Introduction
Ovarian tumors are the leading cause of death among
all gynecologic tumors (1). In the
US, ~22,280 cases of ovarian cancer were estimated to be diagnosed
and it was estimated that 14,240 women died from ovarian cancer in
2016 (2). Despite modern surgical
techniques and chemotherapy, the prognosis of ovarian cancer
remains poor (3). Ovarian tumors
are a common type of neoplasm in women and the histological type is
various and complex. Epithelial ovarian tumors are the most common
type of ovarian tumor and represents 50–70% of all primary ovarian
tumors. According to the characteristics of the tumor cells and the
severity to health, epithelial ovarian tumors can be divided into
three types: benign, borderline and malignant ovarian tumors. In
various cases, benign tumors can develop into a malignant tumor
(4–7), which suggests that benign ovarian
tumors have an increased risk to transform into malignant tumors
due to the changes of various genes (8) or proteins (9). However, extensive research has mainly
focused on malignant epithelial ovarian cancer, and benign ovarian
tumors have not been a principal focus of research. In fact, there
are far more benign lesions occurring in the epithelial ovary, and
these are commonly diagnosed during pregnancy (10). In addition, these are also
associated with an increased risk of malignant epithelial ovarian
cancer. Therefore, a deeper understanding of benign epithelial
ovarian cysts (BEOCs) may not only provide more effective
treatments for BEOCs, but may also reduce the incidence of
malignant epithelial ovarian cancer.
Long non-coding RNAs (lncRNAs) are a type of RNAs
that are longer than 200 nucleotides in length and without coding
protein capacity (11). lncRNAs
have been previously considered as ‘transcriptional noise’ for a
long time (12). Recently, studies
have confirmed that lncRNAs play a critical role in the development
of cancer, including ovarian cancer. Emerging evidence suggests
that lncRNAs are associated with ovarian cancer biological
behaviors such as cell proliferation (13,14),
apoptosis (15,16), and invasion and metastasis (17,18).
Moreover, due to tissue-specificity, some lncRNAs may served as
potential biomarkers for cancer prognosis, including ovarian cancer
(19). However, these studies have
only focused on the regulation of lncRNAs in malignant ovarian
cancer, and little research has been carried out on the
relationship between lncRNAs and benign ovarian tumors. Therefore,
lncRNA expression profiles in BEOCs may help us to better
understand BEOC pathogenesis.
In the present study, we described the distinct
expression profiles of lncRNAs in BEOC and normal ovarian
epithelial tissues. In total, 1,014 transcripts of lncRNAs were
upregulated and 311 transcripts of lncRNAs were downregulated in
BEOCs compared with normal controls [absolute fold-change ≥2, false
discovery rate (FDR) <0.05]. Moreover, we also examined the Gene
Ontology (GO) enrichment of their associated protein-coding genes
and performed pathway analyses to analyze the potential function of
these differentially expressed lncRNAs. The present study may aid
in elucidating the tumorigenesis of ovarian epithelial tissue and
decrease the incidence of malignant transformation in regards to
BEOCs.
Materials and methods
Tissue collection
Samples of BEOCs and normal ovarian tissues were
collected from the Gynecologic Oncology Department of Nanjing
Maternal and Child Health Hospital (Nanjing, China) from 2014 to
2016. No patient received chemotherapy or radiotherapy before
surgery. Informed consent for the use of the tissues was obtained
from all patients. Finally, 10 cases of normal ovarian tissues and
17 cases of BEOCs were collected and immediately stored in
RNAsafety™ and frozen at −80°C before the experiments. All the
tissues were confirmed through histopathological diagnoses. The
present study was approved by the Ethics Review Committee of
Nanjing Maternity and Child Health Care Hospital.
Total RNA extraction
Frozen tissues were dissolved in TRIzol reagent
(Life Technologies, Grand Island, NY, USA). Total RNA was extracted
according to the manufacturer's protocol (Invitrogen, Carlsbad, CA,
USA). NanoDrop and the Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA) were used to check the
quantification and quality of the extracted RNA, respectively. The
extracted RNA samples were stored at −80°C for further experiments.
Complementary DNA (cDNA) was synthesized from 1 µg of total-RNA
using a PrimesScript™ RT Master Mix kit (Applied Takara, Dalian,
China) with random hexamer primers in a final volume of 20 ml. The
condition of reverse-transcription reaction was as follows: 37°C
for 15 min, 85°C for 5 sec and 4°C for 10 min.
Microarray analysis
To screen the global profiling of human lncRNAs and
protein-coding transcripts, we profiled three BEOC tissues and
three normal ovarian epithelial tissues with ArrayStar Human
Microarray V3.0. The lncRNAs were searched for using authoritative
databases such as RefSeq, Ensembl and UCSC Known Genes and related
studies while the mRNAs were collected from RefSeq and GENCODE. To
recognize every individual transcript exactly, each transcript was
described with a specific exon or splice junction probe. Both
positive probes (the housekeeping genes) and negative probes were
printed onto the array for hybridization quality control. The
sample processing and microarray hybridization were performed in
terms of the Agilent One-Color Microarray-Based Gene Expression
Analysis protocol (Agilent Technology). Briefly, 1 mg of total RNA
was obtained for purification by removing the rRNA (mRNA-ONLY
Eukaryotic mRNA Isolation kit; Epicentre, Madison, WI, USA). Then,
each sample was amplified and transcribed into fluorescent cRNA
along the entire length of the transcripts without 3′ bias
utilizing a random priming method. Agilent Quick Amp Labeling kit
was employed to normalize the values, and then, lncRNAs and mRNAs,
for which at least one out of two groups had flags in present or
marginal, were chosen for further data analysis. Additionally,
hierarchical clustering and combined analyses were performed using
homemade scripts.
Quantitative real-time PCR (qPCR)
qPCR was performed to detect the relative gene
expression using Power SYBR-Green PCR Master Mix (2X Applied
Biosystems) according to the standard protocol. GAPDH was taken as
an internal reference. The qPCR reaction conditions were set as
follows: an initial denaturation at 95°C for 30 sec, followed by 40
PCR cycles at 95°C for 5 sec and 60°C for 34 sec. Finally annealing
and extension at 95°C for 15 sec, 60°C for 60 sec and 95°C for 15
sec. Each sample was detected in triplicates. The fold-change was
calculated with the ΔCT method to describe the relative gene
expression in BEOC samples relative to the normal ovarian tissue
samples. All of the primers are presented in Table I.
 | Table I.qPCR primers used in the present
study. |
Table I.
qPCR primers used in the present
study.
| lncRNAs | Forward (5–3′) | Reverse
(5′-3′) |
|---|
| LOC339166 |
GCCTCTCTGGAGCTGAATCG |
CGTGCCAGTGGGTTTCCTAA |
| LOC440214 |
CCACCCCAAAGAAGATGCTG |
ACAGGAGACAAAGCCTTCGC |
| LOC644656 |
AATTAGTTGTGGCCGTTGCG |
ATCTTTAGTCGGCCTGGTGC |
| NENF |
CAGGAGCAGGTTCTTGGGAG |
CCAAGGACAACAGGAGGCAT |
| RP11-471J12.1 |
TCAGCCCACCTGCTCCAA |
TGATCTGTGCCTTCCTGGTACA |
| MEG3 |
CTGCCCATCTACACCTCACG |
CTCTCCGCCGTCTGCGCTAGGGGCT |
| GAPDH |
CCAGAACATCATCCCTGCCT |
CCTGCTTCACCACCTTCTTG |
GO and pathway analyses
Pathway and GO analyses were applied to determine
the potential roles of differentially expressed lncRNAs in
biological pathways or GO terms. The predicted target genes of the
differentially expressed lncRNAs were mapped to GO terms in the
Database for Annotation Visualization and Integrated Discovery
(DAVID) (http://david.abcc.ncifcrf.gov/). Fisher's exact test
was used to ascertain whether true differences existed between the
groups. In addition, we used the Kyoto Encyclopedia of Genes and
Genomes (KEGG) (http://www.kegg.jp/) to confirm the
pathway enrichment analysis. The ontology covers three domains:
biological process (BP), cellular component (CC) and molecular
function (MF). The threshold of significance was defined by
FDR.
Statistical analysis
Differential expression levels of lncRNAs were
selected by fold-change filtering (absolute fold-change >2.0).
Independent samples t-test between two groups was used, and
Fisher's exact test was used in GO and pathway analyses. A value of
FDR <0.05 was considered statistically significant.
Computer-based calculations were conducted using SPSS version 20.0
(SPSS, Inc., Chicago, IL, USA).
Results
Differentially expressed lncRNAs and
mRNAs in BEOCs compared with normal ovarian tissues
Firstly, to explore the altered lncRNAs in the
BEOCs, we determined the lncRNA and mRNA expression profiles using
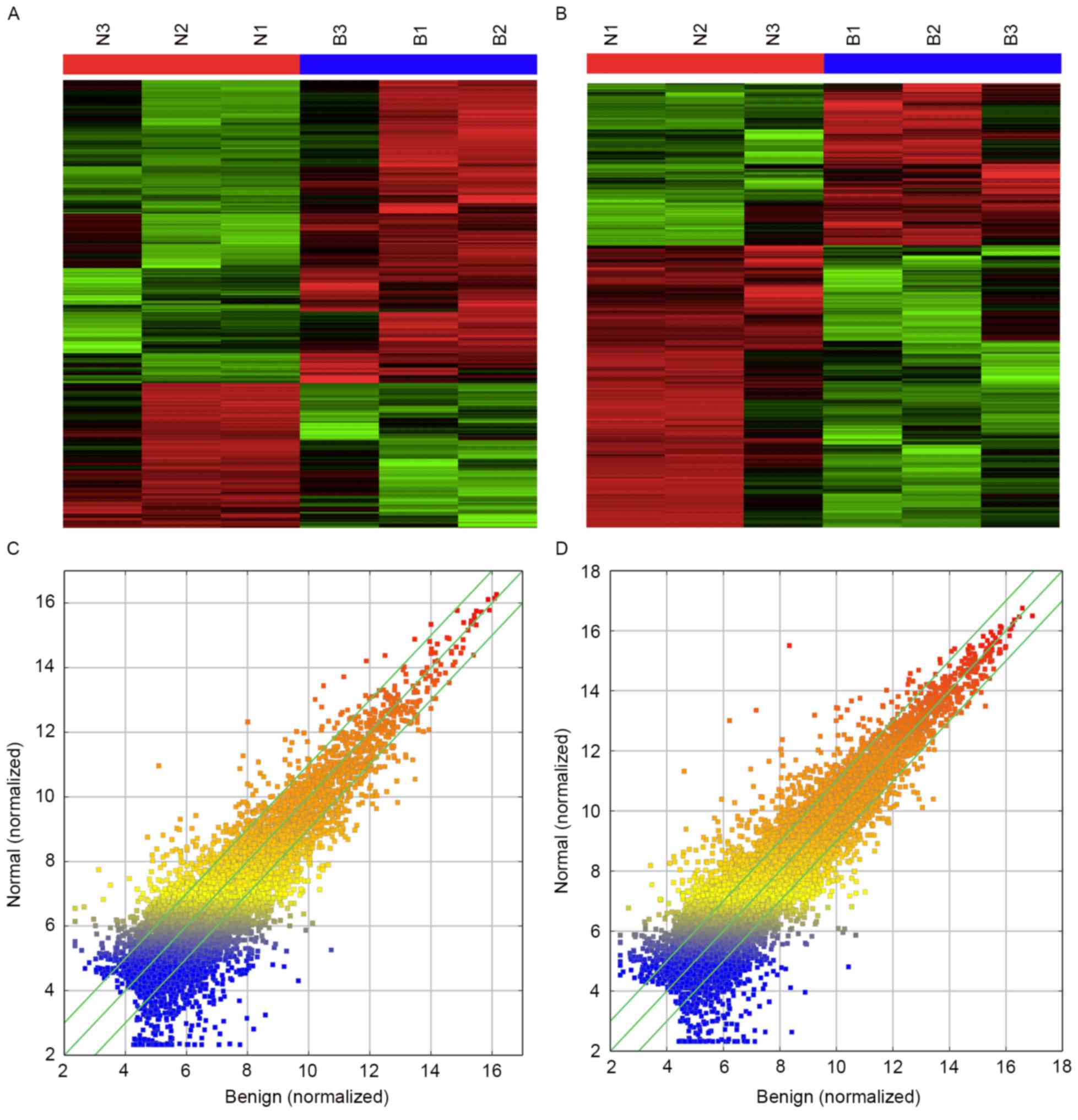
microarray analyses of normal ovarian and BEOC tissues. Heatmaps
and scatter-plots were used to describe the variation in lncRNA
expression among normal ovarian, benign ovarian cysts and malignant
epithelial ovarian cancer tissues (Fig.
1). All lncRNAs and mRNAs with a signal altered by 2-fold and
with a false discovery rate (FDR) <0.05 were identified as
statistically altered. Finally, 1,325 transcripts of lncRNAs (1,014
upregulated and 311 downregulated) and 1,563 mRNAs (613 upregulated
and 950 downregulated) were found to be differentially expressed in
BEOC tissues compared with normal controls. Lists of the top 20
upregulated and downregulated lncRNAs identified in the microarray
analyses are presented in Tables
II and III.
 | Table II.The top 20 upregulated lncRNAs in the
BEOCs compared with the normal ovarian tissues. |
Table II.
The top 20 upregulated lncRNAs in the
BEOCs compared with the normal ovarian tissues.
|
|
|
|
|
|
| Mean
expression |
|---|
|
|
|
|
|
|
|
|
|---|
| Seqname | GeneSymbol | FDR | Fold-change | Source |
Associated-gene | Normal | Benign |
|---|
|
ENST00000502882 | RP11-158J3.2 | 0.017 | 44.223 | GENCODE | HTR1A | 38.428 | 1699.407 |
|
ENST00000534866 | TAS2R64P | 0.022 | 42.733 | GENCODE |
| 5.029 | 214.918 |
| uc010ciy.1 | BC160930 | 0.038 | 41.621 | UCSC_knowngene | RP11-566K11.2 | 7.028 | 292.506 |
| AK021689 |
| 0.017 | 40.678 | NRED |
| 19.848 | 807.381 |
|
ENST00000500487 | RP11-32B5.7 | 0.031 | 40.156 | GENCODE |
| 9.524 | 382.452 |
|
ENST00000419463 | AC019117.1 | 0.018 | 33.387 | GENCODE |
| 5.110 | 170.614 |
|
ENST00000526388 | CTC-497E21.4 | 0.010 | 28.260 | GENCODE |
| 8.881 | 250.981 |
|
ENST00000563752 | SLC25A3P1 | 0.019 | 26.511 | GENCODE |
| 5.029 | 133.333 |
|
ENST00000566892 | RP11-1081M5.2 | 0.027 | 21.685 | GENCODE |
| 26.929 | 583.973 |
| NR_040017 | RNF157-AS1 | 0.020 | 21.544 | RefSeq | FOXJ1 | 6.342 | 136.631 |
| uc001gzl.3 | BC034684 | 0.005 | 17.922 | UCSC_knowngene | CHI3L1 | 5.029 | 90.138 |
|
ENST00000450480 | RP4-797C5.2 | 0.018 | 17.793 | GENCODE | KCND2 | 8.340 | 148.387 |
| TCONS_00026830 | XLOC_013047 | 0.034 | 16.688 | LincRNAs identified
by Cabili et al (12) |
| 7.306 | 121.921 |
| NR_040033 | LOC729950 | 0.023 | 16.490 | RefSeq |
| 37.444 | 617.446 |
|
ENST00000381181 | AP000569.2 | 0.034 | 16.383 | GENCODE |
| 12.908 | 211.472 |
| NR_027309 | LOC148824 | 0.016 | 16.354 | RefSeq | OR2C3 | 68.207 | 1115.456 |
|
ENST00000521558 | RP11-1081M5.1 | 0.025 | 16.292 | GENCODE |
| 58.657 | 955.648 |
| NR_038883 | LINC00649 | 0.017 | 16.086 | RefSeq |
| 28.062 | 451.399 |
| TCONS_00018520 | XLOC_008826 | 0.021 | 15.539 | LincRNAs identified
by Cabili et al (12) |
| 5.811 | 90.301 |
|
chr14:84031800-84050525+ |
chr14:84031800-84050525 | 0.024 | 15.300 | LincRNAs identified
by Khalil et al (26) |
| 5.029 | 76.949 |
 | Table III.The top 20 downregulated lncRNAs in
the BEOCs compared with the normal ovarian tissues. |
Table III.
The top 20 downregulated lncRNAs in
the BEOCs compared with the normal ovarian tissues.
|
|
|
|
|
|
| Mean
expression |
|---|
|
|
|
|
|
|
|
|
|---|
| Seqname | GeneSymbol | FDR | Fold-change | Source |
Associated-gene | Normal | Benign |
|---|
| uc002ejp.1 | MT1JP | 0.016 | 58.627 | UCSC_knowngene |
| 1999.563 | 34.107 |
| uc003xxw.1 | AX747593 | 0.020 | 24.817 | UCSC_knowngene | RP11-664D7.4 | 215.542 | 8.685 |
|
ENST00000584923 | SNORD3A | 0.014 | 20.002 | GENCODE |
| 5117.821 | 255.872 |
|
ENST00000437593 | RP11-500G22.2 | 0.012 | 18.427 | GENCODE | ATE1 | 93.894 | 5.095 |
| TCONS_00023858 | XLOC_011173 | 0.014 | 16.438 | LincRNAs identified
by Cabili et al (12) |
| 432.820 | 26.330 |
|
ENST00000542078 | RP11-392P7.8 | 0.014 | 15.472 | GENCODE | HEBP1 | 96.912 | 6.264 |
|
ENST00000417522 | RP11-38J22.6 | 0.017 | 13.893 | GENCODE | C1orf186 | 70.787 | 5.095 |
|
ENST00000580684 | RP11-835E18.2 | 0.048 | 13.632 | GENCODE |
| 154.759 | 11.353 |
|
ENST00000536029 | RP11-392P7.8 | 0.020 | 11.776 | GENCODE | HEBP1 | 113.762 | 9.660 |
| TCONS_00017618 | XLOC_008306 | 0.011 | 11.503 | LincRNAs identified
by Cabili et al (12) |
| 635.853 | 55.276 |
| uc002lch.1 | AK095045 | 0.018 | 10.279 | UCSC_knowngene |
| 335.169 | 32.609 |
| TCONS_00014161 | XLOC_006144 | 0.023 | 10.191 | LincRNAs identified
by Cabili et al (12) |
| 178.103 | 17.477 |
|
ENST00000499314 | RP11-1277A3.2 | 0.021 | 10.179 | GENCODE |
| 522.572 | 51.336 |
| TCONS_00029245 | XLOC_013980 | 0.031 | 10.076 | LincRNAs identified
by Cabili et al (12) |
| 294.817 | 29.260 |
|
ENST00000417089 | H19 | 0.017 | 9.329 | GENCODE |
| 508.334 | 54.491 |
|
ENST00000555882 | DIO3OS | 0.005 | 9.060 | GENCODE |
| 46.162 | 5.095 |
| NR_026860 | LINC00473 | 0.020 | 8.979 | RefSeq |
| 410.373 | 45.702 |
| uc010rog.2 | NEAT1 | 0.044 | 8.809 | UCSC_knowngene |
| 261.236 | 29.656 |
|
ENST00000520913 | PVT1 | 0.010 | 8.783 | GENCODE |
| 1881.694 | 214.238 |
|
ENST00000447298 | H19 | 0.017 | 8.718 | GENCODE |
| 688.447 | 78.972 |
Validation of candidate lncRNAs by
qPCR
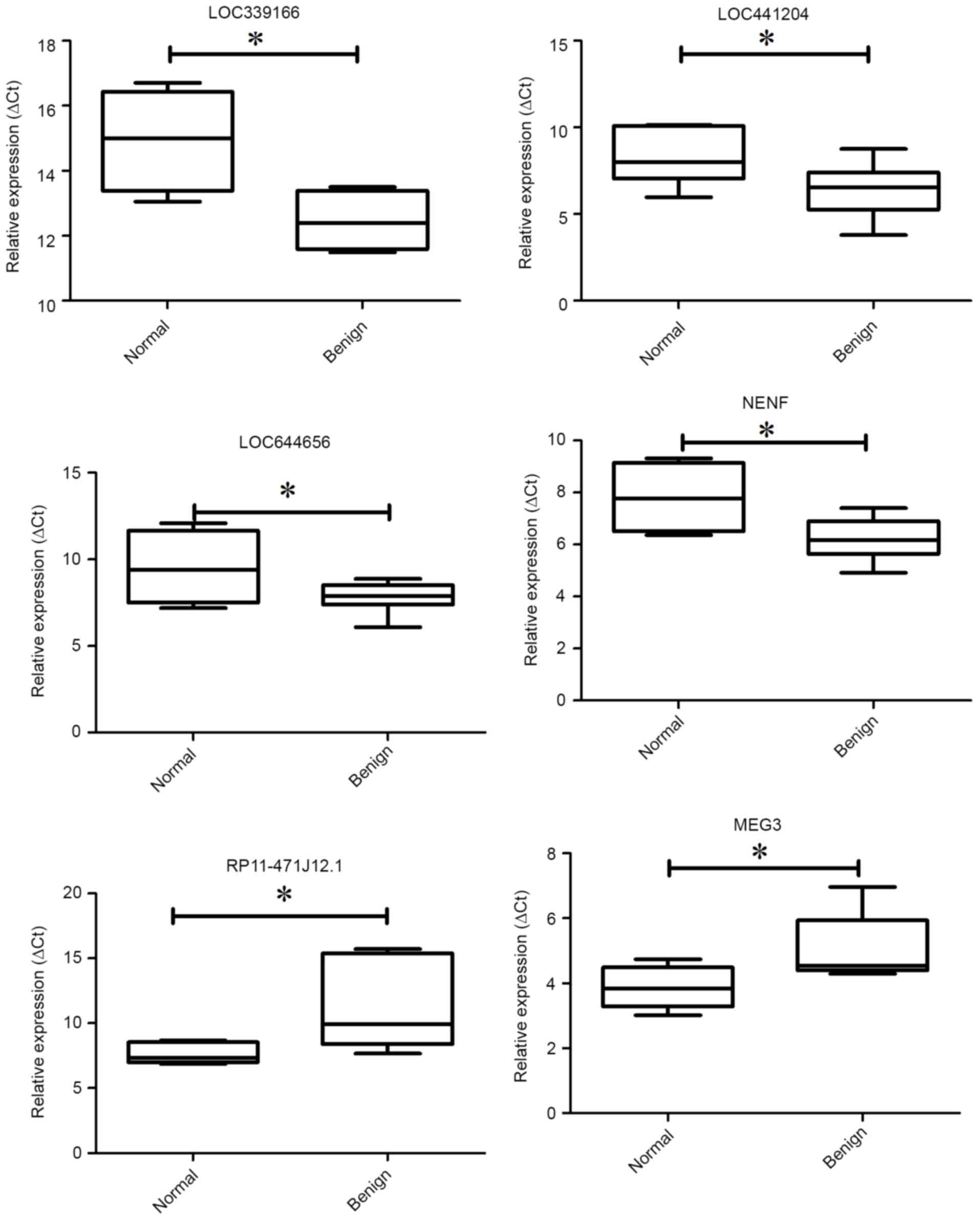
To confirm the validity of the microarray data, we
next conducted qPCR to detect the expression of the lncRNAs. We
randomly selected 6 differentially expressed lncRNAs. Among the 6
lncRNAs, lncRNAs LOC339166, LOC441204, LOC644656 and NENF were
upregulated whereas lncRNAs RP11-471J12.1 and MEG3 were
downregulated in the BEOCs compared with the normal controls
(Fig. 2). The result of qPCR
confirmed that the expression trend of the 6 selected lncRNAs was
consistent with the microarray data.
GO and pathway analyses of the
differentially expressed lncRNAs
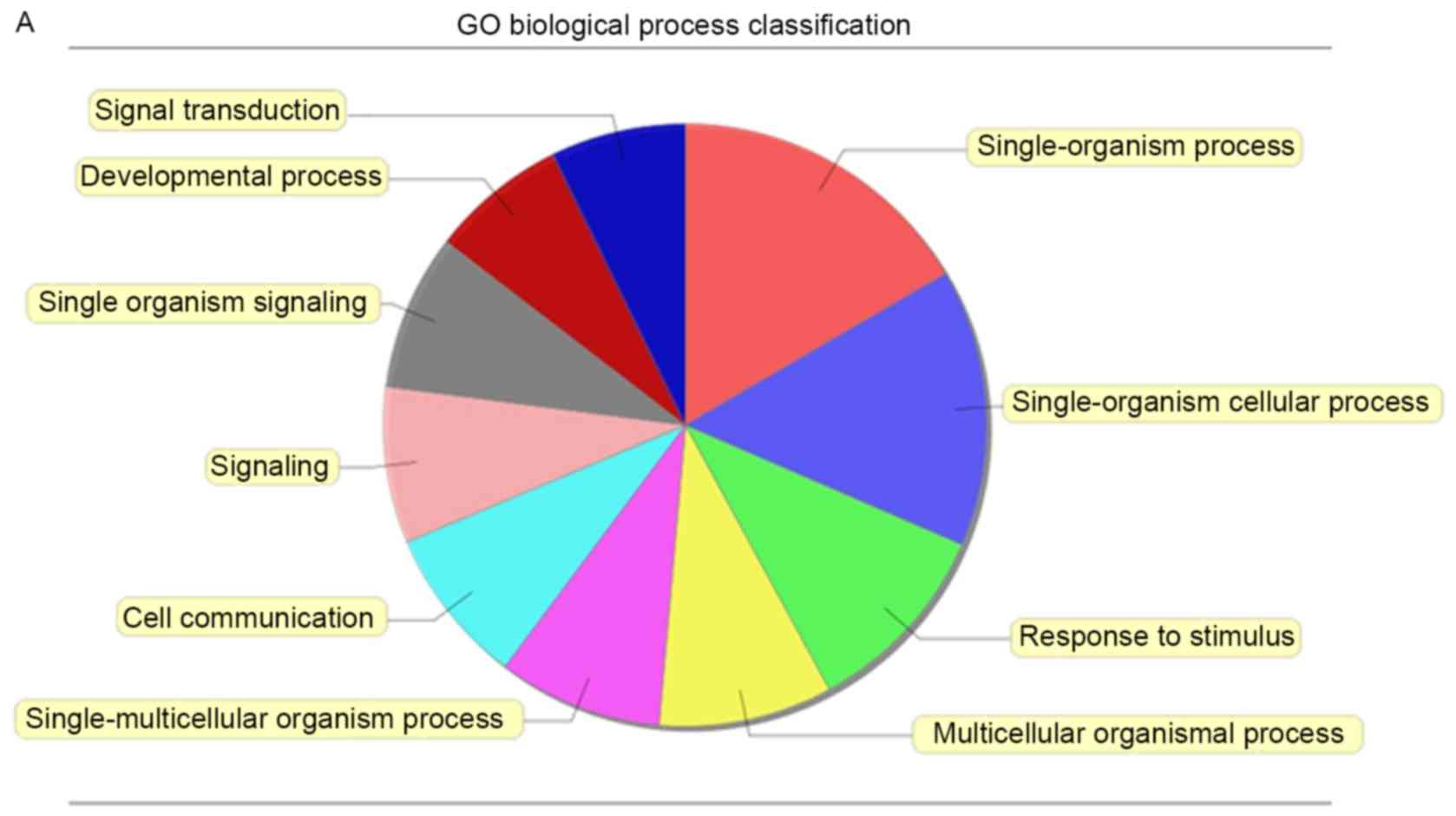
To investigate the function of altered lncRNAs in
the BEOCs, we performed GO analysis which covered the following
three domains: biological processes (BP), cellular components (CC)
and molecular functions (MF). We found that the highest GO
classifications targeted by the upregulated lncRNAs were
single-organism process (Fig. 3A),
membrane (Fig. 3B) and signal
transducer activity (Fig. 3C).
However, the highest GO classifications targeted by downregulated
lncRNAs were cellular process (Fig.
3D), cell part (Fig. 3E) and
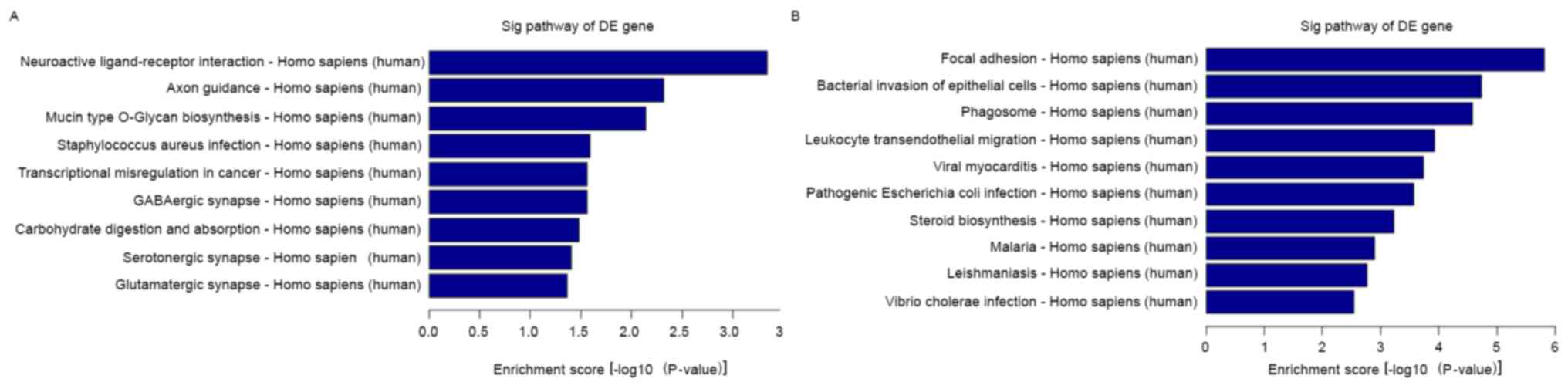
binding, particularly protein binding (Fig. 3F). To map these lncRNAs to pathways,
we also performed pathway analysis. The result indicated that the 9
main pathways corresponding to the upregulated transcripts and the
most enriched network was ‘neuroactive ligand-receptor interaction’
(Fig. 4A). The 9 main pathways are
shown: i) neuroactive ligand-receptor interaction; ii) axon
guidance; iii) mucin type O-glycan biosynthesis; iv)
Staphylococcus areus infection; v) transcriptional
misregulation in cancer; vi) GABAergic synapse; vii) carbohydrate
digestion and absorption; viii) serotonergic synapse; ix)
glutamatergic synapse. We also observed 10 main pathways
corresponding to the downregulated transcripts (Fig. 4B): i) focal adhesion; ii) bacterial
invasion of epithelial cells; iii) phagosome; iv) leukocyte
transendothelial migration; v) viral myocarditis; vi) pathogenic
Escherichia coli infection; vii) steroid biosynthesis; viii)
malaria; ix) leishmaniasis; x) Vibrio cholera infection. The
most enriched network was ‘focal adhesion’ with 37 transcripts
annotated with this term (Fisher P-value=1.53367E-06).
Discussion
BEOCs are the most common form of ovarian tumors in
women, accounting for ~80% of all ovarian masses (20). However, the molecular mechanisms
related to the tumorigenesis of ovarian epithelial cells remains
largely unknown. Increasing studies have claimed that lncRNAs are
highly functional and have a crucial role in malignat ovarian
cancer (13,17,18,21,22).
In our previous study, we identified dysregulation of many lncRNAs
in malignant ovarian cancer compared with benign ovarian cysts and
normal ovary. However, we also found that several lncRNAs, such as
LEMD1-AS1 and AK 125532 were differentially expressed in BEOCs
compared with these in the normal control (23). Thus, we next identified
differentially expressed lncRNAs in BEOCs compared with normal
ovarian tissues. To the best of our knowledge, the present study is
the first to investigate the lncRNA expression profiling in BEOCs
compared with normal ovarian tissues. The present study provides a
better understanding of the molecular mechanisms of BEOCs.
On the basis of the GO analysis, we found that the
BP of upregulated and downregulated lncRNAs was both tightly
associated with single-organism process and single-organism
cellular process. These GO terms are also associated with death and
cell proliferation (24). The
destiny of BEOCs is critically regulated by the cell cycle and
apoptosis process in which lncRNAs may play an important role. For
CC, the top GO term of the upregulated lncRNAs was membrane part
and in downregulated lncRNAs this was cell part. These results
indicate that lncRNAs may primarily regulate various mRNAs which
are located in the cell or on the membrane and exercise their
function. It can also be observed that the highest frequency of the
MF GO terms in the upregulated lncRNAs was molecular transducer
activity. Many studies have reported that some lncRNAs act as a
molecular transducer. For example, lncRNA-TUG1 is overexpressed in
non-small cell lung cancer, and can be regulated by p53 and affect
cell proliferation through HOXB7 expression (25). However, the top MF GO term in the
downregulated lncRNAs was binding, particularly protein binding.
Increasing studies suggest that the primary function of lncRNAs is
the epigenetic regulation of coding gene through proteins or
microRNAs. Many lncRNAs have been reported to recruit and bind to
PRC2 (26) or other
chromatin-associated proteins (27). In addition, accumulating evidence
indicates that lncRNAs act as competing endogenous RNAs (ceRNAs) by
‘sponging’ microRNAs (22,28,29).
In addition, pathway analysis displayed that the
upregulated lncRNAs were mainly correlated with neuroactive
ligand-receptor interaction and axon guidance. Various studies have
reported that the sympathetic nervous system is important in the
tumor microenvironment (30,31),
and autonomic nerve development promotes cancer progression
(32). Therefore, we speculated
that there may be a relationship between ovarian tumorigenesis and
the dysregulation of neural signaling pathways. In contrast, the
downregulated lncRNAs were mainly correlated with focal adhesion.
Focal adhesion is important between two cells or between a cell and
the extracellular matrix. Various coding genes such as E-cadherin
play critical roles in focal adhesion and tumor metastasis when
malignant potential is increased in ovarian cancer cells. Moreover,
some lncRNAs such as H19 (33),
HOTAIR and MALAT1 (34) were found
to promote cancer aggression by regulating E-cadherin.
Research has shown that the expression of H19 is
upregulated in many types of cancers including ovarian cancer. The
ectopic expression of H19 was found to promote cancer cell
proliferation and invasion, suggesting that H19 may function as an
oncogene. In our microarray, the transcript of H19
(ENST00000422826) was overexpressed ~3-fold in BEOCs compared with
that in normal tissues. The results also indicated that BEOCs may
have malignancy potential. However, the transcripts of H19
(ENST00000417089, ENST00000447298, uc001lva.4, uc021qbz.1,
ENST00000442037 and ENST00000446406) were reduced in the BEOCs. The
results of PVT1 were also the same as H19; different transcripts
had differential expression in the BEOCs. The different transcripts
of lncRNAs had differential expression and function. However,
confirmation and elucidation of this finding require further
study.
Due to the important roles of lncRNAs in cancer, a
growing number of studies have focused on the potential
lncRNA-related biomarkers for tumors. lncRNA-related biomarkers aid
clinicians to make accurate diagnoses and treatment decisions. Crea
et al reported that PCAT18, the lncNRA which is specifically
expressed in the prostate, could be a potential therapeutic target
and biomarker for metastatic prostate cancer (35). In addition, similar lncRNA-related
biomarkers with prognostic value have also been identified in other
types of cancers such as breast (36), pancreatic (37) and lung cancer (38,39).
In ovarian cancer, various lncRNAs have been shown to have
potential as prognostic biomarkers, such as HOTAIR (18), H19 (40) and HOXA11AS (41). However, lncRNA-related biomarkers in
benign tumors still have not been reported. The present study may
aid in the identification of lncRNA-related biomarkers for benign
ovarian cysts.
In summary, we profiled the differential expression
of lncRNAs and mRNAs between BEOCs and normal ovarian tissues. In
total, 1,325 transcripts of lncRNAs and 1,563 mRNAs were found to
be differentially expressed between BEOCs and normal ovarian
control tissues (absolute fold-change ≥2, FDR <0.05).
Furthermore, dysregulated lncRNAs were characterized by a
comprehensive examination of GO enrichment and pathway analysis by
their associated protein-coding genes. Collectively, our findings
suggest that lncRNAs play a critical role in the pathological
process of BEOCs.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81402139).
References
|
1
|
Holschneider CH and Berek JS: Ovarian
cancer: Epidemiology, biology, and prognostic factors. Semin Surg
Oncol. 19:3–10. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiang YC, Chen CA, Chiang CJ, Hsu TH, Lin
MC, You SL, Cheng WF and Lai MS: Trends in incidence and survival
outcome of epithelial ovarian cancer: 30-Year national
population-based registry in Taiwan. J Gynecol Oncol. 24:342–351.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Austin RM: Benign to malignant
transformation in epithelial ovarian tumors. Hum Pathol.
24:562–563. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fitzgibbons PL, Henson DE and Hutter RV:
Cancer Committee of the College of American Pathologists: Benign
breast changes and the risk for subsequent breast cancer: An update
of the 1985 consensus statement. Arch Pathol Lab Med.
122:1053–1055. 1998.PubMed/NCBI
|
|
6
|
Dupont WD, Page DL, Parl FF, Vnencak-Jones
CL, Plummer WD Jr, Rados MS and Schuyler PA: Long-term risk of
breast cancer in women with fibroadenoma. N Engl J Med. 331:10–15.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McDivitt RW, Stevens JA, Lee NC, Wingo PA,
Rubin GL and Gersell D: The Cancer and Steroid Hormone Study Group:
Histologic types of benign breast disease and the risk for breast
cancer. Cancer. 69:1408–1414. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Powell DE, Puls L and van Nagell J Jr:
Current concepts in epithelial ovarian tumors: Does benign to
malignant transformation occur? Hum Pathol. 23:846–847. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waldemarson S, Krogh M, Alaiya A, Kirik U,
Schedvins K, Auer G, Hansson KM, Ossola R, Aebersold R, Lee H, et
al: Protein expression changes in ovarian cancer during the
transition from benign to malignant. J Proteome Res. 11:2876–2889.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Haan J, Verheecke M and Amant F:
Management of ovarian cysts and cancer in pregnancy. Facts Views
Vis Obgyn. 7:25–31. 2015.PubMed/NCBI
|
|
11
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Silva JM, Boczek NJ, Berres MW, Ma X and
Smith DI: LSINCT5 is over expressed in breast and ovarian cancer
and affects cellular proliferation. RNA Biol. 8:496–505. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Medrzycki M, Zhang Y, Zhang W, Cao K, Pan
C, Lailler N, McDonald JF, Bouhassira EE and Fan Y: Histone h1.3
suppresses h19 noncoding RNA expression and cell growth of
ovarian cancer cells. Cancer Res. 74:6463–6473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of
ovarian and breast cancer. Clin Cancer Res. 13:5745–5755. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G
and Hua KQ: The long non-coding RNA HOTAIR promotes the
proliferation of serous ovarian cancer cells through the regulation
of cell cycle arrest and apoptosis. Exp Cell Res. 333:238–248.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gloss B, Moran-Jones K, Lin V, Gonzalez M,
Scurry J, Hacker NF, Sutherland RL, Clark SJ and Samimi G:
ZNF300P1 encodes a lincRNA that regulates cell polarity and
is epigenetically silenced in type II epithelial ovarian cancer.
Mol Cancer. 13:32014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
20
|
Pelusi G, Taroni B and Flamigni C: Benign
ovarian tumors. Front Biosci. 2:g5–7. 1997.PubMed/NCBI
|
|
21
|
Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY
and Hua KQ: Long non-coding RNA ANRIL predicts poor prognosis and
promotes invasion/metastasis in serous ovarian cancer. Int J Oncol.
46:2497–2505. 2015.PubMed/NCBI
|
|
22
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J,
Lv M, Gu Y, Zhang J, Hua X, et al: LncRNAs expression profiling in
normal ovary, benign ovarian cyst and malignant epithelial ovarian
cancer. Sci Rep. 6:389832016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang J, Chen L, Kong X, Huang T and Cai
YD: Analysis of tumor suppressor genes based on gene ontology and
the KEGG pathway. PLoS One. 9:e1072022014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales Rivea D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hendrickson GD, Kelley DR, Tenen D,
Bernstein B and Rinn JL: Widespread RNA binding by
chromatin-associated proteins. Genome Biol. 17:282016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a
hidden RNA language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
30
|
Cole SW, Nagaraja AS, Lutgendorf SK, Green
PA and Sood AK: Sympathetic nervous system regulation of the tumour
microenvironment. Nat Rev Cancer. 15:563–572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huan HB, Wen XD, Chen XJ, Wu L, Wu LL,
Zhang L, Yang DP, Zhang X, Bie P, Qian C, et al: Sympathetic
nervous system promotes hepatocarcinogenesis by modulating
inflammation through activation of alpha1-adrenergic receptors of
Kupffer cells. Brain Behav Immun. 59:118–134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Magnon C, Hall SJ, Lin J, Xue X, Gerber L,
Freedland SJ and Frenette PS: Autonomic nerve development
contributes to prostate cancer progression. Science.
341:12363612013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Crea F, Watahiki A, Quagliata L, Xue H,
Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al:
Identification of a long non-coding RNA as a novel biomarker and
potential therapeutic target for metastatic prostate cancer.
Oncotarget. 5:764–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou M, Zhong L, Xu W, Sun Y, Zhang Z,
Zhao H, Yang L and Sun J: Discovery of potential prognostic long
non-coding RNA biomarkers for predicting the risk of tumor
recurrence of breast cancer patients. Sci Rep. 6:310382016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou M, Diao Z, Yue X, Chen Y, Zhao H,
Cheng L and Sun J: Construction and analysis of dysregulated
lncRNA-associated ceRNA network identified novel lncRNA biomarkers
for early diagnosis of human pancreatic cancer. Oncotarget.
7:56383–56394. 2016.PubMed/NCBI
|
|
38
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi
H, Cheng L and Sun J: Relapse-related long non-coding RNA signature
to improve prognosis prediction of lung adenocarcinoma. Oncotarget.
7:29720–29738. 2016.PubMed/NCBI
|
|
40
|
Ma Y, Lu Y and Lu B: MicroRNA and long
non-coding RNA in ovarian carcinoma: Translational insights and
potential clinical applications. Cancer Invest. 34:465–476. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yim GW, Kim HJ, Kim LK, Kim SW, Kim S, Nam
EJ and Kim YT: Long non-coding RNA HOXA11 antisense promotes cell
proliferation and invasion and predicts patient prognosis in serous
ovarian cancer. Cancer Res Treat. Oct 11–2016.(Epub ahead of
print). doi: 10.4143/crt.2016.263. View Article : Google Scholar :
|