer pathogenesis and presenting an alternative
therapeutic method for cancers (5).
Some lncRNAs have been recently identified as
multi-functional gene regulating factors (6). PVT1 is located in the 8q24 region and
likely functions as a highly conserved non-coding RNA in humans
(7). The amplification of the 8q24
region is a common event in cancers; moreover, previous studies
have related the ectopic expressions of local genes in this region
with reduced survival period (8,9). Myc,
an established oncogene, is mapped to 8q24. Myc amplification
likely leads to the pathogenesis of cancers. PVT1 amplification in
8q24 is another frequent event in cancer pathophysiology and has
been observed in colorectal cancers (10), lymphomas (11), serous ovarian and breast cancers
(12). Moreover, PVT1
overexpression is related with the decreased survival duration of
cancer patients.
Ectopic PVT1 expression is associated with various
diseases, particularly malignant tumors, however, the definite
function and intermolecular signal mechanism of PVT1 in breast
cancer remains obscure. Furthermore, the detection of circulating
lncRNA DNA provides valuable information for the treatment and
prognosis of breast cancer patients. Therefore, utilizing
circulating PVT1 DNA as a marker for breast cancer requires further
study.
Materials and methods
Tissue samples and clinical data
collection
Fresh tumor tissue samples, normal adjacent tissue
samples, blood sera and clinical data were gathered from 84
patients with invasive ductal breast cancer aged 30–79 (average
age, 48.3 years) enrolled at the Affiliated Hospital of Weifang
Medical University from July 2009 to July 2011. Negative control
samples were collected from individuals with no breast cancer.
Written informed consent was given by breast cancer patients and
healthy individuals. The present study was approved by the Research
and Ethics Committee of Weifang Medical University.
Cell culture and transient
transfection
The human breast cancer cell lines MDA-MB-231,
MDA-MB-468, SK-BR-3, MDA-MB-435, T47D, MCF-7 and immortalized
normal human mammary epithelial cells MCF-10A were purchased from
the Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). All cells were cultured in Dulbeccos modified
Eagles medium (DMEM; Life Technologies, Gaithersburg, MD, USA)
supplemented with 10% fetal bovine serum (FBS; Hyclone
Laboratories, Inc., Logan, UT, USA). All cell lines were cultured
at 37°C and 5% CO2.
MCF-7 and MDA-MB-231 cells were transfected with
siRNA plasmids using Lipofectamine 2000 (Invitrogen, Grand Island,
NY, USA) following the manufacturers instructions. Negative control
siRNA and PVT-1-specific siRNAs were purchased from Cosmo Bio, Co.,
Ltd. (Tokyo, Japan), the siRNA target sequence for PVT1 are
si-PVT1: sense, 5-CCCAACAGGVAGGACAGCUUTT-3 and antisense,
5-AAGCUGUCVCUCCUGUUGGGTT-3 si-PVT2: sense,
5-GCUUGGAGGVCUGAGGAGUUTT-3 and antisense,
5-AACUCCUCAGVCCUCCAAGCTT-3. Plasmids were tranfected into breast
cancer cells at a concentration of 50 nM. Cells were harvested at
48 h after transfection for further research.
Detection of nucleic acid segments by
qRT-PCR
Total RNA was isolated from serum of breast cancer
patients or breast cancer cells using TRIzol according to the
manufacturers instruction (Invitrogen, Carlsbad, CA, USA), then
reverse transcription was performed using TransScript reverse
transcriptase (TransGen Biotech, Inc., Beijing, China) and random
hexamer primers, then the supernatant was collected after
instantaneous centrifugation. TIANamp Blood DNA kit (Tiangen
Biotech, Co., Ltd., Beijing, China) was used to extracted DNA was
from sera according to the manufacturers instructions. Quantitative
PCR was then implemented using SYBR-Green (Beyotime Institute of
Biotechnology, Haimen, China) to measure the profiles of PVT1.
Forward and reverse primers for PVT1 RNA were 5-CCGACTCTTCCTG
GTGAAGC-3 and 5-GTATGGTCAGCTCAAGCCCA-3; 5-CCGACTCTTCCTGGTGAAGC-3
and 5-CCACATCAT GGCTCCAAATCTG-3 for PVT1 DNA; 5-TGAGCCGCGA
CTGTGATG-3 and 5-GTCTCGGTGACAAAGTCGAAG TT-3 for p21;
5-GAAGGTGAAGGTCGGAGTC-3 and 5-GA AGATGGTGATGGGATTTC-3 for GAPDH;
5-GTAAC CCGTTGAACCCCATT-3 and 5-CCATCCAATCGGTAGT AGCG-3 for 18S
rRNA.
Cell proliferation assay and colony
formation assay
Breast cancer cells were plated at
5×103/well in 96-well plates and cultured for 48 h, cell
density was measured by the Cell Counting kit-8 (CCK-8; Beijing
Solarbio Science and Technology, Co., Ltd., Beijing, China). The
cell growth curves were drawn according to the absorbance at 595
nm. For colony formation assay, 200 cells were seeded into 10 cm
plates and cultured in DMEM contain 10% FBS. Seven days later, the
colonies were fixed and stained by 0.1% crystal violet. The number
of colonies >50 cells was counted. All experiments were
performed in triplicate.
Immunohistochemistry
Immunohistochemical staining was performed on
formalin-fixed and paraffin-embedded tissue sections and incubated
with a primary antibody against p21 (Fuzhou Maixin Biotech, Co.,
Ltd., Fuzhou, China), followed by a horseradish
peroxidase-conjugated secondary antibody (1:500; Fuzhou Maixin
Biotech) and the proteins to be tested were shown with
3,3-diaminobenzidine. Each sample is graded on a standard of 0–9
according to the staining densities (13). All slides were evaluated by two
pathologists blinded to the information of the clinicopathological
characteristics.
Western blot analysis
Total cell proteins were extracted in RIPA buffer;
cell lysates were cleared by centrifugation and prepared in sodium
dodecyl sulfate buffer. A total of 50 µl of cell lysates were
separated by SDS-PAGE. The proteins were then transferred onto PVDF
membranes. After incubation with primary antibodies for p21 or
GAPDH (Fuzhou Maixin Biotech), the blots in horseradish
peroxidase-conjugated goat anti-rabbit IgG for 60 min and enhanced
chemiluminescence detection were used to quantify densitometry.
Statistical analysis
SPSS software version 17.0 was used to perform data
analysis. One-way ANOVA was performed to detect the relationship
between PVT1 and clinicopathological characteristics. Survival
analysis was used for Kaplan-Meier examination. Receiver operating
characteristic (ROC) curves were used to assess the diagnostic
value of PVT1.
Results
The expression of PVT1 in breast
cancer samples and cell lines
Direct reverse transcription followed by qPCR was
performed to determine PVT1 in breast cancer tissues and cell lines
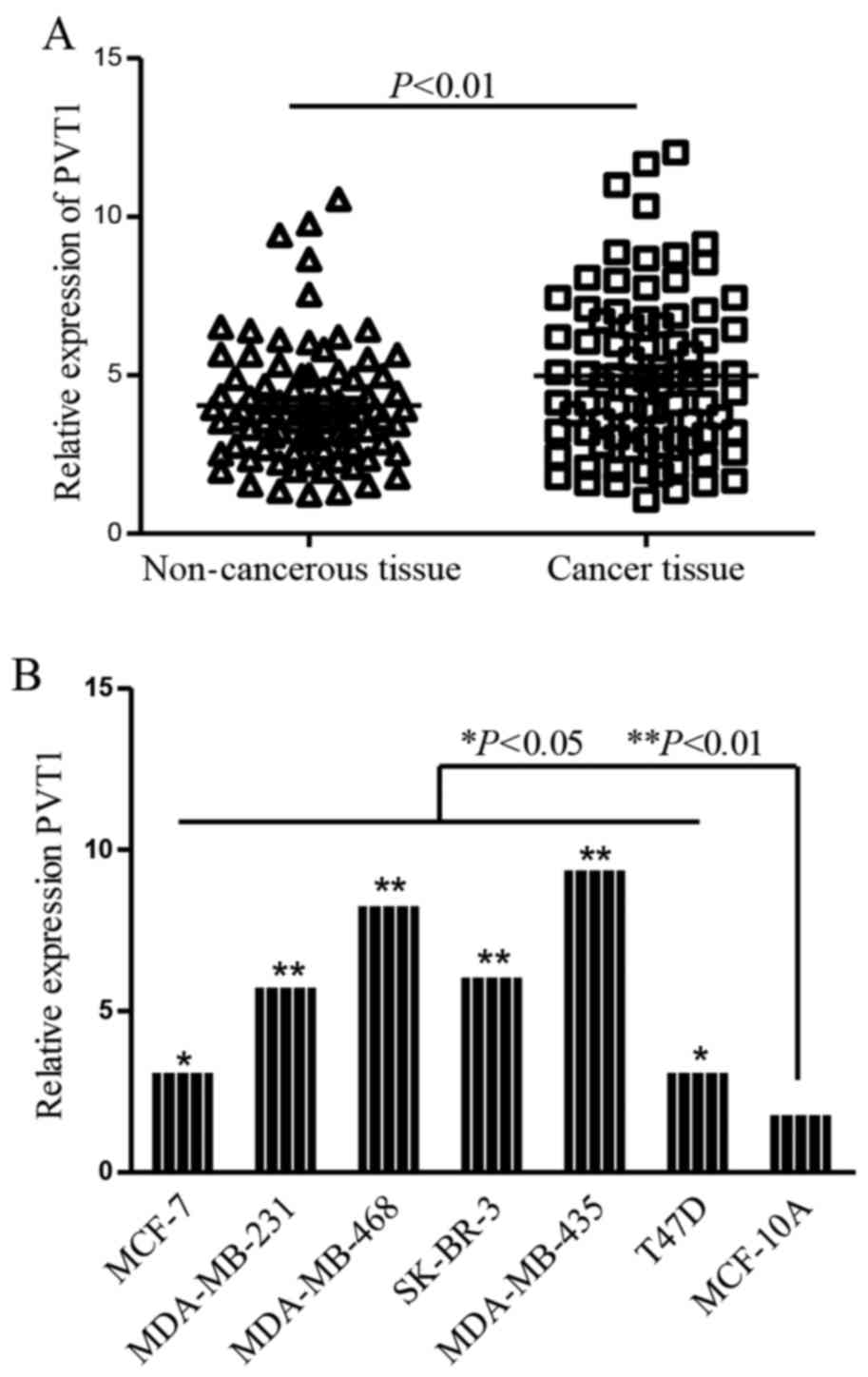
normalizing to 18S rRNA. The results showed that the profiles of
PVT1 were remarkably increased in breast cancer tissues (P<0.01;
Fig. 1A). Moreover, we also detect
the profiles of PVT1 in breast cancer lines and mammary epithelial
cell line MCF-10A, ectopic expression of PVT1 was detected in the
six breast cancer cell lines compared with MCF-10A cells (Fig. 1B).
Association of PVT1 expression with
clinicopathological features
To make clear the clinical relevance of PVT1
expression in breast cancer, according to the median expression of
PVT1 in the breast cancer samples, we divided the 84 breast cancer
patients into PVT1 high-expression group and PVT1 low-expression
group in accordance with the median value of PVT1 expression. As
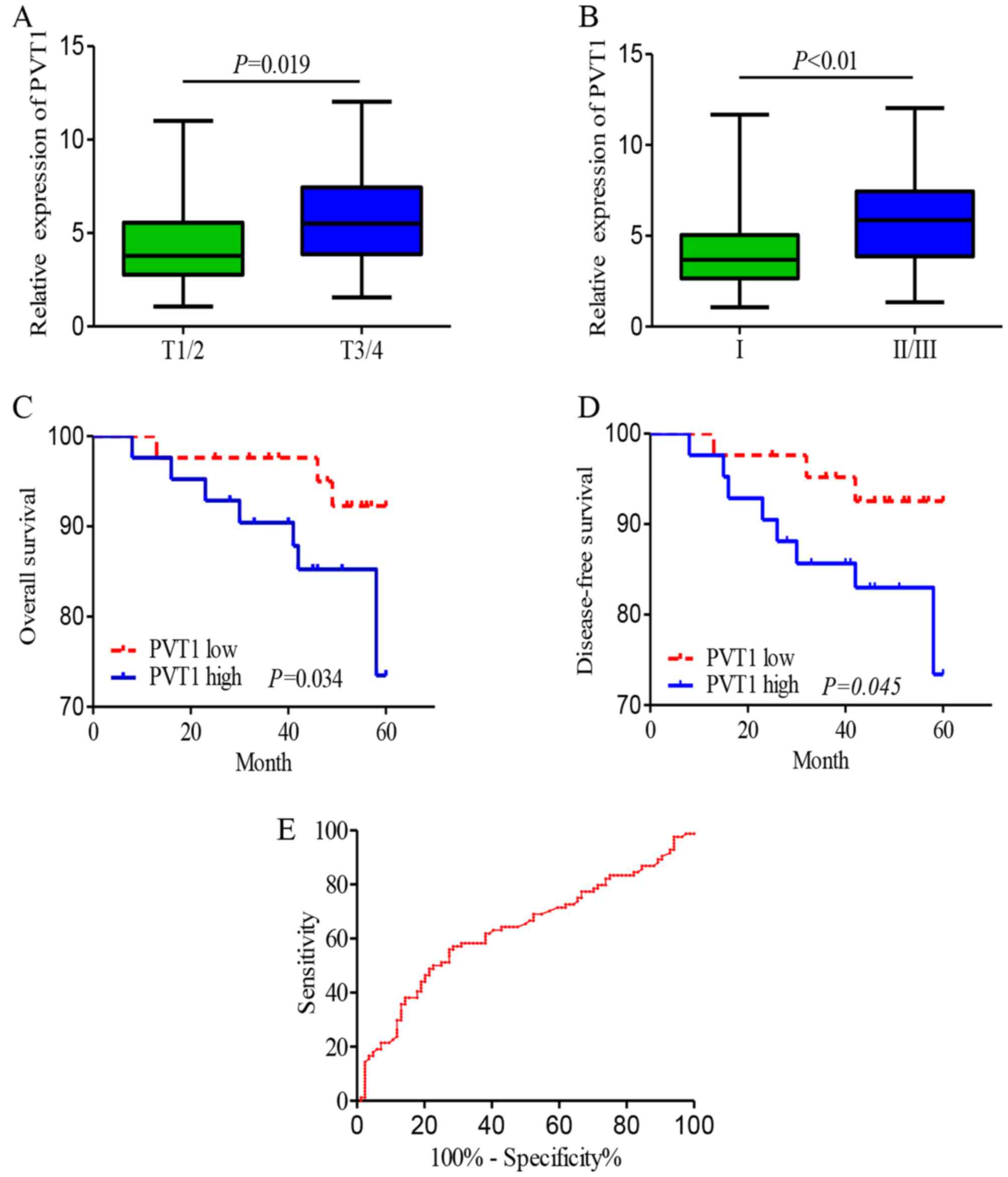
shown in Table I and Fig. 2A and B, the PVT1 level was
associated with cancer histological grade, expression of Ki-67,
tumor size and lymph node metastasis. Whereas, no significant
correlation was found between the expression of PVT1 and other
clinicopathological characteristics of patients, for example, age,
expression of ER, PR and Her-2 in the tissue.
 | Table I.Clinical relevance of PVT1 in breast
cancer. |
Table I.
Clinical relevance of PVT1 in breast
cancer.
|
| PVT1 |
|
|---|
|
|
|
|
|---|
| Features | Low | High | P-value |
|---|
| Age (years) |
|
| 0.617 |
| ≤45 | 33 | 37 |
|
|
>45 | 44 | 42 |
|
| Tumor size (cm) |
|
| 0.010 |
| ≤2 | 42 | 26 |
|
|
>2 | 37 | 53 |
|
| Histological
grade |
|
| 0.013 |
| I | 28 | 17 |
|
| II,
III | 51 | 62 |
|
| Clinical stage |
|
| 0.061 |
| I,
II | 57 | 47 |
|
| III,
IV | 22 | 34 |
|
| Lymph nodes
metastasis |
|
| 0.117 |
|
Positive | 45 | 30 |
|
|
Negative | 34 | 49 |
|
| ER |
|
| 0.143 |
|
Positive | 63 | 55 |
|
|
Negative | 16 | 24 |
|
| PR |
|
| 0.365 |
|
Positive | 53 | 51 |
|
|
Negative | 26 | 18 |
|
| HER-2 |
|
| 0.093 |
|
Positive | 14 | 31 |
|
|
Negative | 65 | 48 |
|
| Ki-67 |
|
| 0.022 |
|
Positive | 14 | 31 |
|
|
Negative | 65 | 48 |
|
Prognostic values of PVT1 for breast
cancer patients
To explore the correlation between PVT1 expression
and prognosis of breast cancer patients, the Kaplan-Meier test and
log-rank test were carried out. The results demonstrated that
breast cancer patients with high PVT1 expression showed significant
shorter 5-year overall survival (OS) compared with patients with
low PVT1 expression (Fig. 2C).
Moreover, the 5-year disease-free survival (DFS) for high PVT1
group was 7.1%, while was 23.8% for low PVT1 group (Fig. 2D). A multivariate Cox proportional
hazards regression analysis was performed to identify independent
predictors of survival, the results showed that PVT1 expression was
significantly correlated with DFS and OS (Table II).
 | Table II.Univariate and multivariate analysis
of clinicopathological parameters influencing prognosis. |
Table II.
Univariate and multivariate analysis
of clinicopathological parameters influencing prognosis.
|
| Overall survival |
| Disease-free
survival |
|
|---|
|
|
|
|
|
|
|---|
| Features | HR | CI 95% | P-value | HR | CI 95% | P-value |
|---|
| Tumor size | 1.461 | 0.534–2.462 | 0.023 | 1.944 | 0.624–2.261 | 0.018 |
| Histologic
grading | 1.279 | 0.915–3.517 | 0.008 | 2.279 | 0.815–2.481 | 0.021 |
| Lymphatic
metastasis | 2.012 | 0.643–1.413 | 0.075 | 1.012 | 0.735–1.214 | 0.258 |
| Ki-67
expression | 1.121 | 0.921–3.276 | 0.041 | 0.926 | 1.845–3.321 | 0.035 |
| Her-2
expression | 0.625 | 1.213–2.016 | 0.152 | 1.142 | 0.663–1.435 | 0.105 |
| PVT1
expression | 2.167 | 1.108–4.265 | 0.015 | 3.167 | 1.416–3.187 | 0.009 |
Receiver operating characteristic (ROC) curve
methodology was used to assess the diagnostic utility of PVT1 for
breast cancers. As Fig. 2E showed,
the best cut-off value for PVT1 in breast cancer was 4.97 with the
sensitivity and specificity at 51.2 and 75.0%, respectively. The
proportion under the ROC curve (AUC) was 0.63 (95% CI, 0.545–0.715,
P=0.004; Fig. 2E), the Youden index
was 0.425.
Association of circulating PVT1 DNA
with clinical stages of breast cancer
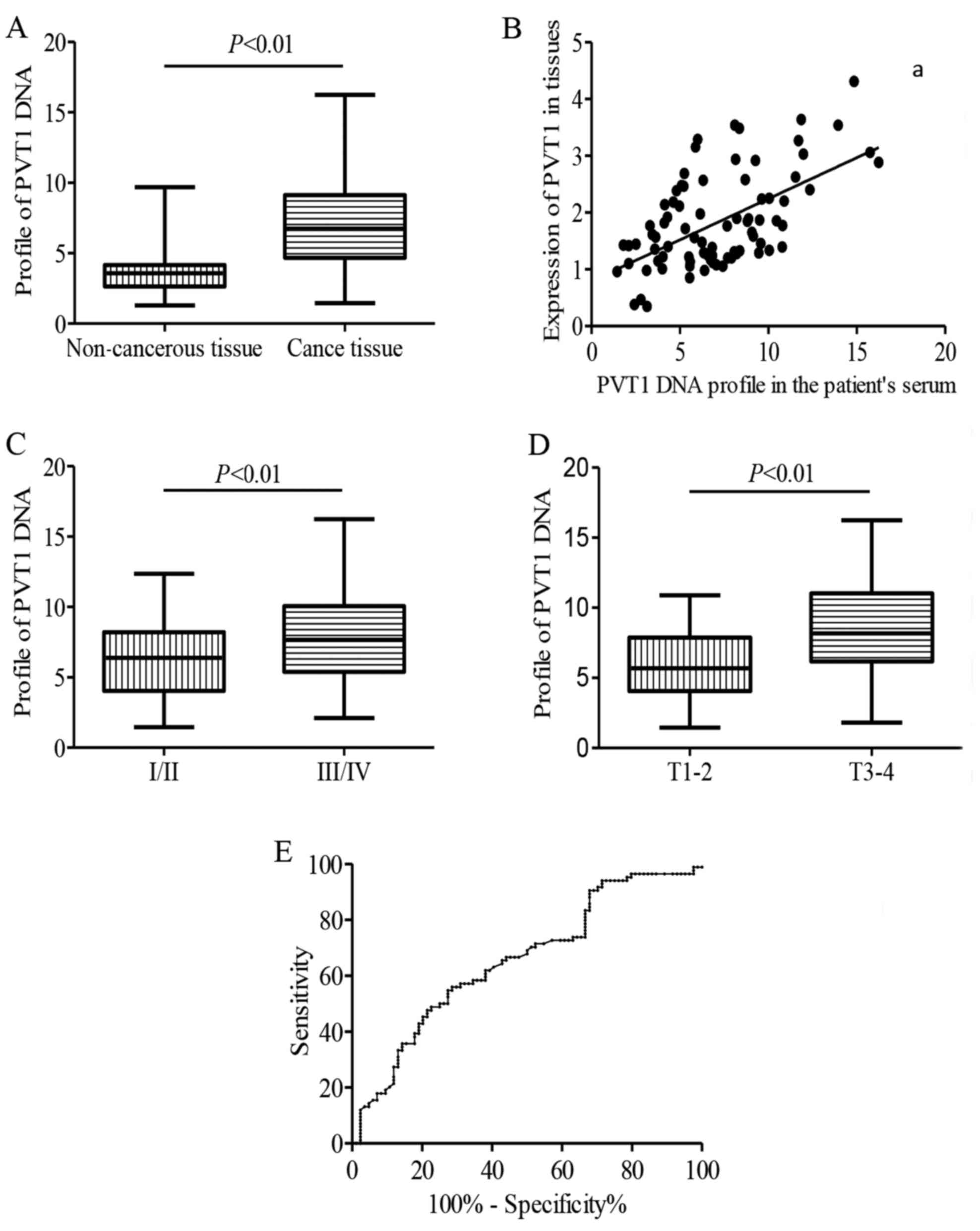
To detect DNA profiles of PVT1 in serum, qPCR
without reverse transcription using serum from the patients and
healthy individuals were performed. The relative contents of PVT1
DNA were higher in the breast cancer group than that in healthy
individuals, corresponding to a fold-change of 2.01 (Fig. 3A). A significant positive
association is showed between the PVT1 in breast cancer tissues and
the PVT1 DNA expression levels in the patient serum
(r2=−0.422, P=0.004; Fig.
3B). Next, the correlation of PVT1 DNA profiles with clinical
features of breast cancer patients were examined further. As shown
in Fig. 3C and D, copy number of
PVT1 DNA was also associated with tumor size (P<0.05) and high
histological grade (P<0.01).
We then assessed the diagnostic performance
circulating DNA level of PVT1 by ROC analysis in breast cancer
patients and healthy control individuals, the AUC was 0.66,
indicating that circulating PVT1 is valuable in discriminating
breast cancer patients from normal individuals (Fig. 3E).
Downregulation of PVT1 inhibits
proliferation and is inversely correlated with p21 in breast
cancer
To achieve cognition into the biological functions
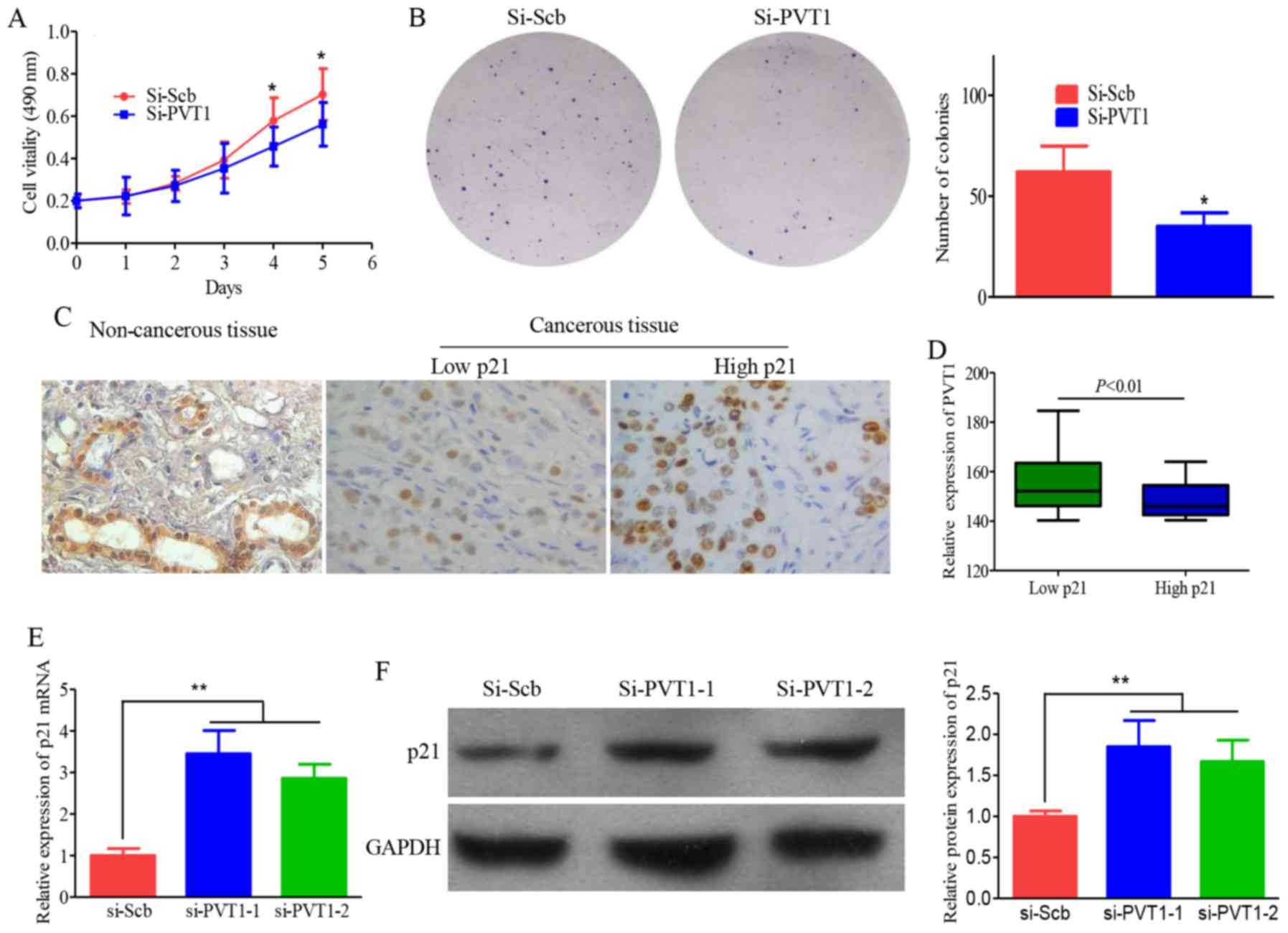
of PVT1 on breast cancer, PVT1 expression was downregulated by
transfecting PVT1 siRNA into the MCF-7 and MDA-MB-231 cells,
employing the scramble siRNA as a negative control. Results of cell
proliferation showed that downregulation of PVT1 expression
inhibited proliferation ability of MCF-7 and MDA-MB-231 cells
significantly (Fig. 4A). Moreover,
colony formation assays exhibited that the clonogenic survival was
significantly declined following inhibition of PVT1 in MCF-7 and
MDA-MB-231 cells compared with control group (Fig. 4B). Above all, our results indicate
that PVT1 plays a pivotal role in proliferation of breast
cancer.
To verify the function role of PVT1 in breast
cancer, IHC was used to determine the expression of p21 protein in
breast cancer and corresponding non-tumorous tissues. Most of the
non tumorous tissues exhibited strongly positive immuno- staining
of p21 protein. In contrast, the corresponding breast cancer
tissues showed negative or weakly positive immunostaining of p21
(Fig. 4C). Further analysis
indicated that the expression of PVT1 was inversely correlated with
p21 in breast tissues (Fig. 4D). To
further understand the controlling relationship between PVT1 and
p21, protein levels of p21 were examined in breast cancer cells
subject to si-PVT1 transfection. Downregulation of PVT1 augmented
p21 mRNA and protein expression (Fig.
4E and F).
Discussion
PVT1 oncogene encodes a long non-coding RNA which
locates to the region 8q24 (7).
Amplification of PVT1 is a common event in various malignant
tumors, such as breast, serous ovarian (12) and colorectal cancer (10). Amplification and upregulation of
PVT1 have been associated with reduced survival duration in
patients.
In the present study, our results illustrated that
PVT1 expression was significantly upregulated in breast cancer
tissues compared with adjacent non-tumorous tissues. Moreover,
higher profiles of PVT1 in breast cancer patients were associated
with lymph node metastasis and tumor size. Additionally, breast
cancer patient with higher PVT1 expression seemed to have a shorter
survival. These results demonstrated that the increase in copy
number contributed to PVT-1 upregulation in breast cancer and
showed that PVT1 might closely connect with the development of
breast cancer, and might be applicable as a new biomarker for
evaluating prognosis of breast cancer patients.
A recent screening for liver oncofetal lncRNAs in a
mouse model for HCC proved the function of PVT1 in regulating
proliferation and the same phenotype was confirmed in human HCC
cell lines (14). Ectopic
expression of PVT1 can contribute to cell proliferation, accelerate
cell cycle, and lead to the achievement of stem cell-like
characteristics by NOP2 (14).
Significantly increased PVT1 blocked the cell cycle in a G1 phase
by regulating expression of p15 and p16 epigenetically via binding
to EZH2 in gastric cancer tissues (15).
p21 is a cell cycle control element and a CDK
inhibitor. p21 could modulate G1 restriction point and G1/S
checkpoint via binding with cyclin-CDK complexes and inhibiting
cyclin-CDK complex kinase activity (16). Expression of p21 is closely
regulated by p53, through this mechanism p21 is involved in the
p53-dependent G1 cell cycle check point resulting from multiple
stress stimuli (17,18). Here, we demonstrated that the
expression of PVT1 was negatively associated with the expression of
p15 and p21 in breast cancer tissues. We also inhibited PVT1 in
breast cancer cells, likewise, the suppression of PVT1
downregulated the expression of p21. These results indicate that
the tumor promoting activity of PVT1 is partially dependent on the
negatively regulation on p21. Considering the combination of PVT1
with EZH2 epigenetically regulated p15 and p16 in Trans (15), and further studies are ongoing to
define the detail regulatory mechanism of PVT1 on the expression of
p21.
During the past decade, cell-free circulating
nucleic acids in plasma, serum and other body fluids have a
potential to change the way we make a diagnosis. Circulating DNA,
miRNA and mRNA in blood may be useful for the detection of various
human diseases (19,20). Circulating DNA in blood may be a
very promising biomarker for precise diagnosis and treatment for
breast cancer (20–22). Because the amplification of
chromosomal 8q24 transcribing PVT1 has been identified in a number
of cancers, including breast cancer, thus, we hypothesized that the
circulating PVT1 DNA might be a useful marker for filtering breast
cancer patients from healthy individuals.
In summary, in the present research, PVT1 was showed
markedly unregulated in breast cancer tissues compared with
adjacent non-tumorous tissues and upregulation of PVT1 might act as
an independent prognostic factor for the survival of breast cancer
patients. Furthermore, PVT1 played a pivotal part on the regulation
of p21 expression. In addition, we detected profiles of PVT1 DNA in
serum; compared with the RNA, DNA is the main form of PVT1-derived
nucleic acid segment in serum, our results showed that circulating
PVT1 DNA was apparently upregulated in breast cancer patients.
Taken together, our results reveal that PVT1 plays a very important
part in the development of breast cancer and might be potentially
used for targeted therapies in breast cancer.
Acknowledgements
The present study was supported by grants from the
National Nature Scientific Foundation of China (81573717) and the
Natural Science Foundation of Shandong Province (ZR2015HL064).
References
|
1
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kunej T, Obsteter J, Pogacar Z, Horvat S
and Calin GA: The decalog of long non-coding RNA involvement in
cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 51:344–357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang GY, Zhu YY and Zhang YQ: The
functional role of long non-coding RNA in digestive system
carcinomas. Bull Cancer. 101:E27–E31. 2014.PubMed/NCBI
|
|
6
|
Song X, Cao G, Jing L, Lin S, Wang X,
Zhang J, Wang M, Liu W and Lv C: Analysing the relationship between
lncRNA and protein-coding gene and the role of lncRNA as ceRNA in
pulmonary fibrosis. J Cell Mol Med. 18:991–1003. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lancaster JM, Dressman HK, Whitaker RS,
Havrilesky L, Gray J, Marks JR, Nevins JR and Berchuck A: Gene
expression patterns that characterize advanced stage serous ovarian
cancers. J Soc Gynecol Investig. 11:51–59. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Popescu NC and Zimonjic DB:
Chromosome-mediated alterations of the MYC gene in human cancer. J
Cell Mol Med. 6:151–159. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ghesquières H, Larrabee BR, Casasnovas O,
Maurer MJ, McKay JD, Ansell SM, Montgomery D, Asmann YW, Farrell K,
Verney A, et al: A susceptibility locus for classical Hodgkin
lymphoma at 8q24 near MYC/PVT1 predicts patient outcome in two
independent cohorts. Br J Haematol. Sep 9–2016.(Epub ahead of
print). doi: 10.1111/bjh.14306. View Article : Google Scholar
|
|
12
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Wang H, Zhang J, Zhai L, Chen W and
Zhao C: miR-199a-5p regulates β1 integrin through Ets-1 to suppress
invasion in breast cancer. Cancer Sci. 107:916–923. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W, et al: Oncofetal long noncoding
RNA PVT1 promotes proliferation and stem cell-like property of
hepatocellular carcinoma cells by stabilizing NOP2. Hepatology.
60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mu C, Jia P, Yan Z, Liu X, Li X and Liu H:
Quercetin induces cell cycle G1 arrest through elevating Cdk
inhibitors p21 and p27 in human hepatoma cell line (HepG2). Methods
Find Exp Clin Pharmacol. 29:179–183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang XQ, Yang CY, Rao XF and Xiong JP:
Plumbagin shows anti-cancer activity in human breast cancer cells
by the upregulation of p53 and p21 and suppression of G1 cell cycle
regulators. Eur J Gynaecol Oncol. 37:30–35. 2016.PubMed/NCBI
|
|
18
|
Liu W, Dai Q, Lu N, Wei L, Ha J, Rong J,
Mu R, You Q, Li Z and Guo Q: LYG-202 inhibits the proliferation of
human colorectal carcinoma HCT-116 cells through induction of G1/S
cell cycle arrest and apoptosis via p53 and p21WAF1/Cip1
expression. Biochem Cell Biol. 89:287–298. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eng C: Circulating DNA biomarkers: A
primer for metastatic colorectal cancer? Lancet Oncol. 16:878–879.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hällström TM, Puhka M and Kallioniemi O:
Circulating tumor DNA in early-stage breast cancer: Personalized
biomarkers for occult metastatic disease and risk of relapse? EMBO
Mol Med. 7:994–995. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fackler MJ, Bujanda Z Lopez, Umbricht C,
Teo WW, Cho S, Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C,
et al: Novel methylated biomarkers and a robust assay to detect
circulating tumor DNA in metastatic breast cancer. Cancer Res.
74:2160–2170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schwarzenbach H and Pantel K: Circulating
DNA as biomarker in breast cancer. Breast Cancer Res. 17:1362015.
View Article : Google Scholar : PubMed/NCBI
|