Introduction
Gastric cancer is one of the most common
malignancies worldwide and causes one million cancer-related deaths
each year (1). Advanced disease
stage and metastasis are often present at diagnosis due to the lack
of specific symptoms and limitation of early diagnostic methods,
resulting in a poor prognosis with a less than 10% 5-year survival
rate (2). As an effective means of
treatment for gastric cancer, molecular-targeted therapy is
important both in clinical research and practice.
Epidermal growth factor receptor (EGFR) is a crucial
member of the HER/erbB family of receptor tyrosine kinases (RTKs),
which includes HER1 (EGFR/erbB1), HER2 (neu, erbB2), HER3 (erbB3)
and HER4 (erbB4) (3). The EGFR
signaling pathway is initiated by the binding of EGF, one of the
major ligands, to EGFR, leading to the activation of downstream
signaling cascades, and finally to the proliferation, invasion and
metastasis of tumor cells, the inhibition of apoptosis of tumor
cells, and the promotion of angiogenesis of tumors, making it a
paramount target in various types of cancer (4–6).
Studies have revealed a high level of expression of EGFR in lung
(7), breast (8), gastric (9), prostate (10) and head and neck cancer (11), and a high expression indicates a
worse prognosis (12). There are
two types of anti-EGFR agents designed to inhibit the activity of
EGFR: small-molecule tyrosine kinase inhibitors (TKIs) and
monoclonal antibodies. But, unlike their fruitful efficacy in lung
cancer, neither type of agent has brought patients with gastric
cancer survival benefits according to the present clinical research
(13). Thus, further research in
anti-EGFR targeted therapy in gastric cancer is warranted.
MicroRNAs have been known to play important roles in
the tumorigenesis and progression of various types of cancer.
Studies have shown that aberrant expression is one of the
mechanisms of carcinogenesis, invasion and metastasis of gastric
cancer (14,15). Therefore, further investigation of
miRNAs in gastric cancer could help us to better understand the
initiation and the development of gastric cancer and may provide
new approaches in targeted therapy. Dysregulation of EGFR and
miRNAs has been known to be interrelated with the carcinogenesis
and development of gastric cancer. But few studies have revealed
the relationship between miRNAs and EGFR in gastric cancer
(16,17).
In this study, bioinformatic prediction and
experimental evidence were provided for the specific binding
between miR-455 and EGFR. Then the suppression of EGFR protein
expression by miR-455 was evaluated. The tumor-promoting effect of
EGFR was further assessed in gastric cancer cells. Our findings
provide evidence of the function of miR-455 in gastric cancer and
help to better understand the modulation of signaling pathways in
gastric cancer.
Materials and methods
Human tissues
Histologically confirmed gastric cancer tissues and
paired adjacent non-cancerous tissues were obtained from patients
who underwent partial or total gastrectomy at Tianjin Medical
University Cancer Institute and Hospital (Tianjin, China). Each
cancer tissue was confirmed as adenocarcinoma pathologically.
Written informed consent was provided by each patient, and all
aspects of this study were approved by the Ethics Committee of
Tianjin Medical University Cancer Institute and Hospital. Tissues
were immediately frozen in liquid nitrogen after surgery and were
stored at −80°C.
Cell line and culture
The human gastric cancer cell line SGC-7901 and
human embryo kidney epithelial cell line HEK293T were resuscitated
from the Cancer Cell Biological Laboratory of Tianjin Medical
University Cancer Institute and Hospital. SGC-7901 cells were
cultured in RPMI-1640 medium (Gibco, Rockville, MD, USA) and
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium
(DMEM) (Gibco), both containing 10% fetal bovine serum (FBS)
(Gibco) and 1% penicillin/streptomycin (Solarbio Science &
Technology Co., Ltd., Beijing, China) in a humidified incubator at
37°C with 5% CO2. Cells were grown in sterilized culture
dishes.
Cell transfection
Cell transfection was conducted through
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) method after the
cells were seeded in appropriate plates. miR-455 mimics and miR-455
inhibitor were purchased with the corresponding controls (RiboBio
Co., Ltd., Guangzhou, China) and were used to upregulate or
downregulate the level of miR-455. The overexpressing lentivirus
and the control lentivirus were purchased from GenePhama (Shanghai,
China) and were used to upregulate or downregulate the expression
of EGFR. Both the overexpression and control lentivirus contained
green fluorescence for the identification of the infection
efficiency after the cells were infected for 36–72 h. The siRNA
sequence targeting human EGFR and a control siRNA were purchased
from Santa Cruz Biotechnology (sc-29301). The cells were harvested
after transfection or infection to isolate total RNA and total cell
lysates for qPCR and western blot analysis, respectively.
RNA isolation and quantitative
RT-PCR
TRIzol reagent (Invitrogen) was used to isolate the
total RNA of the cultured SGC-7901 cells and the obtained tissues
according to the manufacturer's protocol. Nanodrop 1000
spectrophotometer (Thermo Fisher Scientific, USA) was used to
confirm the quality and the concentration of the isolated RNA.
First-strand cDNA was synthesized from 1 µg of total
RNA through reverse transcription reaction. The reaction was
carried out as follows: 16°C for 15 min, 42°C for 60 min, 85°C for
5 min and maintained at 4°C. The cDNA was stored at −20°C after the
reaction.
The level of miR-455 was detected by TaqMan miRNA
probes (Applied Biosystems, USA). Gene-specific PCR products were
assayed using a CFX96 real-time PCR system. The reaction conditions
were initiated by a 5-min hold at 95°C, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing/extension at 60°C for
1 min. All the reactions were conducted three times. U6 snRNA was
used as an internal control for miRNA. The mRNA levels of EGFR were
analyzed by SYBR-Green fluorescent method and the final results
were normalized to glyceraldehyde-3-phosphate dehydrogenase
(GAPDH). The reaction was as follows: 95°C for 30 sec following 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The relative levels
of the miRNA and mRNA were calculated by ΔCt (cycle threshold) and
normalized to the control using the equation 2−ΔCt, ΔCt
= Ctgene - Ctcontrol.
Protein extraction and western blot
analysis
Lysates were obtained after the cells and tissues
were lysed in RIPA buffer with freshly added PMSF. Subsequently,
~50 µg of protein was separated using SDS-PAGE gels and transferred
to PVDF membranes. Immunoblotting was then conducted at 4°C
overnight with monoclonal anti-EGFR antibodies (1:100,000; Abcam)
and monoclonal anti-GAPDH antibodies (1:2,000; Santa Cruz
Biotechnology) after blocking with 2% BSA. An enhanced
chemiluminescence system kit (Millipore, USA) was used to visualize
the membranes after incubation with HRP-coupled anti-mouse/rabbit
IgG (1:2,000; Santa Cruz Biotechnology, USA) at 37°C for 1 h with
the secondary antibody.
Luciferase reporter assay
The amplified PCR products of human wild-type EGFR
and a mutant EGFR in which the predicted 3′-untranslated region
(3′UTR) miR-455 targeting regions were inserted into the reporter
plasmid (Ambion, USA). Lipofectamine 2000 (Invitrogen) method was
used for cell transfection with a reporter plasmid, β-galactosidase
expression vector (Ambion), miR-455 mimics, miR-455 inhibitors, or
a corresponding negative control RNA.
Cell proliferation assay
EdU (RiboBio) was added to the cell culture medium
at a final concentration of 50 µM for a 5-h incubation at 37°C
after transfection or infection. After fixation in 4%
paraformaldehyde and treatment with 0.5% Triton-X for 15 min, the
cells were incubated at room temperature in the dark with Apollo
after treatment with 4% paraformaldehyde for fixation and 0.5%
Triton-X for permeabilization. Nuclei were stained by Hoechst. Five
random fields were selected to calculate the ratio between
EdU-labeled cells and the total cells.
Cell migration assay
Cells (105) were seeded on a Boyden
chamber with 200 µl of serum-free medium after transfection and
infection. Medium (600 µl) with 10% serum was added to the lower
chamber for chemotaxis. The membranes of the Boyden chambers were
fixed and stained after 24 h of incubation. Five random visual
fields were selected for analysis.
Wound scratch assay
SGC-7901 cells that had undergone different
treatments were seeded in 6-well plates. Each well was scraped with
a 20 µl pipette tip to create 1 or 2 linear regions devoid of cells
after the cells reached 90% confluence. RPMI-1640 medium with 2%
FBS (both from Gibco) were used subsequently for cell culture. We
monitored wound healing at 0, 12, and 24 h after scraping. Five
random fields of each well were selected for analysis.
Immunohistochemical assay
Paraffin-embedded specimens of gastric cancer
tissues and the paired non-cancerous tissues were incubated with an
anti-EGFR monoclonal antibody (1:100; Abcam) following antigen
retrieval. The DAB system (Zhongshan Jinqiao, China) was used to
identify the positive reaction. Five random fields were selected
for each specimen.
Bioinformatics prediction of miRNA
target
TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRanda (http://www.microrna.org/) were combined for the
prediction of the target of miR-455.
Statistical analysis
Each experiment was performed at least three times.
P<0.05 was considered as statistically significant and
differences were assessed using the Student's t-test. Data are
expressed as the median values ± SE and analyzed using the
Student's t-test. In this study, ‘*’ indicates P<0.05, ‘**’
indicates P<0.01, and ‘***’ indicates P<0.001.
Results
EGFR protein levels are upregulated in
gastric cancer tissues
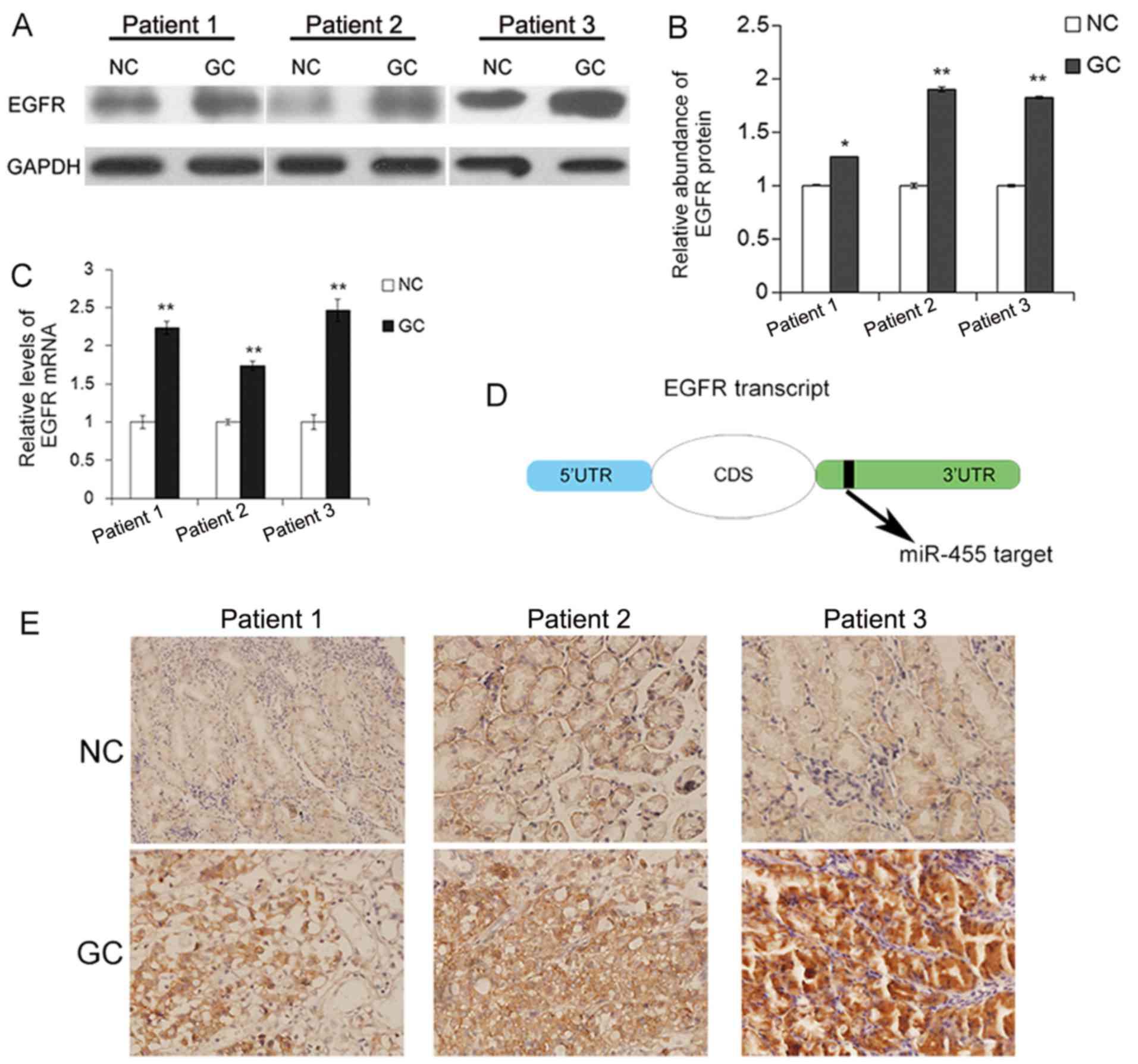
The expression pattern of EGFR in human gastric
cancer tissues was first evaluated by western blot analysis. As
previously reported, the EGFR protein level was significantly
upregulated in the gastric cancer tissues (Fig. 1A and B). As is well known, an
increase in EGFR copies is one of the causes for the upregulation
of EGFR. Here, the mRNA level was upregulated in the cancerous
tissues (Fig. 1C), which may be due
to a variety of factors. The predicted target of miR-455 on the
transcript of EGFR is shown in Fig.
1D. IHC assays revealed that EGFR distributed in the cytoplasm
and cancer tissue specimens exhibited a higher positive rate,
compared with paired non-cancerous tissues, which was consistent
with previous studies (Fig.
1E).
miR-455 acts as a potential upstream
regulator of EGFR
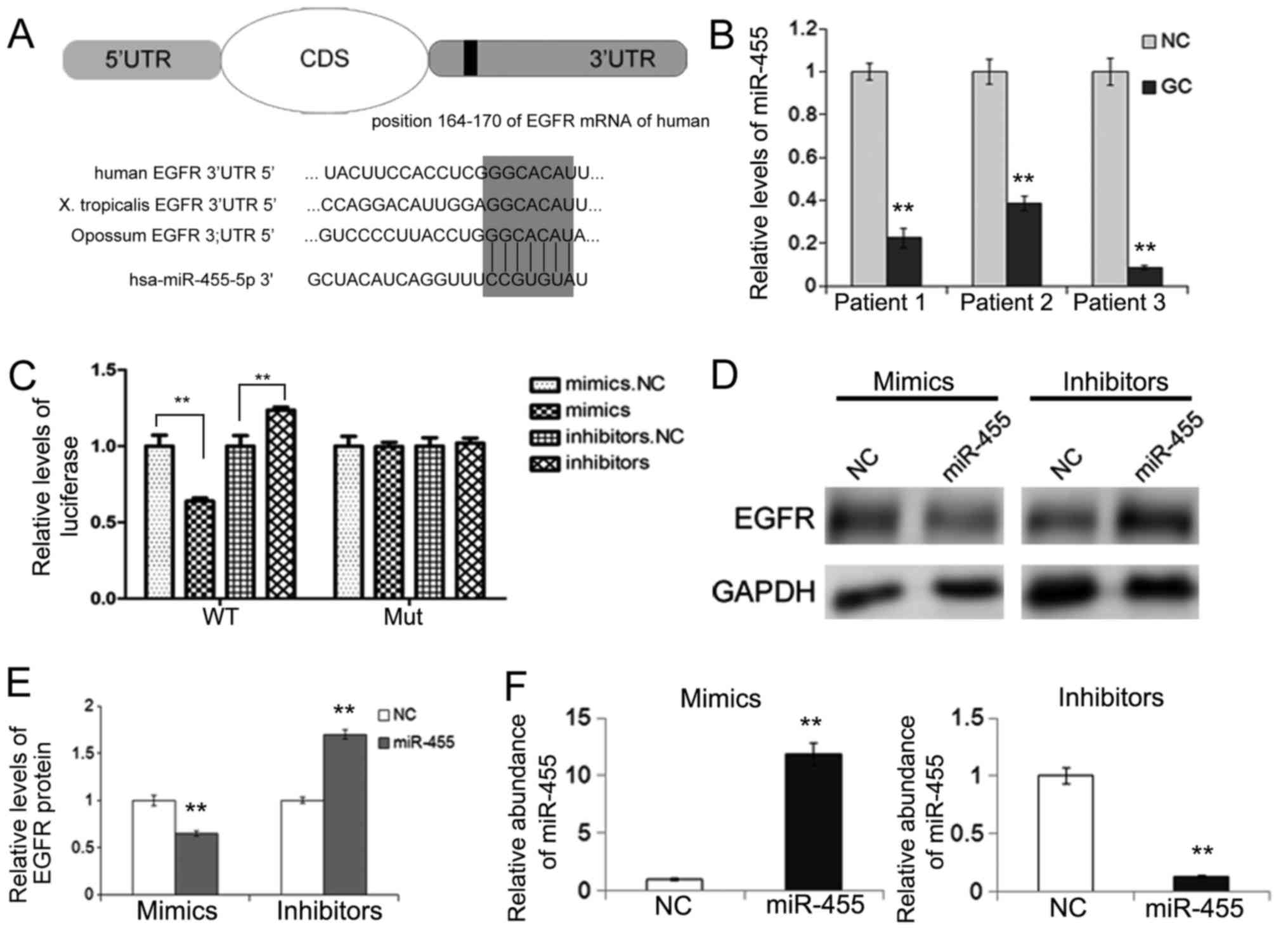
Bioinformatic prediction indicated a direct binding
of miR-455 to the region in the 3′UTR of EGFR mRNA. The binding
sites are conserved among several species (Fig. 2A). The evaluation of miRNA levels
using real-time PCR revealed that compared with para-carcinoma
tissues, miR-455 was markedly downregulated in the gastric cancer
tissues (Fig. 2B). The inverse
expression patterns of the EGFR protein and miR-455 prompted
further investigation into the potential effect of miR-455 on EGFR.
Therefore, miR-455 was chosen for further experiments to identify
its role in gastric cancer.
Validation of EGFR as a direct target
of miR-455
Direct evidence of the combination of miR-455 and
EGFR was needed despite the bioinformatic prediction and the
inverse correlation between miR-455 and the EGFR protein level. A
dual luciferase reporter assay was conducted to evaluate the direct
interaction between miR-455 and EGFR. The relative luciferase
activity was clearly suppressed by the co-transfection of miR-455
mimics and the luciferase reporters containing the predicted
binding region of the wild-type 3′UTR of EGFR. However, the
suppressive effect was lost when the binding site was mutated.
Furthermore, the co-transfection of miR-455 inhibitors and the
reporter plasmid with the wild-type EGFR 3′UTR resulted in an
increase in luciferase activity (Fig.
2C).
The expression of EGFR protein and mRNA were
evaluated when SGC-7901 cells were transfected with miR-455 mimics
or inhibitors, which was confirmed by qRT-PCR (Fig. 2F). As shown in Fig. 2D and E, upregulation of miR-455 led
to a sharp decrease in the EGFR protein, whereas downregulation of
miR-455 enhanced the expression of EGFR in the gastric cancer
cells.
The results indicated that miR-455 regulated EGFR by
directly binding with the specific region of the 3′UTR of EGFR
mRNA, thus rendering it as an important regulator of EGFR.
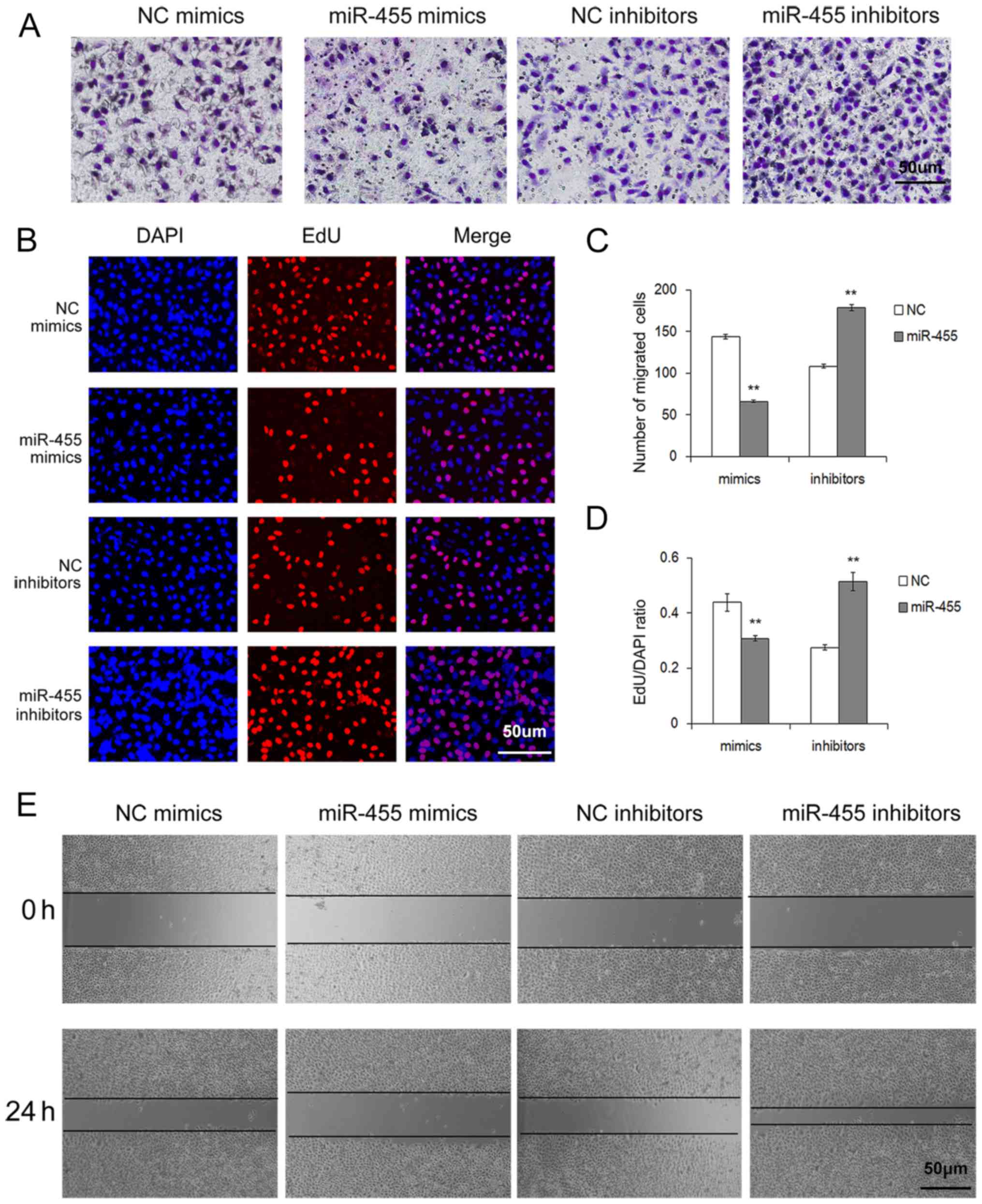
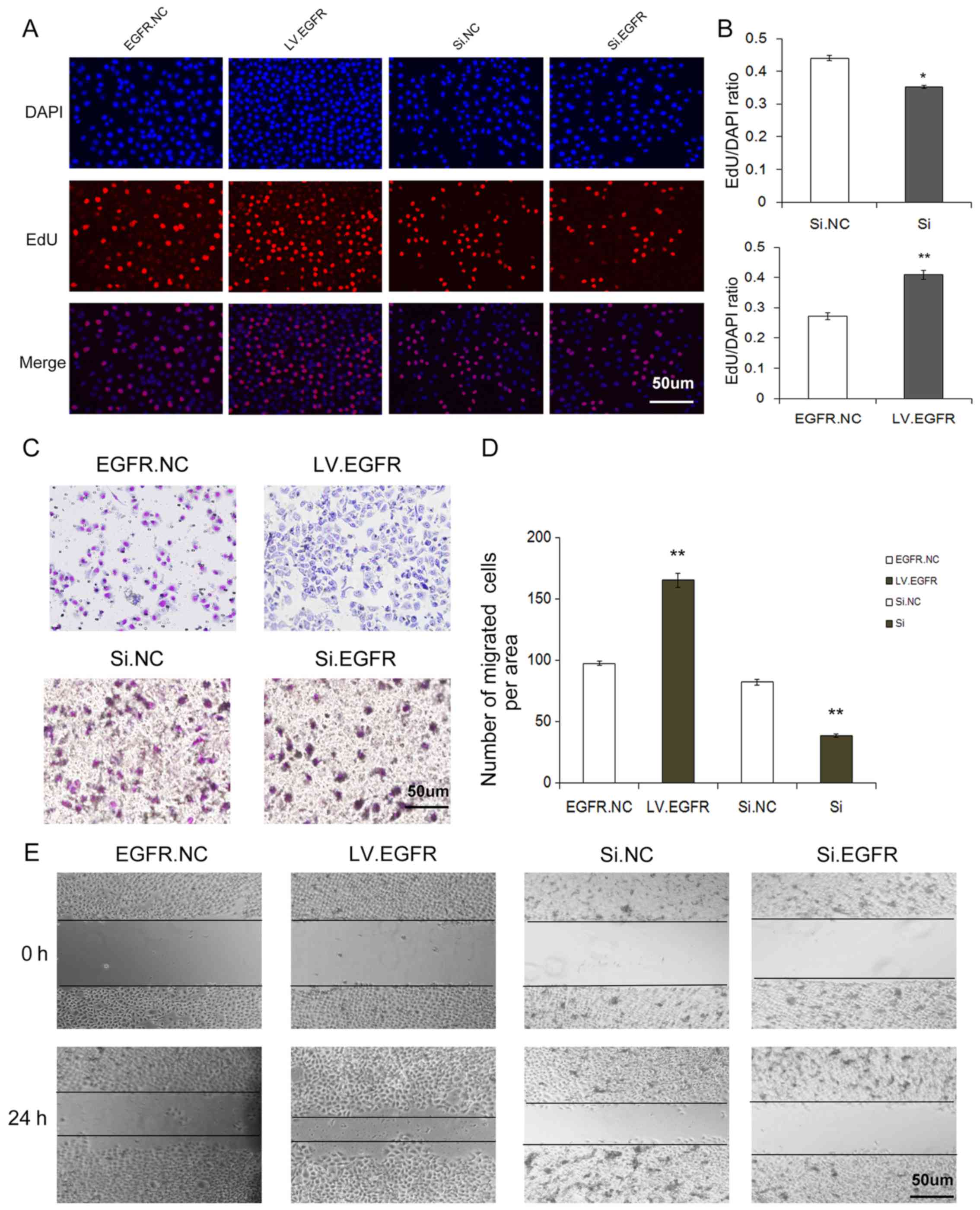
Upregulation of miR-455 inhibits the
proliferation and migration of SGC-7901 cells
To further assess the role of miR-455 in gastric
cancer cells, EdU cell proliferation assay, Transwell migration
assay and wound healing assay were performed respectively.
The cell proliferation rate via the Cell-Light EdU
DNA cell kit was used to identify the proliferation ability of
SGC-7901 cells after treatment with miR-455 mimics or inhibitors.
As shown in Fig. 3B and D,
upregulation of miR-455 resulted in a sharp decrease in cell
proliferation, whereas downregulation of miR-455 led to an increase
in cell proliferation.
The effects of miR-455 on cell migration were
evaluated through Transwell assays. As was expected, upregulation
of miR-455 expression strongly inhibited the migration of gastric
cancer cells, whereas the downregulation of miR-455 expression
promoted cell migration (Fig. 3A and
C).
A wound-healing assay was also conducted to further
examine the migration ability of SGC-7901 cells. Similar to the
Transwell assay result, upregulation of miR-455 significantly
suppressed the migration of gastric cancer cells (Fig. 3E).
These aforementioned in vitro results
indicated that miR-455 exerts a cancer-suppressive role in gastric
cancer cells and provides evidence that miRNA participates in the
processes of gastric cancer.
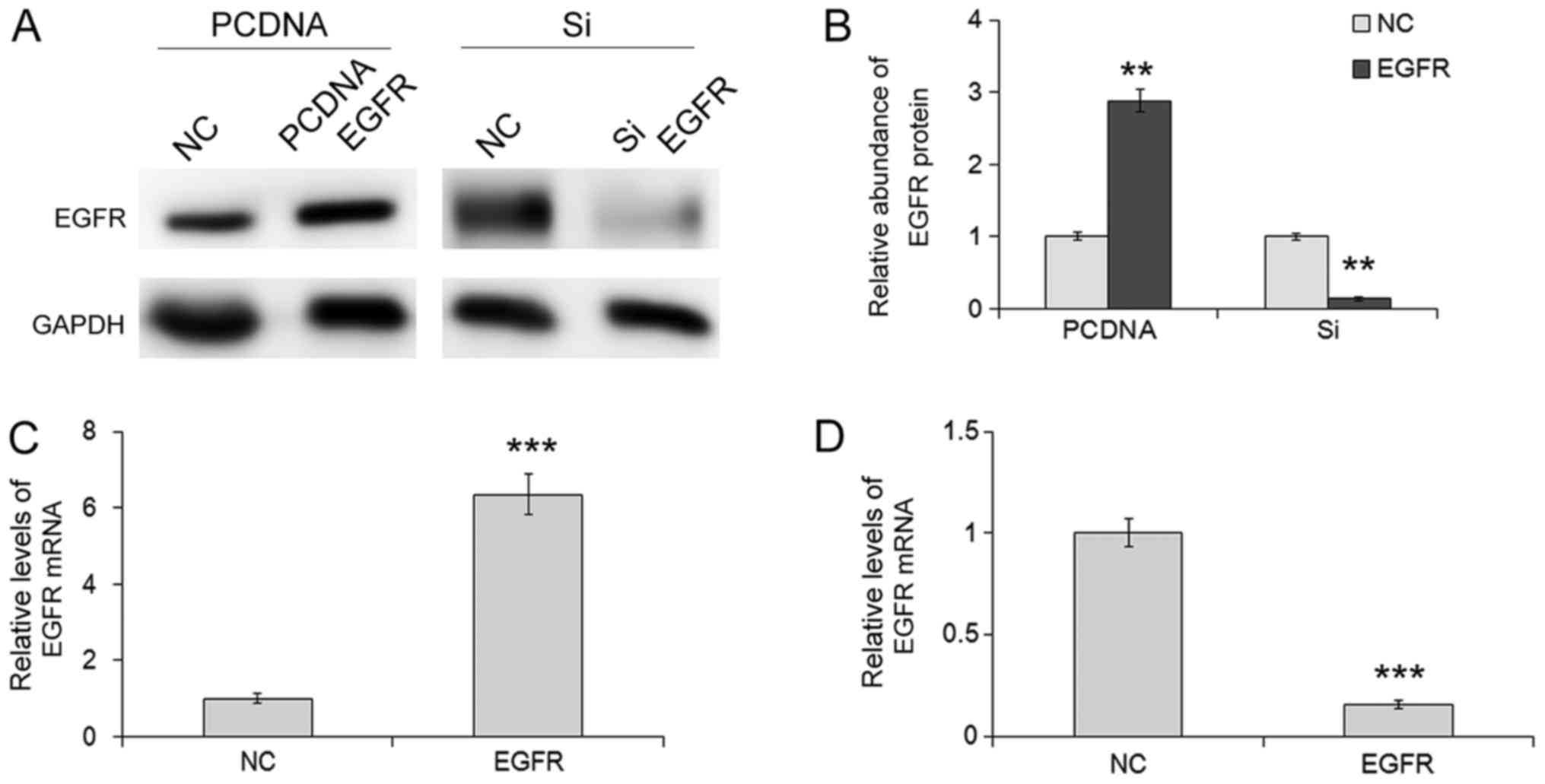
Overexpression of EGFR promotes
proliferation and migration of SGC-7901 cells
Next, we investigated the effects of EGFR on cell
proliferation and migration by overexpressing and silencing EGFR.
An overexpression lentivirus was constructed to overexpress EGFR,
and the siRNA sequence targeting human EGFR was designed to knock
down the expression of EGFR. It was revealed in our study that
infection of the EGFR overexpression lentivirus led to a
significant increase or siRNA led to a decrease in protein and mRNA
levels in SGC-7901 cells, compared with the control (Fig.4). Knockdown of EGFR by siRNA
downregulated the protein expression and suppressed the
proliferation and migration, whereas overexpression of EGFR
resulted in a higher rate of proliferation and migration. In
addition, we used EdU assay, Transwell assay and wound-healing
assay to assess SGC-7901 cells in which EGFR was knocked down and
the cells exhibited a significantly lower rate of proliferation and
decreased migration, whereas the cells overexpressing EGFR
exhibited a significantly higher rate of proliferation and
decreased migration (Fig. 5).
Discussion
A substantial number of studies on certain miRNAs or
miRNA patterns have been published in recent years, indicating that
miRNAs play an important role in the initiation and development of
gastric cancer. For example, miR-1, miR-206, miR-34a and miR-144
directly target the MET gene and downregulate its expression, thus
inhibiting gastric cancer cell proliferation and migration
(18–21). miR-196a/-196b promotes cell
metastasis by targeting radixin in gastric cancer (22). miRNAs are also relevant to the
chemosensitivity of cancer cells. For example, miR-23b-3p inhibited
autophagy by targeting ATG12 and HMGB2 and made gastric cancer
cells sensitive to chemotherapy (23). miR-135 can reverse resistance to
chemotherapy and promote apoptosis by negative regulation of MCL1
(24). Therefore, identifying the
function of miRNA can help us to better understand the
carcinogenesis and progression of gastric cancer.
In the present study, bioinformatic prediction for
upstream miRNAs of EGFR and an inverse expression pattern of
predicted miR-455 and EGFR in human gastric cancer and
para-carcinoma tissues indicated a potential binding of miR-455
with the 3′UTR of EGFR mRNA. Then further verification through a
reporter assay provided direct evidence for the specific binding.
The subsequent experiments revealed that miR-455 could suppress the
proliferation and migration of gastric cancer cells by negatively
regulating the expression of EGFR. Furthermore, the
cancer-promoting function of EGFR was further confirmed in gastric
cancer cells through the silencing and overexpression of EGFR. It
was demonstrated in our study that miR-455 was significantly
downregulated in gastric cancer tissues and exhibited a
cancer-suppressive function in gastric cancer.
EGFR is a transmembrane protein with cytoplasmic
kinase activity. Homodimerization and/or heterodimerization induced
by ligands causes autophosphorylation of the cytoplasmic domain of
the receptor, thus activating downstream cell signaling pathways,
such as the RAS-RAF-MEK-MAPK pathway (25,26),
the PI3K-PTEN-AKT pathway (27,28)
and the STAT pathway (29,30), and finally facilitating the
proliferation, invasion and metastasis and the suppression of
apoptosis of cancer cells. Thence, suppression of EGFR is a notable
method for the treatment of cancers.
There are two types of anti-EGFR targeting
therapeutic agents based on the aforementioned theory: monoclonal
antibodies that bind to extracellular domain of EGFR thereby
preventing EGFR from binding with its endogenous ligands and
tyrosine kinase inhibitors that target the cytoplasmic TK domain
(31). An EXPAND study revealed
that, in contrast to the favorable results in colorectal, head and
neck and lung cancer, cetuxiumab, a type of monoclonal antibody,
combined with chemotherapy brought no survival benefit to gastric
cancer patients (32). Another
monoclonal antibody, panitumumab induced a decrease in the survival
of patients with gastric cancer, according to a REAL-3 study
(33). Moreover, some phase II
trials with EGFR TKIs, such as gefitinib or erlotinib revealed only
modest benefits when used as monotherapy or combined with
chemotherapy (34–36). Thus, more exploration is warranted
in the targeted therapy against EGFR in gastric cancer, and
negative regulation of miRNAs on target genes provide a new
approach for targeted therapy. The current study demonstrated the
cancer-promoting function of EGFR in gastric cancer cells and a
post-transcriptional regulation strategy for EGFR mediated by
miR-455, which may become a new targeted therapy for anti-EGFR
therapy.
To conclude, we demonstrated that miR-455 as a tumor
suppressor inhibits cell proliferation and migration in gastric
cancer. In addition, our study offers a potential targeted
therapeutic method against EGFR in gastric cancer mediated with the
use of miR-455, which effectively inhibits the gene expression of
EGFR via regulation of its 3′UTR mRNA. Future studies should be
focused on the exploration of agents that can deliver anticancer
miRNAs in vivo to suppress the expression of oncogenes.
Acknowledgements
This study was funded by grants from the National
Natural Science Foundation of China (nos. 81372394, 81602158,
81572321 and 81602156) and Tianjin Health and Family Planning
Commission Foundation of Science and Technology (15KG142). This
study was also funded by Tianjin Science Foundation (no.
15JCYBJC28200) and Doctoral Foundation of Tianjin Medical
University Cancer Institute and Hospital (B1502).
Glossary
Abbreviations
Abbreviations:
|
EGFR
|
epidermal growth factor receptor
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
3UTR
|
3-untranslated region
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hashim D, Boffetta P, La Vecchia C, Rota
M4, Bertuccio P, Malvezzi M and Negri E: The global decrease in
cancer mortality: Trends and disparities. Ann Oncol. 27:926–933.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zahonero C, Sepúlveda JM and Sánchez-Gómez
P: Epidermic growth factor receptor (EGFR) in glioblastomas: the
mechanism of tumorigenesis and its role as a therapeutic target.
Rev Neurol. 61:85–93. 2015.(In Spanish). PubMed/NCBI
|
|
5
|
Weber KL, Doucet M, Price JE, Baker C, Kim
SJ and Fidler IJ: Blockade of epidermal growth factor receptor
signaling leads to inhibition of renal cell carcinoma growth in the
bone of nude mice. Cancer Res. 63:2940–2947. 2003.PubMed/NCBI
|
|
6
|
Mendelsohn J: The epidermal growth factor
receptor as a target for cancer therapy. Endocr Relat Cancer.
8:3–9. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Y, Liu H, Shi X and Song Y: Can EGFR
mutations in plasma or serum be predictive markers of
non-small-cell lung cancer? A meta-analysis. Lung Cancer.
88:246–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee HJ, Seo AN, Kim EJ, Jang MH, Kim YJ,
Kim JH, Kim SW, Ryu HS, Park IA, Im SA, et al: Prognostic and
predictive values of EGFR overexpression and EGFR copy number
alteration in HER2-positive breast cancer. Br J Cancer.
112:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagatsuma AK, Aizawa M, Kuwata T, Doi T,
Ohtsu A, Fujii H and Ochiai A: Expression profiles of HER2, EGFR,
MET and FGFR2 in a large cohort of patients with gastric
adenocarcinoma. Gastric Cancer. 18:227–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weber DC, Tille JC, Combescure C, Egger
JF, Laouiti M, Hammad K, Granger P, Rubbia-Brandt L and Miralbell
R: The prognostic value of expression of HIF1α, EGFR and VEGF-A, in
localized prostate cancer for intermediate- and high-risk patients
treated with radiation therapy with or without androgen deprivation
therapy. Radiat Oncol. 7:662012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Numico G, Russi EG, Colantonio I, Lantermo
RA, Silvestris N, Vitiello R, Comino A, Abrate M, Zavattero C,
Melano A, et al: EGFR status and prognosis of patients with locally
advanced head and neck cancer treated with chemoradiotherapy.
Anticancer Res. 30:671–676. 2010.PubMed/NCBI
|
|
12
|
Normanno N, Maiello MR and De Luca A:
Epidermal growth factor receptor tyrosine kinase inhibitors
(EGFR-TKIs): Simple drugs with a complex mechanism of action? J
Cell Physiol. 194:13–19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ayyappan S, Prabhakar D and Sharma N:
Epidermal growth factor receptor (EGFR)-targeted therapies in
esophagogastric cancer. Anticancer Res. 33:4139–4155.
2013.PubMed/NCBI
|
|
14
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang QX, Zhu YQ, Zhang H and Xiao J:
Altered miRNA expression in gastric cancer: A systematic review and
meta-analysis. Cell Physiol Biochem. 35:933–944. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang XT, Zhang Z, Xin YN, Ma XZ and Xuan
SY: Impairment of growth of gastric carcinoma by miR-133-mediated
Her-2 inhibition. Tumour Biol. 36:8925–8930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu
YP, Gui QJ, Zhang L and Li GQ: miR-7 inhibits the invasion and
metastasis of gastric cancer cells by suppressing epidermal growth
factor receptor expression. Oncol Rep. 31:1715–1722.
2014.PubMed/NCBI
|
|
18
|
Han C, Zhou Y, An Q, Li F, Li D, Zhang X,
Yu Z, Zheng L, Duan Z and Kan Q: MicroRNA-1 (miR-1) inhibits
gastric cancer cell proliferation and migration by targeting MET.
Tumour Biol. 36:6715–6723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng Z, Yan D, Chen X, Huang H, Chen K,
Li G, Zhou L, Zheng D, Tu L and Dong XD: MicroRNA-206: Effective
inhibition of gastric cancer progression through the c-Met pathway.
PLoS One. 10:e01287512015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng Y, Guo JJ, Liu YM and Wu XL:
MicroRNA-34A inhibits the growth, invasion and metastasis of
gastric cancer by targeting PDGFR and MET expression. Biosci Rep.
34:pii: e001122014. View Article : Google Scholar
|
|
21
|
Liu J, Xue H, Zhang J, Suo T, Xiang Y,
Zhang W, Ma J, Cai D and Gu X: MicroRNA-144 inhibits the metastasis
of gastric cancer by targeting MET expression. J Exp Clin Cancer
Res. 34:352015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsai MM, Wang CS, Tsai CY, Chen CY, Chi
HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY, et al:
MicroRNA-196a/-196b promote cell metastasis via negative regulation
of radixin in human gastric cancer. Cancer Lett. 351:222–231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An Y, Zhang Z, Shang Y, Jiang X, Dong J,
Yu P, Nie Y and Zhao Q: miR-23b-3p regulates the chemoresistance of
gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis.
6:e17662015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou L, Qiu T, Xu J, Wang T, Wang J, Zhou
X, Huang Z, Zhu W, Shu Y and Liu P: miR-135a/b modulate cisplatin
resistance of human lung cancer cell line by targeting MCL1. Pathol
Oncol Res. 19:677–683. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giehl K, Skripczynski B, Mansard A, Menke
A and Gierschik P: Growth factor-dependent activation of the
Ras-Raf-MEK-MAPK pathway in the human pancreatic carcinoma cell
line PANC-1 carrying activated K-ras: Implications for cell
proliferation and cell migration. Oncogene. 19:2930–2942. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Troiani T, Napolitano S, Vitagliano D,
Morgillo F, Capasso A, Sforza V, Nappi A, Ciardiello D, Ciardiello
F and Martinelli E: Primary and acquired resistance of colorectal
cancer cells to anti-EGFR antibodies converge on MEK/ERK pathway
activation and can be overcome by combined MEK/EGFR inhibition.
Clin Cancer Res. 20:3775–3786. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Davis NM, Sokolosky M, Stadelman K, Abrams
SL, Libra M, Candido S, Nicoletti F, Polesel J, Maestro R, D'Assoro
A, et al: Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in
breast cancer: Possibilities for therapeutic intervention.
Oncotarget. 5:4603–4650. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lim HJ, Crowe P and Yang JL: Current
clinical regulation of PI3K/PTEN/Akt/mTOR signalling in treatment
of human cancer. J Cancer Res Clin Oncol. 141:671–689. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang L and Fu L: Mechanisms of resistance
to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B. 5:390–401.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu N, Wang SQ, Tan D, Gao Y, Lin G and Xi
R: EGFR, Wingless and JAK/STAT signaling cooperatively maintain
Drosophila intestinal stem cells. Dev Biol. 354:31–43. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Janmaat ML and Giaccone G: The epidermal
growth factor receptor pathway and its inhibition as anticancer
therapy. Drugs Today (Barc). 39 Suppl C:61–80. 2003.PubMed/NCBI
|
|
32
|
Lordick F, Kang YK, Chung HC, Salman P, Oh
SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, et
al: Arbeitsgemeinschaft Internistische Onkologie and EXPAND
Investigators: Capecitabine and cisplatin with or without cetuximab
for patients with previously untreated advanced gastric cancer
(EXPAND): A randomised, open-label phase 3 trial. Lancet Oncol.
14:490–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Waddell T, Chau I, Cunningham D, Gonzalez
D, Okines AF, Okines C, Wotherspoon A, Saffery C, Middleton G,
Wadsley J, et al: Epirubicin, oxaliplatin, and capecitabine with or
without panitumumab for patients with previously untreated advanced
oesophagogastric cancer (REAL3): A randomised, open-label phase 3
trial. Lancet Oncol. 14:481–489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang WP, Wang KN, Gao Q and Chen LQ: Lack
of EGFR mutations benefiting gefitinib treatment in adenocarcinoma
of esophagogastric junction. World J Surg Oncol. 10:142012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dragovich T, McCoy S, Fenoglio-Preiser CM,
Wang J, Benedetti JK, Baker AF, Hackett CB, Urba SG, Zaner KS,
Blanke CD, et al: Phase II trial of erlotinib in gastroesophageal
junction and gastric adenocarcinomas: SWOG 0127. J Clin Oncol.
24:4922–4927. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cappetta A, Lonardi S, Pastorelli D,
Bergamo F, Lombardi G and Zagonel V: Advanced gastric cancer (GC)
and cancer of the gastro-oesophageal junction (GEJ): Focus on
targeted therapies. Crit Rev Oncol Hematol. 81:38–48.
2012.PubMed/NCBI
|