Introduction
Head and neck cancer is the term given to a variety
of malignant tumors that develop in the oral cavity, laryngeal,
pharynx and salivary glands. They are predominantly squamous cell
carcinomas. Head and neck squamous cell carcinoma (HNSCC) is the
6th most common malignancy worldwide (1). The incidence rates in the world are
higher in the most developed countries (2). It was estimated that there were
approximately 550,000 new head and neck cancer cases in the world
in 2008. The expected number of deaths was approximately 300,000.
Despite progress in therapeutic procedures, patients with HNSCC
have a high risk of early locoregional relapse. The five-year
survival rate for HNSCC has shown only modest improvement over the
last decades (3,4). Most of them are oral squamous cell
carcinomas (OSCC) (5). Not only the
delayed detection but also the difficulties to monitor the
treatment and outcome are responsible for the high morbidity rate
of HNSCC. Therefore, early detection of disease progression remains
a challenging task mainly due to the lack of adequate
biomarkers.
A variety of promising biomarkers has been
described, nearly all of them can be detected immunohistochemically
(6). In addition, saliva is
preferentially studied for such biomarkers, because it has many
advantages in terms of collection, storage, shipping, and
voluminous sampling; all of these processes can be carried out very
economically compared with serum or urine. All investigations focus
on an early detection of OSCC (7).
Moreover, salivary biomarkers are of great value for
differentiation of an OSCC from potentially malignant oral
disorders (8). However, the main
advantages of saliva as a diagnostic tool for HNSCC is that oral
cancer cells are immersed in the salivary milieu. Therefore saliva
provides more direct information regarding the disease status.
Saliva bears different biological components potentially capable
for diagnosis and monitoring of HNSCC including the small nucleic
acids designated as microRNAs (9).
Several studies indicated the potential of saliva miRNAs for early
oral cancer diagnosis (10–13).
MicroRNAs (miRNAs) are small (18–25 bp) non-coding
RNAs of endogenous origin, which regulate gene expression
post-transcriptionally by translation inhibition and increased
decay of their target mRNAs. They recognize their target mRNA by
binding of 6 nucleotide sequences in the 3′-UTR (‘seed’ sequence),
which is specific for each individual microRNA species. To date,
2,588 individual microRNA sequences have been identified in human
(miRBase database, release 21: June 2014), which are estimated to
regulate the expression of 60% of human protein-coding genes
(14). MicroRNAs are essential
regulators of cell proliferation, differentiation and apoptosis
(reviewed in refs. 15–17). MiRNAs were recently shown to be
abundant and stable in different body fluids such as serum, urine
or saliva (18–20). Furthermore, altered expression of
several microRNAs has already been described to be associated with
the genesis and/or progression of oral cancer.
Among others, miR-93, miR-125a, miR-142-3p,
miR-200a, miR-203, miR-213 (designated by actual miRBase
terminology as miR-181a-3p), were found to be associated with
HNSCC. miR-93, miR-125a, miR-142-3p and miR-200a were concordantly
expressed in saliva of 12 HNSCC patients and 12 healthy controls
(21). Furthermore, miR-125a and
miR-200a expression was significantly lower in the saliva of a
cohort of 50 HNSCC patients than in corresponding healthy controls
(21). MicroRNA miR-203 was
significantly downregulated in 23 samples of oral cavity SCC
compared to control tissue (22).
MicroRNA miR-213 was upregulated >3.0-fold in patients samples
of squamous cell carcinoma of the tongue (23). Furthermore, miR-213 targets Nanog
mRNA in a subpopulation of CD34+ cells in peripheral
blood, driving differentiation of components of the immune system
(24). Despite their global role in
oncogenic cell transformation members of the let-7 family (let-7a,
let-7b, let-7g and let-7i) have been shown to be associated with
radiation sensitivity and therefore may monitor radiotherapy in
HNSCC (25). In detail, let-7a and
−7b were downregulated by radiation, while let-7g and let-7i were
upregulated and inhibition of let-7g by antagomirs led to increased
radiosensitivity of lung carcinoma cells (26).
Radical surgical resection added with radiation and
chemotherapy is the established curative treatment for HNSCC
(27–29). The post-operative radiotherapy is
intended to improve loco-regional control and therefore it is
inevitable. Unfortunately, hyposalivation is the most common oral
complication of radiation therapy (RT) for head and neck cancer
patients (30). In addition to
decreased saliva volume, RT also causes changes in saliva
consistency, buffering capacity, and pH-value (31). However, in recent years several
improvements, such as three-dimensional (3D) conformal radiation
therapy (3D-CRT) and intensity-modulated radiation therapy (IMRT)
have proven to be capable of sparing the salivary glands and
thereby to prevent hyposalivation (32).
Hence, it is still unknown whether and to which
extent microRNAs associated with HNSCC and radiosensitivity are
detectable in oral fluid during and after radiotherapy and if they
may act as salivary biomarkers for tumor monitoring. In this study,
we screened a HNSCC- and radiation-associated panel of microRNAs
for their feasibility as indicators of a tumor-free state of the
oral cavity. It was the aim of the present investigation to analyze
the prognostic impact of salivary microRNA levels of miR-93,
miR-125a, miR-142-3p, miR-200a, miR-203, miR-213, let-7a, let-7b,
let-7g and let-7i in patients receiving 3D-CRT treatment.
Materials and methods
Study population
Saliva samples were collected between 06/2002 and
10/2008 within the scope of our previously described study, wherein
we examined the parotid gland recovery after radiotherapy (33). The experimental protocol, consent
form, and the present retrospective analysis of the saliva samples
were approved by the intern institutional review board, the ethics
committee of the Martin-Luther-University Halle-Wittenberg, in
accordance to the Helsinki declaration. All patients gave written
detailed informed consent.
Treatment planning and determination
of the parotid gland doses
All patients received 3D-CRT or IMRT treatment
planning as previously described (33). Patients were immobilized with
individual thermoplastic head-neck-shoulder masks. The planning
goal was, while maintaining a homogeneous dose distribution in the
target volumes, to minimize mean dose in the contra-lateral parotid
gland. No effort was undertaken to spare the submandibular, the
sublingual or minor salivary glands. All patients were treated with
continuous conventional fractionation and received 2.0-Gy
fractions, 1 fraction per day, 5 fractions per week, for 7 weeks.
The mean dose and the partial volumes receiving specified doses
were determined for each gland from dose-volume histogram (DVH).
Based on an algorithm initially proposed by Lyman the DVHs were
transformed to single step DVHs (34). Afterwards, mean doses of the
ipsilateral and contralateral parotid glands were calculated for
every patient in Gy.
All patients underwent saliva collection at
different time points: within one week before radiation treatment
(baseline), during treatment and 6 and 12 months after the end of
RT. All salivary samples were collected at least one hour after a
meal at a standardized time of the day (9:00 am to 11:00 pm).
Patients were asked to rinse the mouth and swallow any residual
saliva. Then, the patients were instructed to chew on a paraffin
pellet (Ivoclar Vivadent, Schaan, Liechtenstein) for 5 min. After 5
min samples were collected with the patients expectorating all
saliva into sterile test tubes. The samples were cryopreserved
immediately after collection at −80°C.
RNA isolation
Saliva (200 µl) was thawed to room temperature and
immediately centrifuged (2000 × g, 10 min) to remove remaining
cells and cellular debris. RNA was isolated by phenol/chloroform
extraction using TRIzol (Invitrogen, Karlsruhe, Germany) according
to the manufacturer's instructions. Briefly, saliva supernatant was
mixed with 750 µl of TRIzol reagent and 200 µl of chloroform. After
centrifugation, the aequous phase containing the RNA was separated
and remaining DNA was digested by 5 units DNase (Qiagen, Hilden,
Germany). RNA was precipitated with 500 µl of ethanol overnight and
washed several times with ethanol (96% and 70%). RNA concentration
was analyzed after elution in 50 µl RNase-free H2O using
spectrometry (Eppendorf, Hamburg, Germany).
MicroRNA-specific cDNA synthesis
In this study, saliva expression of the following
microRNAs was analyzed using TaqMan microRNA primer kits (Applied
Biosystems, Darmstadt, Germany): miR-93, miR-125a, miR-142-3p,
miR-200a, miR-203, miR-213, let-7a, let-7b, let-7g, let-7i and U18
snoRNA as reference gene. RNA (10 ng) were applied for each
microRNA cDNA synthesis using specific RT primer (Applied
Biosystems). RNA was incubated with RNAse Inhibitor, Buffer, dNTPs
(20 mM) and MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot,
Germany) at 16°C for 30 min and 42°C for 30 min. Afterwards,
reverse transcriptase was inactivated by a 5 min 85°C step. cDNA
was stored at −20°C upon quantitative real-time analysis.
RT-qPCR analyses
cDNA of each individual patient sample was measured
in RT-qPCR on a RotorGene 6000 (LTF Labortechnik, Wasserburg,
Germany). For the reaction mix, buffer, dNTPS (20 mM),
HotStartTaq-Polymerase (Qiagen) and microRNA-specific TaqMan primer
(Applied Biosystems) were applied. Product accumulation was
detected in fluorescence increase per cycle and quantified by ∆∆Cq
method according to Livak and Schmittgen (35). U18 snoRNA expression served as
reference gene.
Statistical evaluation
Data were analyzed using SPSS software version 20
(IBM Inc., Chicago, IL, USA). Differences in microRNA expression
between the patients groups were visualized by box-plots and
analyzed by non-parametric tests (Mann-Whitney U test,
Kruskal-Wallis test), and bivariate correlation analyses according
to Spearman-Rho. P-values <0.05 were considered statistically
significant.
Results
Longitudinal saliva samples were available for 17
patients: 3 women and 14 men (42 samples, termed Coh 1). Overall
mean age was 57.8 years (range: 38–79 years). There were 4 patients
with oropharyngeal squamous cell carcinomas, 5 patients with oral
squamous cell carcinoma, and 8 patients with laryngeal squamous
cell carcinoma. The majority were advanced tumors with regional
node involvement at the time of treatment. None had distant
metastases, whereas it was unknown in two subjects (Table I). On average, patients received a
mean dose of 27.7 Gy of the contralateral parotid gland.
 | Table I.Patient demographics and tumor
statistics. |
Table I.
Patient demographics and tumor
statistics.
| Subject | Gender | Age (yrs.) | Tumor site | Stage |
|---|
|
|
|
|
|
|
|---|
| 1 | Female | 69 | Oral, scc | T1 | N0 | Mx |
| 2 | Male | 63 | Laryngeal, scc | T3 | N0 | M0 |
| 3 | Male | 68 | Oral, scc | T3 | N0 | M0 |
| 4 | Male | 56 | Laryngeal, scc | T2 | N2a | M0 |
| 5 | Male | 48 | Oropharynegeal,
scc | T3 | N1 | M0 |
| 6 | Male | 55 | Laryngeal, scc | T2 | N0 | M0 |
| 7 | Male | 63 | Laryngeal, scc | T1 | N1 | M0 |
| 8 | Female | 51 | Oral, scc | T2 | N0 | M0 |
| 9 | Male | 40 | Oral, scc | T2 | N0 | Mx |
| 10 | Male | 38 | Oral, scc | T1 | N1 | M0 |
| 11 | Male | 58 | Oropharynegeal,
scc | T2 | N2b | M0 |
| 12 | Male | 78 | Laryngeal, scc | T2 | N2a | M0 |
| 13 | Male | 54 | Oropharynegeal,
scc | T3 | N0 | M0 |
| 14 | Male | 64 | Laryngeal, scc | T3 | N2c | M0 |
| 15 | Female | 47 | Laryngeal, scc | T2 | N2b | M0 |
| 16 | Male | 57 | Oropharynegeal,
scc | T2 | N2b | M0 |
| 17 | Male | 79 | Laryngeal, scc | T2 | N1 | M0 |
Additional saliva samples at non-fixed intervals
were collected from 16 patients: 3 women and 13 men (41 samples,
termed Coh 0), which were analysed only for the determination of
the effects of cryopreservation on the stability of microRNAs.
Measurement of saliva microRNAs
We were able to isolate RNA in detectable amounts
from every saliva sample analyzed (Coh 0 + 1). In 95% of the
samples analyzed, RNA concentrations ranged from 50 to 180 ng/µl.
Expression levels of the selected microRNAs were measurable in the
saliva samples to a different extent. miR-93 expression could be
detected in 79 samples (95.2%), miR-125a in 71 samples (85.5%),
miR-142-3p in 70 samples (84.3%), miR-200a in 80 samples (96.4%),
mir-203 in 67 samples (80.7%), miR-213 in 73 samples (88.0%),
let-7a in 78 samples (94.0%), let-7b in 68 samples (81.9%), let-7g
in 56 samples (67.5%) and let-7i in 80 samples (96.4%). U18 snoRNA
was detectable in each saliva sample and was used as reference
gene.
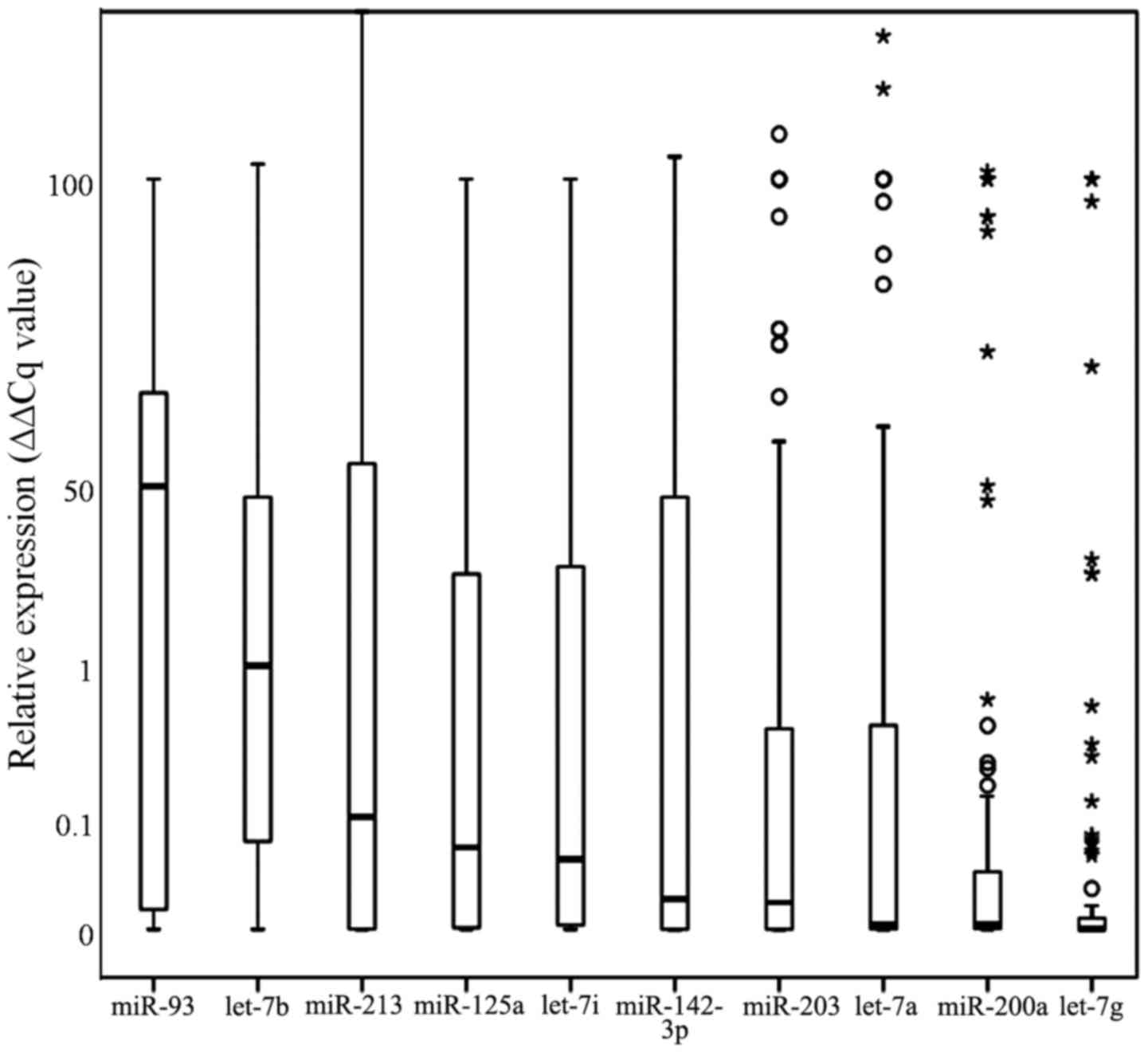
MicroRNA expression between the single species was
significantly different, ranging from a median relative expression
of 58.3 (∆∆Cq value) for miR-93 to a median relative expression of
0.0005 (∆∆Cq value) for let-7g. The specific median relative
expression values were as follows: miR-93: 0.583, let-7b: 0.103,
miR-213: 0.018, miR-125a: 0.011, let-7i: 0.009, miR-142-3p: 0.0032,
miR-203: 0.0028, miR-200a: 0.00004, let-7a: 0.00004, let-7g:
0.000005, respectively (Fig.
1).
In bivariate correlation analyses according to
Spearman-Rho, especially miR-93 expression exhibited several highly
significant associations to other microRNA expression levels,
namely miR-125a, miR-142-3p, miR-203, miR-213, let-7a and b (all
positive correlations, Table II).
On the other hand, miR-213 and let-7i expression levels as well as
let-7a and let-7g expression levels were significantly negatively
correlated (rs= −0.31 and rs= −0.39, p=0.005
and p=0.0003, respectively), indicating that there is no general
microRNA overexpression in saliva of HNSCC patients.
 | Table II.Bivariate correlation analyses
according to Spearman-Rho. |
Table II.
Bivariate correlation analyses
according to Spearman-Rho.
| miR
correlations | rs | p-value | n |
|---|
|
|
|
|
|---|
| miR-93
expression | miR-125a
expression | 0.34 | 0.002 | 83 |
|
| miR-142-3p
expression | 0.52 |
6.4×10−7 | 83 |
|
| miR-203
expression | 0.41 | 0.0001 | 83 |
|
| miR-213
expression | 0.31 | 0.005 | 83 |
|
| let-7a
expression | 0.34 | 0.002 | 83 |
|
| let-7b
expression | 0.31 | 0.005 | 83 |
| miR-125a
expression | miR-213
expression | 0.37 | 0.001 | 83 |
| miR-213
expression | let-7a
expression | 0.41 | 0.0001 | 83 |
|
| let-7i
expression | −0.31 | 0.005 | 83 |
| let-7a
expression | let-7g
expression | −0.39 | 0.0003 | 83 |
MicroRNA expression pre- and
post-radiotherapy
A subset of our cohort with comparable samples (Coh
1) was analyzed for distinct miRNA expression changes in our
selected microRNA panel between baseline sample and post-treatment
samples. MicroRNAs miR-142-3p (16 of 25 samples), miR-93 (15 of 25
samples) and miR-125a (15 of 25 samples) exhibited an increased
level after radiotherapy, while microRNAs let-7a (17 of 25 samples)
and let-7i (13 of 25 samples) showed a decrease in their expression
after radiotherapy (Fig. 2).
However, a consistently down- or up-regulated microRNA could not be
identified in this first attempt.
In a more robust statistical encounter, we evaluated
whether there is a significantly different expression of one or
several microRNAs before (n=21), during (n=31) or after (n=31)
radiotherapy without stratification according to the individual
patients. In Mann-Whitney U tests, there was no significant
alteration between during and post-treatment samples in the
expression of the microRNAs tested (p-values ranging from p=0.14
for miR-200a and p=0.2 for miR-93 to p=0.97 for miR-203).
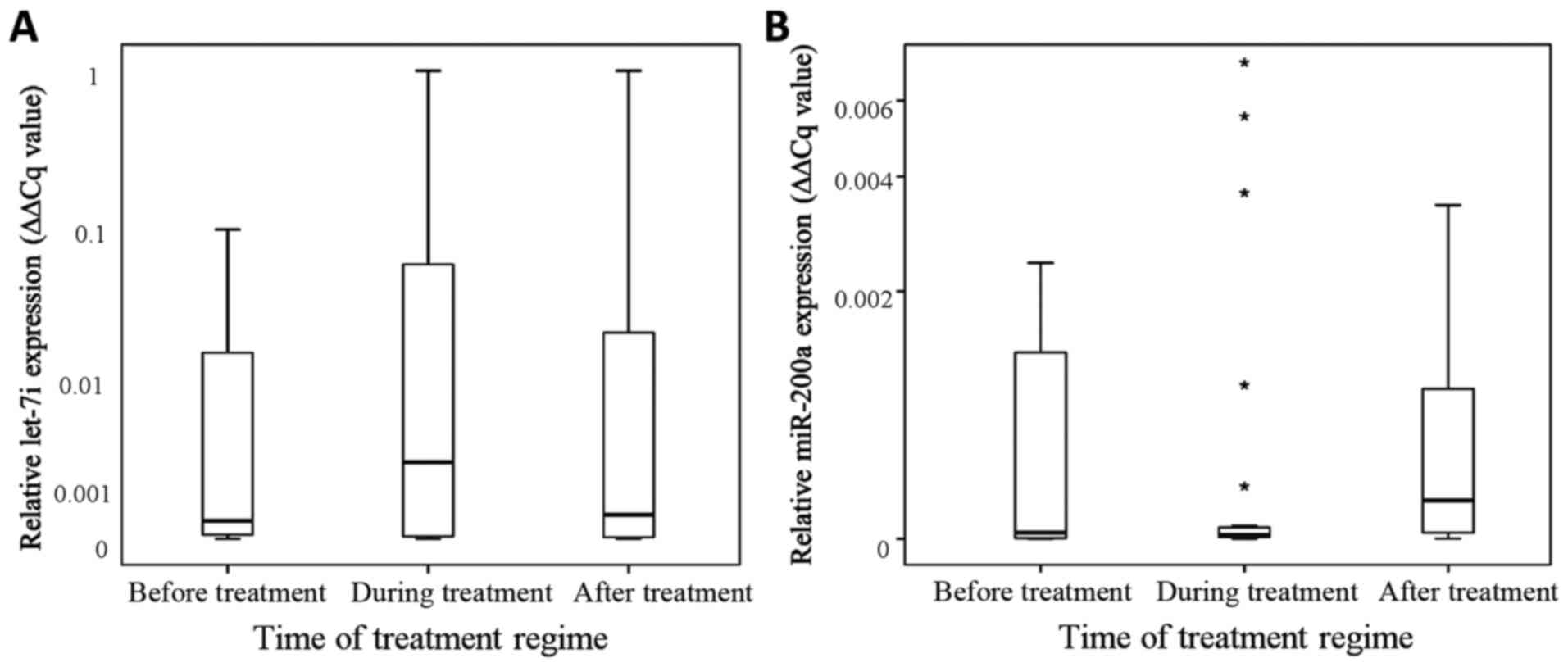
MicroRNA let-7i exhibited an increase in salivary
expression during radiotherapy in comparison to the levels before
radiotherapy (p=0.058, Mann-Whitney U test) and decreased after
treatment in comparison to the levels during radiotherapy (p=0.12,
Mann-Whitney U test; Fig. 3A).
However, Kruskal-Wallis analyses of the distribution of the
specific microRNA expression between the phases pre-, during and
post-radiotherapy revealed an attenuation for let-7i (p=0.87),
whereas miR-200a remains unequally distributed over the different
treatment phases and increases after radiotherapy (p=0.04, Fig. 3B).
Longitudinal distribution of microRNA
expression post radiotherapy
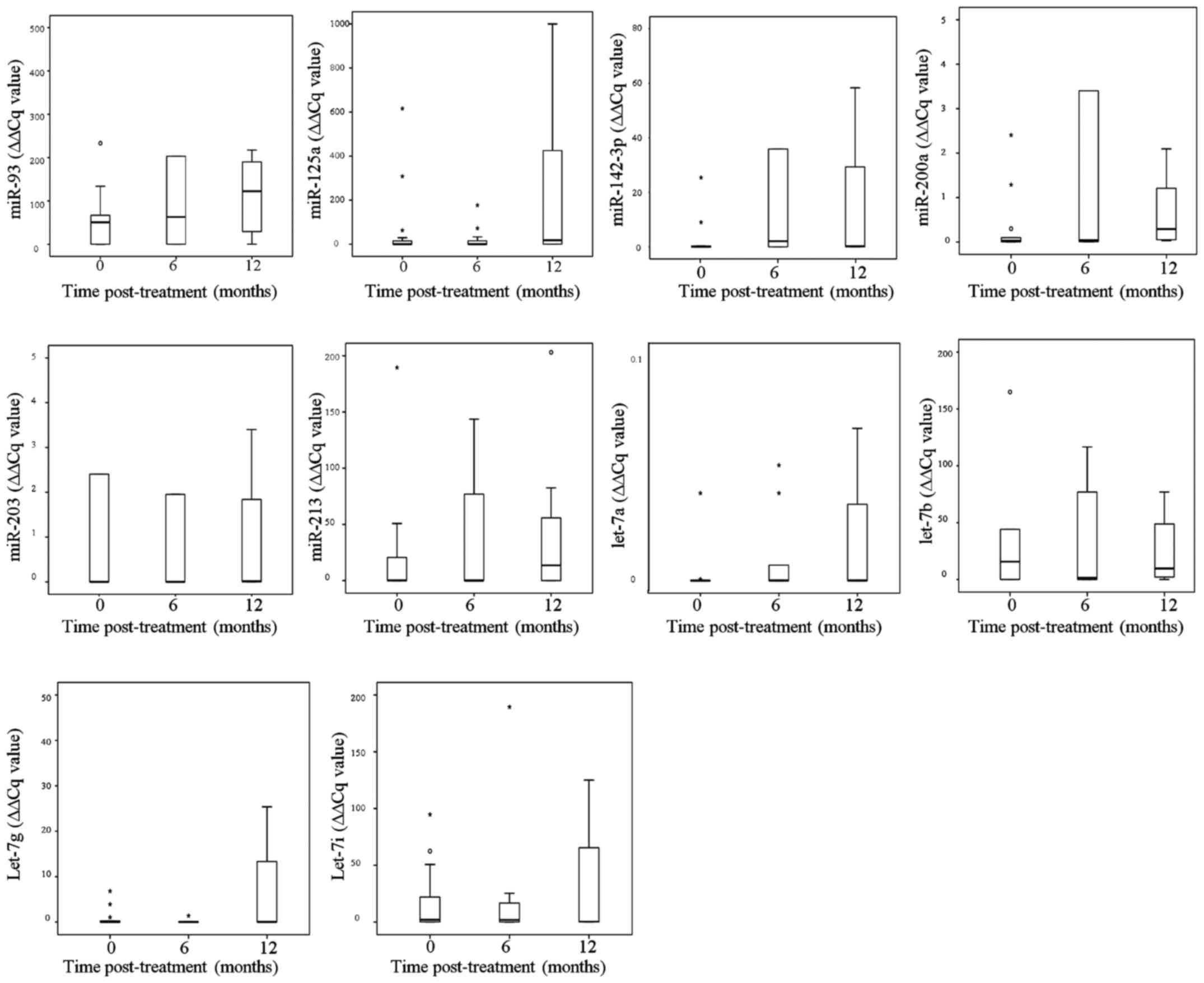
Next, we evaluated the statistical distribution of
relative expression levels of the selected microRNAs according to
the individual patients with saliva samples from defined time
points post-radiotherapy - baseline (n=17), 6 months post-therapy
(n=14), and 12 months post-therapy (n=8). Of note, especially
median miR-93 relative expression increases post-therapy in a
time-dependent manner (Fig. 4). In
Kruskal-Wallis analyses, no significant alterations of relative
microRNA expression distribution were detected over the three time
points (ranging from p=0.09 for miR-200a to p=0.98 for let-7i).
Comparing miR-93 and miR-200a expression at baseline (n=17) with
those at 12 months of post-therapy (n=8), both microRNAs were
significantly higher expressed (p=0.047 and p=0.036 in Mann-Whitney
U test, respectively).
MicroRNA expression in association to
functional saliva gland parameter
Taking into account, that saliva production and
secretion after radiotherapy is heavily dependent on saliva gland
regeneration, we included the different saliva flow rates (in ml
saliva/minute) of the patients at the different time points of
collection (by calculating the ratio of microRNA expression/saliva
flow rate) in our analyses. Noteworthy, in bivariate correlation
according to Spearman-Rho miR-93 and let-7g expression were both
associated to saliva flow rate (miR-93: rs= −0.337;
p=0.036; let-7g: rs=0.334; p=0.037). The formerly
detected associations between microRNA expression and time point of
collection (baseline against 12 months post-treatment) remained
after inclusion of saliva flow rates (p=0.049 for miR-93 and
p=0.012 for miR-200a in Mann-Whitney U tests, respectively).
However, in bivariate correlation analyses miR-93 expression was
significantly positively correlated to miR-125a
(rs=0.59; p=0.000077), miR-142-3p (rs=0.46;
p=0.003) and let-7b (rs=0.44; p=0.005), while miR-200a
expression was only significantly correlated to let-7a expression
(rs=0.37; p=0.02). Furthermore, in a Kruskal-Wallis test
the miR-200a expression/saliva flow ratio was significantly
differently expressed at the end of treatment and 6 and 12 months
after radiotherapy (p=0.036).
Discussion
The result of this study indicate that salivary
microRNAs associated with HNSCC and radiosensitivity stay
detectable in oral fluid during and after 3D-CRT. Therefore,
salivary miRNA analysis still represents a potentially promising
approach to monitor therapy response and recurrence of HNSCC in
patients treated with radiotherapy. Firstly, we detected a
significant increased expression of miR-200a comparing expression
at baseline with that at 12 months post-radiotherapy. miR-200a is
significantly less expressed in the saliva of OSCC patients in
comparison to healthy controls and is considered as putative marker
for tumor monitoring (21).
Furthermore, low expression of miR-200a is associated with a worse
outcome for ovarian carcinoma (36)
and cervical cancer patients (37).
In terms of pro-oncogenic action, decreased expression of miR-200a
promotes the acceleration of meningioma cell growth (38) or the epithelial-mesenchymal
transition and invasive potential of anaplastic thyroid carcinoma
cells (39). miR-200a
overexpression induced inhibition of nasopharyngeal carcinoma cell
line growth via translation inhibition of ZEB2 and CTNNB1, while
downregulation of miR-200a triggers the epithelial-mesenchymal
transition of nasopharyngeal carcinoma cell lines also by ZEB2
activation (40). The increase of
miR-200a detected in our cohort was significant in saliva samples
at 12 months after end of treatment and may be a hint for wound
healing processes and the return to a physiological state.
Furthermore, we detected a significant increased
expression of miR-93. miR-93 is also significantly less expressed
in the saliva of OSCC patients in comparison to healthy controls
and it is a member of the miR-106b-25 cluster. miR-93 plays a role
in cell proliferation and anchorage-independent cell growth in
hepatocellular carcinoma by regulating for instance the
transcription factor E2F1 (41) or
the tumor suppressor gene FUS1 (42). In addition, miR-93 promotes tumor
growth and angiogenesis in cocultured glioblastoma cell lines,
pointing towards the oncogenic potential of the miR-106b-25 cluster
(43). On the other hand, miR-93
overexpression inhibits proliferation and colony formation in colon
cancer cells (44) and acts
differentially in several osteosarcoma cell lines (45). In gastric adenocarcinoma, elevated
expression of miR-93 was detected and high miR-93 expression was
significantly associated with an advanced disease stage and a
decreased survival (46). Moreover,
downregulated miR-93 expression was detected in gastric carcinoma,
and low miR-93 expression was significantly correlated with an
advanced tumor stage and an unfavorable outcome for patients
(47). Altogether, these facts show
a multi-faceted role of miR-93 in tumor genesis and progression. In
our study, miR-93 re-induction and increased expression during
post-treatment follow-up was demonstrated, a hint toward
tumorsuppressive action of miR-93 in HNSCC progression.
let-7 family was demonstrated as clearly oncogenic
transformation associated in several tumor entities such as lung
cancer (48), gastric cancer
(49) and breast cancer (50). The tumor suppressor roles of let-7
family in HNSCC were consistently demonstrated across studies.
let-7 family is also a crucial modulator of stemness and HNSCC
progression (51). In this study,
the let-7 family members were not clearly correlated with HNSCC
treatment follow-up. Only let-7i showed a weak significant
association with the application of ionizing radiation within the
radiation therapy. However, this could be due to the diverging
expression patterns and sometimes synonymous functions of this
microRNA family.
These findings were limited by several factors. Most
samples had a storage time over more than 5 years at −80°C and had
not been stored by specialized protocols or additives. Extensive
degradation of samples over time might be possible. However, in the
light of several mostly forensic approaches it is likely that
degradation was minimal. MicroRNA miR-15b, miR-16 and miR-24 have
been proven to be stable in plasma at room temperature for at least
24 h, although introduction of a synthetic microRNA showed high
RNAse activity (52). Furthermore,
the analyzed microRNAs were still stable after samples underwent up
to 5 freeze-thaw cycles (52).
Stabilization of microRNA patterns in human whole saliva for up to
two days at room temperature was achieved by specialized protocols
(53). Noteworthy, mRNA detection
of saliva-specific marker in dried saliva stains stored at ambient
temperature for up to 6 years and microRNA detection in saliva
stains stored at the same conditions up to 1 year was possible in a
forensic context (19,54).
Another limitation of the present study is the high
standard deviations of the microRNA expression levels due to the
small sample size. Thus, prospective analyses in an extended cohort
are needed. Furthermore, the patients are only a subpopulation of
patients from a previous study with a mean dose of 27.7 Gy of the
contralateral parotid gland.
In conclusion, in this study we have shown for the
first time that tumor monitoring with salivary microRNAs remains
possible even in the radiation-related changes of saliva compounds.
In addition to the results of Salazar and coworkers (55) we also developed a reliable method to
isolate salivary miRNA at different times under radiotherapy of
OSCC. Analysis of salivary microRNA expression in HNSCC patients
after RT might potentially yield monitor markers, urgently asked
for in the existing literature (56). Salivary miR-200a and miR-93 both
were identified to be differentially expressed after radiotherapy
and further validation of their role as well as other
HNSCC-associated microRNAs such as miR-31 (57) in disease progression is warranted.
Furthermore, larger studies about the potential of these microRNAs
as tumor or therapy markers even in multicenter studies are
possible, as we were able to show a stable detection of microRNA
patterns after long-time storage at −80°C.
Acknowledgements
We wish to thank Gabriele Thomas and Katrin Theile
for excellent technical support. The study involving the collection
of HNSCC patient saliva samples was supported by the German Cancer
Aid e.V. grant no. 106386 and grant no. 108429. This study was
supported by a Wilhelm-Roux Grant of the Medical Faculty of the
Martin-Luther-University Halle-Wittenberg (no. 23/03) to J.H.
References
|
1
|
Curado MP and Boyle P: Epidemiology of
head and neck squamous cell carcinoma not related to tobacco or
alcohol. Curr Opin Oncol. 25:229–234. 2013.PubMed/NCBI
|
|
2
|
Sturgis EM and Cinciripini PM: Trends in
head and neck cancer incidence in relation to smoking prevalence:
An emerging epidemic of human papillomavirus-associated cancers?
Cancer. 110:1429–1435. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mork J: Forty years of monitoring head and
neck cancer in Norway - no good news. Anticancer Res. 18:3705–3708.
1998.PubMed/NCBI
|
|
4
|
Brunin F, Mosseri V, Jaulerry C, Point D,
Cosset JM and Rodriguez J: Cancer of the base of the tongue: Past
and future. Head Neck. 21:751–759. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Listl S, Jansen L, Stenzinger A, Freier K,
Emrich K, Holleczek B, Katalinic A, Gondos A and Brenner H: GEKID
Cancer Survival Working Group: Survival of patients with oral
cavity cancer in Germany. PLoS One. 8:e534152013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eckert AW, Kappler M, Schubert J and
Taubert H: Correlation of expression of hypoxia-related proteins
with prognosis in oral squamous cell carcinoma patients. Oral
Maxillofac Surg. 16:189–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Michailidou E, Tzimagiorgis G,
Chatzopoulou F, Vahtsevanos K, Antoniadis K, Kouidou S, Markopoulos
A and Antoniades D: Salivary mRNA markers having the potential to
detect oral squamous cell carcinoma segregated from oral
leukoplakia with dysplasia. Cancer Epidemiol. 43:112–118. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gleber-Netto FO, Yakob M, Li F, Feng Z,
Dai J, Kao HK, Chang YL, Chang KP and Wong DT: Salivary biomarkers
for detection of oral squamous cell carcinoma in a Taiwanese
population. Clin Cancer Res. 22:3340–3347. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fábryová H and Celec P: On the origin and
diagnostic use of salivary RNA. Oral Dis. 20:146–152. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wiklund ED, Gao S, Hulf T, Sibbritt T,
Nair S, Costea DE, Villadsen SB, Bakholdt V, Bramsen JB, Sørensen
JA, et al: MicroRNA alterations and associated aberrant DNA
methylation patterns across multiple sample types in oral squamous
cell carcinoma. PLoS One. 6:e278402011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brinkmann O and Wong DT: Salivary
transcriptome biomarkers in oral squamous cell cancer detection.
Adv Clin Chem. 55:21–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu CJ, Lin SC, Yang CC, Cheng HW and
Chang KW: Exploiting salivary miR-31 as a clinical biomarker of
oral squamous cell carcinoma. Head Neck. 34:219–224. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matse JH, Yoshizawa J, Wang X, Elashoff D,
Bolscher JG, Veerman EC, Bloemena E and Wong DT: Discovery and
prevalidation of salivary extracellular microRNA biomarkers panel
for the noninvasive detection of benign and malignant parotid gland
tumors. Clin Cancer Res. 19:3032–3038. 2013.PubMed/NCBI
|
|
14
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guarnieri DJ and DiLeone RJ: MicroRNAs: A
new class of gene regulators. Ann Med. 40:197–208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hunter P: The great leap forward. Major
evolutionary jumps might be caused by changes in gene regulation
rather than the emergence of new genes. EMBO Rep. 9:608–611. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanson EK, Lubenow H and Ballantyne J:
Identification of forensically relevant body fluids using a panel
of differentially expressed microRNAs. Anal Biochem. 387:303–314.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zubakov D, Boersma AW, Choi Y, van Kuijk
PF, Wiemer EA and Kayser M: MicroRNA markers for forensic body
fluid identification obtained from microarray screening and
quantitative RT-PCR confirmation. Int J Legal Med. 124:217–226.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Park NJ, Zhou H, Elashoff D, Henson BS,
Kastratovic DA, Abemayor E and Wong DT: Salivary microRNA:
Discovery, characterization, and clinical utility for oral cancer
detection. Clin Cancer Res. 15:5473–5477. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boldrup L, Coates PJ, Wahlgren M, Laurell
G and Nylander K: Subsite-based alterations in miR-21, miR-125b,
and miR-203 in squamous cell carcinoma of the oral cavity and
correlation to important target proteins. J Carcinog. 11:182012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mintz PJ, Saetrom P, Reebye V, Lundbaek
MB, Lao K, Rossi JJ, Gaensler KM, Kasahara N, Nicholls JP, Jensen
S, et al: MicroRNA-181a* targets Nanog in a subpopulation of CD34
(+) cells isolated from peripheral blood. Mol Ther Nucleic Acids.
1:e342012. View Article : Google Scholar :
|
|
25
|
Weidhaas JB, Babar I, Nallur SM, Trang P,
Roush S, Boehm M, Gillespie E and Slack FJ: MicroRNAs as potential
agents to alter resistance to cytotoxic anticancer therapy. Cancer
Res. 67:11111–11116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boyerinas B, Park S-M, Hau A, Murmann AE
and Peter ME: The role of let-7 in cell differentiation and cancer.
Endocr Relat Cancer. 17:F19–F36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pignon JP, Bourhis J, Domenge C and
Designé L: Chemotherapy added to locoregional treatment for head
and neck squamous-cell carcinoma: Three meta-analyses of updated
individual data. MACH-NC Collaborative Group. Meta-Analysis of
Chemotherapy on Head and Neck Cancer. Lancet. 355:949–955. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shah JP and Gil Z: Current concepts in
management of oral cancer - surgery. Oral Oncol. 45:394–401. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Forastiere AA, Goepfert H, Maor M, Pajak
TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, et
al: Concurrent chemotherapy and radiotherapy for organ preservation
in advanced laryngeal cancer. N Engl J Med. 349:2091–2098. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cooper JS, Fu K, Marks J and Silverman S:
Late effects of radiation therapy in the head and neck region. Int
J Radiat Oncol Biol Phys. 31:1141–1164. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Möller P, Perrier M, Ozsahin M and Monnier
P: A prospective study of salivary gland function in patients
undergoing radiotherapy for squamous cell carcinoma of the
oropharynx. Oral Surg Oral Med Oral Pathol Oral Radiol Endod.
97:173–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hey J, Setz J, Gerlach R, Janich M,
Sehlleier S, Schaller HG, Gernhardt CR and Kuhnt T:
Parotid-gland-sparing 3D conformal radiotherapy in patients with
bilateral radiotherapy of the head and neck region - results in
clinical practice. Oral Oncol. 45:e11–e17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hey J, Setz J, Gerlach R, Janich M,
Hildebrandt G, Vordermark D, Gernhardt CR and Kuhnt T: Parotid
gland-recovery after radiotherapy in the head and neck region - 36
months follow-up of a prospective clinical study. Radiat Oncol.
6:1252011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lyman JT and Wolbarst AB: Optimization of
radiation therapy, IV: A dose-volume histogram reduction algorithm.
Int J Radiat Oncol Biol Phys. 17:433–436. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu X, Macdonald DM, Huettner PC, Feng Z,
El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN and Wang X:
A miR-200 microRNA cluster as prognostic marker in advanced ovarian
cancer. Gynecol Oncol. 114:457–464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu X, Schwarz JK, Lewis JS Jr, Huettner
PC, Rader JS, Deasy JO, Grigsby PW and Wang X: A microRNA
expression signature for cervical cancer prognosis. Cancer Res.
70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saydam O, Shen Y, Würdinger T, Senol O,
Boke E, James MF, Tannous BA, Stemmer-Rachamimov AO, Yi M, Stephens
RM, et al: Downregulated microRNA-200a in meningiomas promotes
tumor growth by reducing E-cadherin and activating the
Wnt/beta-catenin signaling pathway. Mol Cell Biol. 29:5923–5940.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Braun J, Hoang-Vu C, Dralle H and
Hüttelmaier S: Downregulation of microRNAs directs the EMT and
invasive potential of anaplastic thyroid carcinomas. Oncogene.
29:4237–4244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Y, Tan W, Neo TW, Aung MO, Wasser S,
Lim SG and Tan TM: Role of the miR-106b-25 microRNA cluster in
hepatocellular carcinoma. Cancer Sci. 100:1234–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Du L, Schageman JJ, Subauste MC, Saber B,
Hammond SM, Prudkin L, Wistuba II, Ji L, Roth JA, Minna JD, et al:
miR-93, miR-98, and miR-197 regulate expression of tumor suppressor
gene FUS1. Mol Cancer Res. 7:1234–1243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW, et al: MicroRNA miR-93
promotes tumor growth and angiogenesis by targeting integrin-β8.
Oncogene. 30:806–821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu XF, Zou J, Bao ZJ and Dong J: miR-93
suppresses proliferation and colony formation of human colon cancer
stem cells. World J Gastroenterol. 17:4711–4717. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Montanini L, Lasagna L, Barili V, Jonstrup
SP, Murgia A, Pazzaglia L, Conti A, Novello C, Kjems J, Perris R,
et al: MicroRNA cloning and sequencing in osteosarcoma cell lines:
Differential role of miR-93. Cell Oncol (Dordr). 35:29–41. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen L, Jiang M, Yuan W and Tang H:
Prognostic value of miR-93 overexpression in resectable gastric
adenocarcinomas. Acta Gastroenterol Belg. 75:22–27. 2012.PubMed/NCBI
|
|
47
|
Xiao ZG, Deng ZS, Zhang YD, Zhang Y and
Huang ZC: Clinical significance of microRNA-93 downregulation in
human colon cancer. Eur J Gastroenterol Hepatol. 25:296–301. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J, et al: let-7 regulates self
renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tu HF, Lin SC and Chang KW: MicroRNA
aberrances in head and neck cancer: Pathogenetic and clinical
significance. Curr Opin Otolaryngol Head Neck Surg. 21:104–111.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Patel RS, Jakymiw A, Yao B, Pauley BA,
Carcamo WC, Katz J, Cheng JQ and Chan EK: High resolution of
microRNA signatures in human whole saliva. Arch Oral Biol.
56:1506–1513. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zubakov D, Kokshoorn M, Kloosterman A and
Kayser M: New markers for old stains: Stable mRNA markers for blood
and saliva identification from up to 16-year-old stains. Int J
Legal Med. 123:71–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Salazar C, Nagadia R, Pandit P,
Cooper-White J, Banerjee N, Dimitrova N, Coman WB and Punyadeera C:
A novel saliva-based microRNA biomarker panel to detect head and
neck cancers. Cell Oncol (Dordr). 37:331–338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Murdoch-Kinch CA, Russo N, Griffith S,
Braun T, Eisbruch A and D'Silva NJ: Recovery of salivary epidermal
growth factor in parotid saliva following parotid sparing radiation
therapy: A proof-of-principle study. Oral Surg Oral Med Oral Pathol
Oral Radiol Endod. 111:64–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW
and Lin SC: Increase of microRNA miR-31 level in plasma could be a
potential marker of oral cancer. Oral Dis. 16:360–364. 2010.
View Article : Google Scholar : PubMed/NCBI
|