Introduction
Colorectal cancer (CRC) is one of the most common
cancers with a high incidence, and one of the leading causes of
cancer-related deaths both in men and women worldwide (1). A large-scale report showed that
metastatic CRC is refractory to systemic therapy (2). Although there are some therapies for
CRC, such as anticancer drug treatment and antiangiogenic
therapies, the overall survival of CRC patients is always very poor
(3). Identifying the function of
cancer-related genes in the biology of CRC is a critical step in
finding the effective therapeutic approaches for patients with CRC
(4). Recent attention has focused
on the molecular function and mechanism of CRC and reports have
indicated that several members of the Krüppel-like family play a
role in multiple types of cancer, including glioma (5), gastric (6), lung cancer (7) and CRC (8,9).
The KLF (Krüppel-like factors) family which was
named due to their homology with Krüppel, is an evolutionarily
conserved sequence-specific DNA-binding transcriptional regulator
(10,11). KLF family members recognize similar
consensus sites because the members contain three conserved zinc
fingers (12). KLFs play various
and important functions in a number of cellular processes, such as
cell differentiation, pluripotency, inflammation, growth,
proliferation, apoptosis and migration (13–16).
Also, ample evidence suggests that KLF members are involved in the
pathobiology of many human diseases, such as metabolic disorders,
cardiovascular disease and cancer (17).
KLF2 is one of the KLF family members and has a high
expression in fetal and adult lungs, as well as expression in
several other organs, including skeletal muscle, heart, spleen and
kidney (18,19). It has been well documented that KLF2
plays an important role in numerous cellular physiological
processes, including adipogenesis (20), cell differentiation and cell
apoptosis (18). Also, KLF2 has
been associated with many types of cancers, such as non-small cell
lung (21), gastric (22), oral (23), hepatocellular carcinoma (23) and breast cancer (24). Wu and Lingrel (18) revealed that KLF2 also has expression
in CRC cells; however, its specific function is still unknown.
A recent study found that KLF2 could inhibit
hypoxia-inducible factor 1α (HIF-1α) expression and also impair its
function (25). HIF-1α is a central
regulator of hypoxic response in many cell types, including cancer
cells (26). There is convincing
evidence that hypoxia plays an important role in tumor progression,
angiogenesis, distant metastasis and cancer therapy (27,28).
HIF-1α could mediate multiple oncogenes for cancer progression,
including CRC, and inhibition of HIF-1α has been shown to inhibit
the proliferation of human CRC cells (29) and reverses multidrug resistance in
CRC cells (30). Also, Wang et
al (31) showed that HIF-1α can
bind to the Notch target gene to modulate its signaling in cancer
stem cells. The Notch-1 signal also promotes tumorigenesis in CRC
and protects cells from apoptosis (32). Thus, we propose that the
HIF-1α/Notch-1 signal pathway is implicated in the function of KLF2
in CRC cells. We overexpressed KLF2 in CRC cells to examine the
effects of KLF2 on CRC cells growth, and the function of
HIF-1α/Notch-1 signal pathway. The present study demonstrates that
overexpression of KLF2 inhibits colorectal cancer growth via
regulating the HIF-1α/Notch-1 signal pathway.
Materials and methods
Cell culture and transfection
Human SW480 HT29, SW620 and HCT116 CRC cell lines
and the normal colon epithelium cell line FHC were obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA). CRC
cell lines were cultured in Dulbeccos modified Eagles medium
(DMEM), supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin. FHC cells were grown in DMEM/F12 complete
medium. All cells were maintained in a humidified atmosphere of 5%
CO2 at 37°C. SW480 and HCT116 cells were transfected by
PcDNA3.1-KLF2, PcDNA3.1/HIF-1α or PcDNA3.1-Notch-1 recombinant
plasmid by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA)
according to the manufacturers instructions.
Cell survival assay
Human SW480 and HCT116 CRC cells were seeded into
96-well plates at a density of 1×104 cells/well. Cells
were transfected with PcDNA3.1-KLF2, PcDNA3.1/HIF-1α or
PcDNA3.1-Notch-1 plasmid. After 24, 48, 72 and 96 h, CellTiter-Blue
Cell Viability assay (Promega, Madison, WI, USA) was used to detect
the viability of SW480 and HCT116 cells at each time-point
according to the manufacturers instructions.
MTS assay
SW480 and HCT116 CRC cells were seeded into 96-well
plates with the appropriate culture medium and transfected with
PcDNA3.1-KLF2, PcDNA3.1/HIF-1α or PcDNA3.1-Notch-1 plasmid for 48
h. MTS [3,4-(5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy
phenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt] assays was used to
determine cell proliferation according to the manufacturers
protocol (Promega). Cell proliferation was determined as
absorbance, which was measured by a microplate reader at OD490.
Cell apoptosis assay
SW480 cells were seeded in 96-well plates and
treated as described above. Apoptosis was measured after 48 h by
the Cell Death Detection ELISA plus kit (Roche Diagnostics,
Manheim, Germany) according to the manufacturers protocol.
Histone-associated DNA fragments (nucleosomes) were determined
according to the method previously described (33). The concentration of nucleosome for
each group was normalized for total protein.
Caspase-3/7 activity
SW480 cells were cultured and transfected as
described above. Caspase-3/7 activity in cells was measured by
using a Caspase-Glo 3/7 assay kit (Promega) following the
manufacturers protocol.
RNA extraction and quantitative
real-time PCR
KLF2, HIF-1α and Notch-1 mRNA expression was
determined by quantitative real-time PCR (RT-PCR). Total protein
was isolated from SW480 cells by using the RNeasy Plus Mini kit
(Qiagen, Germantown, MD, USA). CDNA was synthesized by reverse
transcription using reverse transcription reagents (Bio-Rad
Laboratories, Hercules, CA, USA). The gene-specific primers used in
the present study are as follows: KLF2 (34), 5-AGACCTACACCAAGAGTTCGCATC-3 (F) and
5-ATC GCACAGATGGCACTGGAATG-3 (R); HIF-1α (28), 5-GT GTTATCTGTCGCTTTGAGTC-3 (F) and
5-GTCTGGCTG CTGTAATAATGTT-3 (R); Notch-1 (28), 5-AAGCTGCATC CAGAGGCAAAC-3 (F) and
5-TGGCATACACACTCCGA GAACAC-3 (R); β-actin (28), 5-CACCCACTCCTCCACCT TTG-3 (F) and
5-CCACCACCCTGTTGCTGTAG-3.
Western blotting
Western blotting was performed according to the
method previously described (28).
Total cellular protein was extracted from SW480 cells by using a
RIPA buffer (150 mM NaCl, 1% NP40, 50 mM Tris, 0.5% sodium
deoxycholate and 0.1% SDS). Protein extractions were separated on
10% SDS-PAGE and then the proteins were transferred to
nitrocellulose (NC) membranes (Sigma-Aldrich, St. Louis, MO, USA).
Rabbit anti-human KLF2, HIF-1α and Notch-1 antibody (Abcam,
Cambridge, MA, USA) was used as a primary antibody and HRP goat
anti-rabbit IgG antibody (Abcam) was used as a secondary antibody.
β-actin was used as the internal control.
Statistical analysis
All data in the present study are presented as means
± SD from at least three experiments in triplicate. Statistical
analyses were performed using SPSS 19.0 statistical software.
Statistical significance for comparisons between groups was carried
out by using Students t-test as well as ANOVA. Test results of
P<0.05 were considered statistically significant.
Results
KLF2 expression is markedly decreased
in CRC cell lines
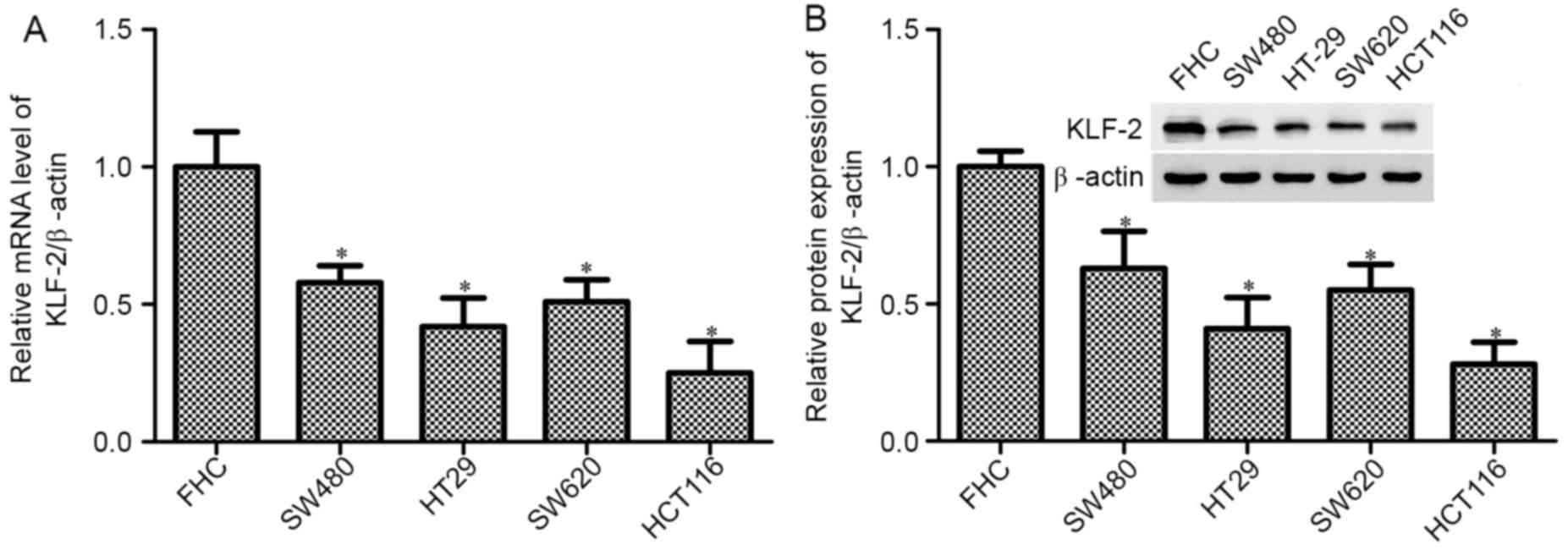
KLF2 has been shown to decrease in various types of
cancer and to act as a tumor suppressor. This study aimed to detect
the function and mechanism of KLF2 in CRC cell lines. Firstly, we
used RT-PCR and western blot analysis to measure the level of KLF2
in controlled normal human colon epithelial cell line FHC, and CRC
cell lines SW480, HT29, SW620 and HCT116. As shown in Fig. 1, the mRNA and protein expression was
significantly reduced in CRC cell lines compared with the control
FHC cell (P<0.05). Between the CRC cell lines, HCT116 cell lines
have obvious effects in the decrease of KLF2, and the SW480 cell
line has the highest level.
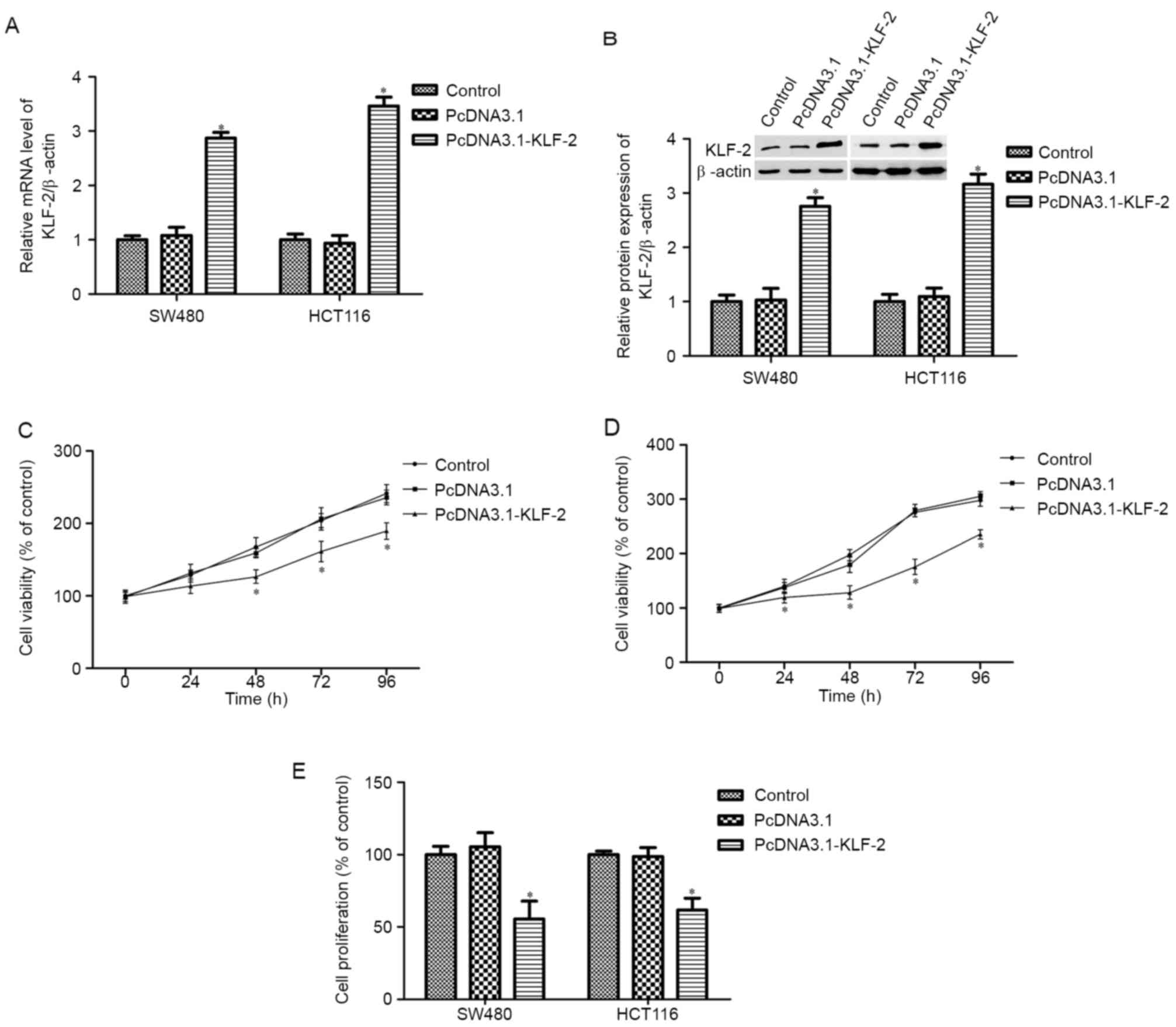
KLF2 overexpression inhibits CRC cells
survival and proliferation
To further examine the effects of KLF2 on CRC cell
biological action, KLF2 overexpression SW480 and HCT116 cells were
created by transfection with PcDNA3.1 KLF2 recombinant plasmid.
After transfection for 48 h, KLF2 mRNA and protein expression
increased markedly (Fig. 2A and B);
KLF2 expression also increased after transfection for 24, 72 and 96
h (data not shown). Next, SW480 and HCT116 cell viability was
detected at each time-point. The results indicated that KLF2
overexpression inhibits SW480 and HCT116 cell viability compared
with the control and PcDNA3.1 empty plasmid group (P<0.05;
Fig. 2C and D). Cell proliferation
was measured by the MTS method; the results showed that KLF2
overexpression for 48 h remarkably suppressed SW480 and HCT116 cell
proliferation (P<0.05; Fig.
2E).
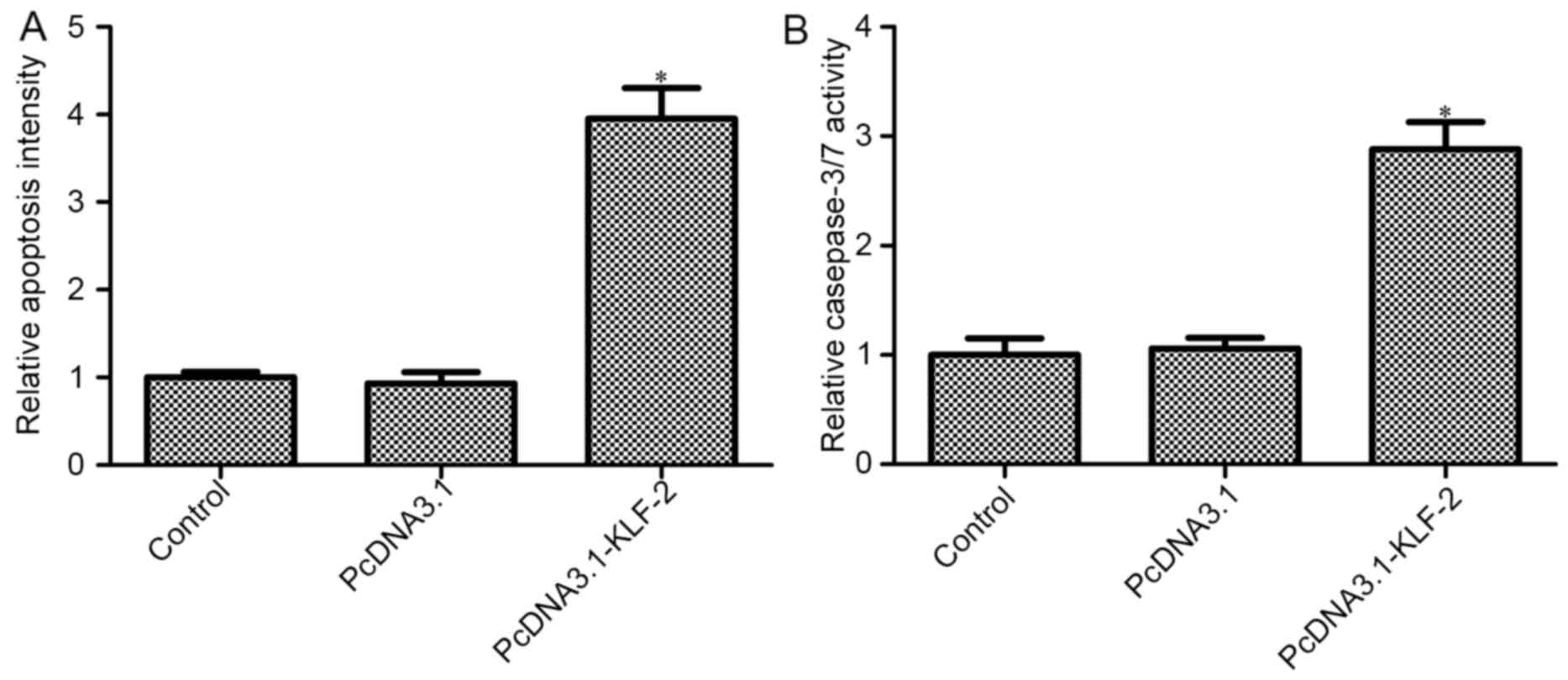
KLF2 overexpression promotes CRC cell
apoptosis
We further studied the effects of KLF2
overexpression on HCT116 cell apoptosis, and histone-associated DNA
fragments (nucleosomes) in HCT116 cells were measured. The level of
nucleosomes increased in PcDNA3.1-KLF2 transfected cells compared
with the control group (P<0.05; Fig.
3A), whereas the level of nucleosomes in the PcDNA3.1 group had
no obvious change compared with the control group (P>0.05;
Fig. 3A). The results indicated
that KLF2 overexpression promotes HCT116 cell apoptosis. We also
demonstrated the results by determining the level of caspase-3/7.
The results of the Caspase-Glo 3/7 assay showed that KLF2
overexpression increased caspase-3/7 activity compared with the
control and PcDNA3.1 empty plasmid group (P<0.05; Fig. 3B).
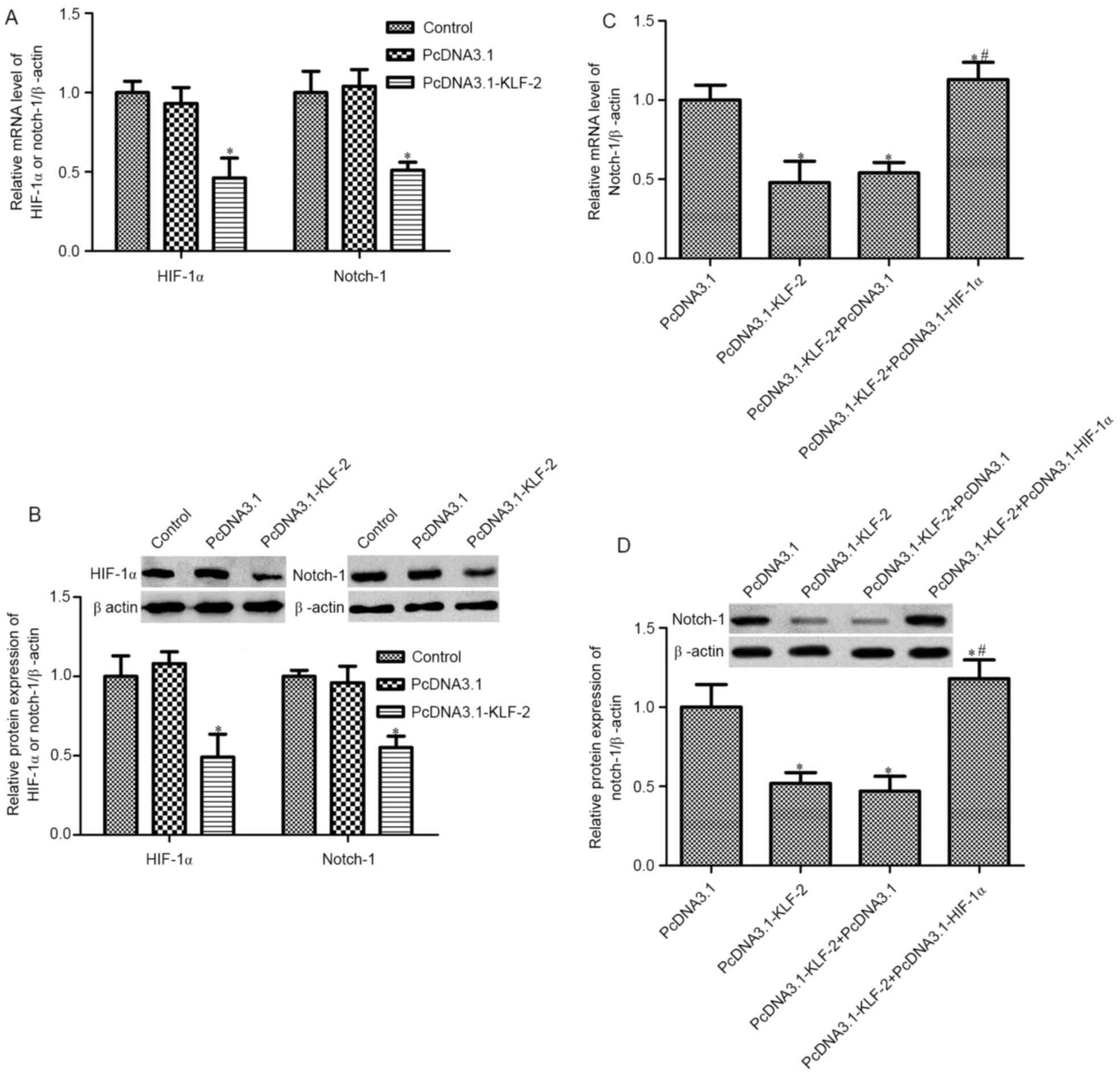
KLF2 inhibit Notch-1 expression via
inhibiting HIF-1α
In order to verify the mechanism of KLF2 in HCT116
cell biological action, further studies focused on the effects of
KLF2 on HIF-1α/Notch-1 expression. RT-PCR and western blot analysis
was used to detect the mRNA and protein level of HIF-1α and
Notch-1, respectively. A reduction in HIF-1α and Notch-1 mRNA and
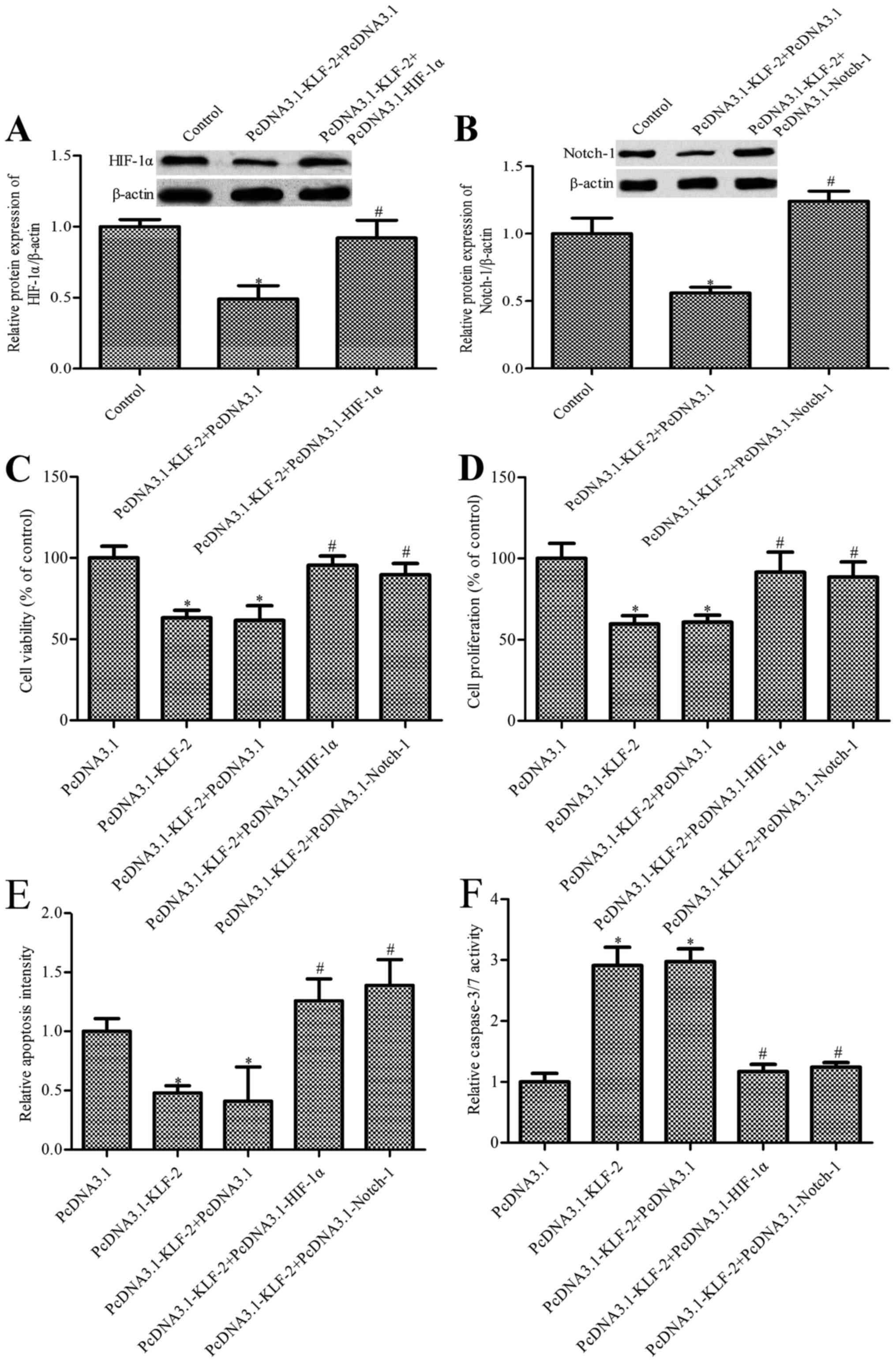
protein expression was observed by KLF2 overexpression (Fig. 4A and B). Furthermore, we constructed
PcDNA3.1-KLF2 and PcDNA3.1-HIF-1α co-transfected HCT116 cells; the
expression of Notch-1 was measured. As shown in Fig. 4C and D, PcDNA3.1-KLF2 and
PcDNA3.1-HIF-1α co-transfected impaired the decrease of Notch-1
expression induced by PcDNA3.1-KLF2 only, both in mRNA and protein
expression. These results indicated that KLF2 inhibits Notch-1
expression via inhibiting HIF-1α in HCT116 cells.
KLF2 overexpression regulates HCT116
cell growth via HIF-1α/Notch-1 signal pathway
To further depict the function and mechanism of KLF2
overexpression in HCT116 cells, we constructed PcDNA3.1-KLF2 and
PcDNA3.1-HIF-1α co-transfected cells, and PcDNA3.1-KLF2 and
PcDNA3.1-Notch-1 co-transfected cells. The results in Fig. 5A and B have showed that compared
with the PcDNA3.1-KLF2 + PcDNA3.1 group, the level of HIF-1α or
Notch1 was markedly increased in the PcDNA3.1-KLF2 +
PcDNA3.1-HIF-1α group or PcDNA3.1-KLF2 + PcDNA3.1-Notch1 group,
respectively. Cell biology including viability, proliferation and
apoptosis were measured. As shown in Fig. 5C and D, PcDNA3.1-KLF2 and
PcDNA3.1-HIF-1α co-transfection or PcDNA3.1-KLF2 and
PcDNA3.1-Notch-1 co-transfection strengthened HCT116 cell survival
and proliferation compared with the PcDNA3.1-KLF2 group. We also
found that HIF-1α overexpression or Notch-1 overexpression impaired
the decrease of PcDNA3.1-KLF2 induced CRC cell apoptosis (Fig. 5E). Finally, caspase-3/7 level was
determined, similarly to the above results; caspase-3/7 level in
PcDNA3.1-KLF2 and PcDNA3.1-HIF-1α co-transfected or PcDNA3.1-KLF2
and PcDNA3.1-Notch-1 co-transfected group markedly decreased
compared with the KLF2 overexpression group (Fig. 5F). The above results suggested that
KLF2 overexpression regulates HCT116 cell growth, and apoptosis is
dependent on the silencing of the HIF-1α/Notch-1 signal
pathway.
Discussion
The main findings in the present study are as
follows. First, KLF2 decreased in various CRC cell lines (SW480,
HT29, SW620 and HCT116) compared with the control human colon
epithelial cell line FHC. Recently, numerous pieces of evidence
show that members of the Krüppel-like factor family play essential
roles cancers, including CRC (7,9). It is
now well established that the KLF proteins with Cys2/His2
zinc-finger domains play a pivotal role in cell proliferation,
differentiation and could be characterized as suppressors or
activators in various cell types, including cancer cells (15). KLF2 is a tumor-suppressor associated
with cancer pathogenesis. The KLF member KLF2 has shown a generally
low expression in many malignancies, such as prostate (35) and ovarian (36) cancer. Consistent with these reports,
the present study showed that KLF2 is diminished in CRC cell lines,
thus, we supposed that dys-regulated expression of KLF2 may affect
the biofunction of CRC cells.
Overexpression of KLF2 in SW480 and HCT116 cells
remarkably inhibits SW480 and HCT116 cell survival and
proliferation; moreover, overexpression of KLF2 promotes HCT116
cell apoptosis and caspase-3 activity. KLF2 has been shown to
function as a tumor suppressor, such as inhibiting cell growth,
increasing DNA-damage-associated apoptosis, anti-angiogenesis
properties and the induction of cell quiescence (37). Nie and colleagues (21) showed that silencing KLF2 expression
promotes non-small cell lung cancer cell proliferation and inhibits
apoptosis. Wu and Lingrel (18)
showed that KLF2 could inhibit Jurkat T leukemia cell growth via
mediated p21WAF1/CIP1 expression. Consistent with these
reports, our results provide preliminary data regarding the
inhibition role of KLF2 in CRC cell growth and the promotion role
in CRC apoptosis.
KLF2 could inhibit the Notch-1 level via inhibiting
HIF-1α expression; furthermore, a HIF-1α/Notch-1 signal was
implicated in KLF2 action in CRC cells. KLF members were associated
with a burst of cancer progression by regulating various oncogenes
(5). Also, evidence has suggested
that KLF2 could mediate the level of HIF-1α (25). There is convincing evidence that
HIF-1α functions as an oncogene in CRC. Zhang et al
(38) showed that HIF-1α is linked
to the angiogenesis and epithelial-mesenchymal transition, which is
mediated by LRG1 in colorectal cancer. Huynh and colleagues
(39) showed that glaucarubinone
suppresses CRC progression via downregulating the expression of
HIF-1α. Moreover, it is well accepted that Notch-1 is associated
with HIF-1α induced-cancer progression (40). In the present study, we demonstrated
that KLF2 inhibits CRC cell growth via mediating the HIF-1α/Notch-1
signal pathway.
Collectively, the present results provide the first
evidence that KLF2 inhibits CRC cell growth via inhibiting the
HIF-1α/Notch-1 signal pathway. KLF2 is characterized as a tumor
suppressor in CRC, suggesting it may provide new targets for the
biology of CRC.
Glossary
Abbreviations
Abbreviations:
|
KLF2
|
Krüppel-like factor 2
|
|
CRC
|
colorectal cancer
|
|
HIF-1α
|
hypoxia-inducible factor 1α
|
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davies JM and Goldberg RM: First-line
therapeutic strategies in metastatic colorectal cancer. Oncology
(Williston Park). 22:1470–1479. 2008.PubMed/NCBI
|
|
3
|
Lu J, Ye X, Fan F, Xia L, Bhattacharya R,
Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, et al:
Endothelial cells promote the colorectal cancer stem cell phenotype
through a soluble form of Jagged-1. Cancer Cell. 23:171–185. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai C, Ashktorab H, Pang X, Zhao Y, Sha W,
Liu Y and Gu X: MicroRNA-211 expression promotes colorectal cancer
cell growth in vitro and in vivo by targeting tumor suppressor
CHD5. PLoS One. 7:e297502012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang S, Wang C, Yi Y, Sun X, Luo M, Zhou
Z, Li J, Cai Y, Jiang X and Ke Y: Krüppel-like factor 9 inhibits
glioma cell proliferation and tumorigenicity via downregulation of
miR-21. Cancer Lett. 356:547–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang H, Sun L, Xiao X, Xie R, Liu C, Wang
Y, Wei Y, Zhang H and Liu L: Krüppel-like factor 8 contributes to
hypoxia-induced MDR in gastric cancer cells. Cancer Sci.
105:1109–1115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fadous-Khalifé MC, Aloulou N, Jalbout M,
Hadchity J, Aftimos G, Paris F and Hadchity E: Krüppel-like factor
4A new potential biomarker of lung cancer. Mol Clin Oncol. 5:35–40.
2016.PubMed/NCBI
|
|
8
|
Kim SH, Park YY, Cho SN, Margalit O, Wang
D and DuBois RN: Krüppel-like factor 12 promotes colorectal cancer
growth through early growth response protein 1. PLoS One.
11:e01598992016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brown AR, Van TT, Simmen RC and Simmen FA:
The potential tumor-suppressive role of Krüppel-like factor 9 in
colorectal cancer. Cancer Res. 73(8 Suppl): 1969. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bieker JJ: Krüppel-like factors: Three
fingers in many pies. J Biol Chem. 276:34355–34358. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Preiss A, Rosenberg UB, Kienlin A, Seifert
E and Jäckle H: Molecular genetics of Krüppel, a gene required for
segmentation of the Drosophila embryo. Nature. 313:27–32. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eaton SA, Funnell AP, Sue N, Nicholas H,
Pearson RC and Crossley M: A network of Krüppel-like Factors
(Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J
Biol Chem. 283:26937–26947. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tetreault MP, Yang Y and Katz JP:
Krüppel-like factors in cancer. Nat Rev Cancer. 13:701–713. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dang DT, Pevsner J and Yang VW: The
biology of the mammalian Krüppel-like family of transcription
factors. Int J Biochem Cell Biol. 32:1103–1121. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and Krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaczynski J, Cook T and Urrutia R: Sp1-
and Krüppel-like transcription factors. Genome Biol. 4:2062003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McConnell BB and Yang VW: Mammalian
Krüppel-like factors in health and diseases. Physiol Rev.
90:1337–1381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu J and Lingrel JB: KLF2 inhibits Jurkat
T leukemia cell growth via upregulation of cyclin-dependent kinase
inhibitor p21WAF1/CIP1. Oncogene. 23:8088–8096. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang N, Miao H, Li YS, Zhang P, Haga JH,
Hu Y, Young A, Yuan S, Nguyen P, Wu CC, et al: Shear stress
regulation of Krüppel-like factor 2 expression is flow
pattern-specific. Biochem Biophys Res Commun. 341:1244–1251. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Banerjee SS, Feinberg MW, Watanabe M, Gray
S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H and Jain MK: The
Krüppel-like factor KLF2 inhibits peroxisome proliferator-activated
receptor-gamma expression and adipogenesis. J Biol Chem.
278:2581–2584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia
R, Liu YW, Liu XH, Zhang EB, Lu KH, et al: Long noncoding RNA ANRIL
promotes non-small cell lung cancer cell proliferation and inhibits
apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther.
14:268–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen
WM, Huang MD and Shu YQ: SP1-induced upregulation of the long
noncoding RNA TINCR regulates cell proliferation and apoptosis by
affecting KLF2 mRNA stability in gastric cancer. Oncogene.
34:5648–5661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Uchida D, Onoue T, Begum NM, Kuribayashi
N, Tomizuka Y, Tamatani T, Nagai H and Miyamoto Y: Vesnarinone
downregulates CXCR4 expression via upregulation of Krüppel-like
factor 2 in oral cancer cells. Mol Cancer. 8:622009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ebert R, Zeck S, Meissner-Weigl J, Klotz
B, Rachner TD, Benad P, Klein-Hitpass L, Rudert M, Hofbauer LC and
Jakob F: Krüppel-like factors KLF2 and 6 and Ki-67 are direct
targets of zoledronic acid in MCF-7 cells. Bone. 50:723–732. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kawanami D, Mahabeleshwar GH, Lin Z,
Atkins GB, Hamik A, Haldar SM, Maemura K, Lamanna JC and Jain MK:
Kruppel-like factor 2 inhibits hypoxia-inducible factor 1alpha
expression and function in the endothelium. J Biol Chem.
284:20522–20530. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen G, Li X, Jia YF, Piazza GA and Xi Y:
Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol Sin.
34:336–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian Q, Xue Y, Zheng W, Sun R, Ji W, Wang
X and An R: Overexpression of hypoxia-inducible factor 1α induces
migration and invasion through Notch signaling. Int J Oncol.
47:728–738. 2015.PubMed/NCBI
|
|
29
|
Cárdeno A, Sánchez-Hidalgo M, Rosillo MA
and Alarcón de la Lastra C: Oleuropein, a secoiridoid derived from
olive tree, inhibits the proliferation of human colorectal cancer
cell through downregulation of HIF-1α. Nutr Cancer. 65:147–156.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Ding Z, Peng Y, Pan F, Li J, Zou
L, Zhang Y and Liang H: HIF-1alpha inhibition reverses multidrug
resistance in colon cancer cells via downregulation of
MDR1/P-glycoprotein. PLoS One. 9:920142014.
|
|
31
|
Wang Y and Liu Y, Malek SN, Zheng P and
Liu Y: Targeting HIF1α eliminates cancer stem cells in
hematological malignancies. Cell Stem Cell. 8:399–411. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng RD, Shelton CC, Li YM, Qin LX,
Notterman D, Paty PB and Schwartz GK: gamma-Secretase inhibitors
abrogate oxaliplatin-induced activation of the Notch-1 signaling
pathway in colon cancer cells resulting in enhanced
chemosensitivity. Cancer Res. 69:573–582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thacker SA, Robinson P, Abel A and Tweardy
DJ: Modulation of the unfolded protein response during hepatocyte
and cardiomyocyte apoptosis in trauma/hemorrhagic shock. Sci Rep.
3:11872013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Macari ER, Schaeffer EK, West RJ and
Lowrey CH: Simvastatin and t-butylhydroquinone suppress KLF1 and
BCL11A gene expression and additively increase fetal hemoglobin in
primary human erythroid cells. Blood. 121:830–839. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duhagon MA, Hurt EM, Sotelo-Silveira JR,
Zhang X and Farrar WL: Genomic profiling of tumor initiating
prostatospheres. BMC Genomics. 11:3242010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang F, Zhu Y, Huang Y, McAvoy S, Johnson
WB, Cheung TH, Chung TK, Lo KW, Yim SF, Yu MM, et al:
Transcriptional repression of WEE1 by Kruppel-like factor 2 is
involved in DNA damage-induced apoptosis. Oncogene. 24:3875–3885.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Taniguchi H, Jacinto FV, Villanueva A,
Fernandez AF, Yamamoto H, Carmona FJ, Puertas S, Marquez VE,
Shinomura Y, Imai K, et al: Silencing of Kruppel-like factor 2 by
the histone methyltransferase EZH2 in human cancer. Oncogene.
31:1988–1994. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang J, Zhu L, Fang J, Ge Z and Li X:
LRG1 modulates epithelial-mesenchymal transition and angiogenesis
in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer
Res. 35:016–0306. 2016. View Article : Google Scholar
|
|
39
|
Huynh N, Beutler JA, Shulkes A, Baldwin GS
and He H: Glaucarubinone inhibits colorectal cancer growth by
suppression of hypoxia-inducible factor 1α and β-catenin via a p-21
activated kinase 1-dependent pathway. Biochim Biophys Acta.
1853:157–165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qiang L, Wu T, Zhang HW, Lu N, Hu R, Wang
YJ, Zhao L, Chen FH, Wang XT, You QD, et al: HIF-1α is critical for
hypoxia-mediated maintenance of glioblastoma stem cells by
activating Notch signaling pathway. Cell Death Differ. 19:284–294.
2012. View Article : Google Scholar : PubMed/NCBI
|