Introduction
Gliomas account for 30 to 40% of all intracranial
tumors. Approximately half of all gliomas in adults are
glioblastoma (GBM) the most aggressive subtype with a 5-year
survival rate of less than 5% (1,2).
Currently, some advances have been achieved in regards to
multimodal treatments, including surgical extirpation, local
irradiation and conventional chemotherapy. However, the overall
survival of most glioma patients remains poor, particularly for GBM
patients (3,4). Mounting efforts have been made to
explore the molecules and signaling pathways involved in glioma
cell proliferation, migration and invasion (5,6).
However, the mechanisms are still poorly understood, and the
identification of key molecules that show a potential effect on
glioma development is still imperative.
Long non-coding RNAs (lncRNAs) are defined as
endogenous cellular RNAs more than 200 nucleotides long, which lack
a functional open reading frame (7). Accumulating evidence suggests that
lncRNAs are pivotal regulatory molecules that are implicated in
diverse biological processes, including epigenetic, transcriptional
and post-transcriptional regulatory mechanisms (8–10). It
has been found that numerous lncRNAs play central roles in the
tumor-related gene regulatory system, and dysregulation of their
expression is thought to contribute to tumor cell proliferation,
invasion and metastasis (11–13).
Several lines of evidence point to the etiologic role of
dysregulated lncRNAs in glioma, including CRNDE, CASC2, HOTAIR,
GAS5 and MEG3 (14–18).
Recently, Lin et al reported that long
intergenic non-coding RNA for kinase activation (LINK-A) is
critical to the growth factor-induced normoxic HIF1α signaling
pathway in triple-negative breast cancer (19). However, far less is known concerning
the role of LINK-A in glioma as well as the underlying mechanisms.
To expand our knowledge regarding the biological function of LINK-A
in glioma cells, the present study was designed in an attempt to
identify the contribution of LINK-A to the proliferation and
invasion of glioma cells, thereby providing novel therapeutic
strategies for gliomas.
Materials and methods
Cell culture procedures
U87 and U251 glioma cells, and normal human
astrocytes (HAs) were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). Cells were cultured in
Dulbeccos modified Eagles medium (DMEM), supplemented with 10%
heat-inactivated fetal bovine serum (FBS) and 100 U/ml
penicillin/streptomycin. Cell cultures were maintained at 37°C in a
humidified atmosphere of 5% CO2.
RNA interference (RNAi) analysis and
plasmid construction
The following short hairpin RNA (shRNA) was used to
target human LINK-A: 5′-TTACTGAGGTTGAATATGT-3′. Recombinant
lentiviruses expressing sh-LINK-A or sh-control were produced. The
lactate dehydrogenase A (LDH-A) sequences were synthesized and
subcloned into the pCDNA3.1 vector. The pCDNA constructs or the
empty vector were transfected into glioma cells cultured on 6-well
plates according to the manufacturer's instructions.
Real-time PCR analysis
Total RNA was extracted from glioma cells using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The first-strand
cDNA was synthesized from total RNA using the ThermoScript RT-PCR
system. In brief, each PCR reaction mixture containing 10 µl of 2X
SYBR-Green Master Mix, 1 µl of sense and antisense primers (5
µmol/µl) and 1 µl of cDNA (10 ng), was run for 45 cycles with
denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec and
extension at 72°C for 30 sec in a total volume of 20 µl. For
relative quantification, 2−ΔΔCt was calculated and used
as an indication of the relative expression levels, which were
calculated by subtracting CT values of the control gene from the CT
values of LINK-A and LDH-A. The primer sequences for PCR
amplification were: LINK-A, 5′-TTCCCCCATTTTTCCTTTTC-3′ and
5′-CTCTGGTTGGGTGACTGGTT-3′; LDH-A, 5′-TGTGCCTGTATGGAGTGGAA-3′ and
5′-AGCACTCTCAACCACCTGCT-3′. GAPDH was applied as an internal
control. The primer sequences of GAPDH were:
5′-AGCAAGAGCACAAGAGGAAG-3′ and 5′-GGTTGAGCACAGGGTACTTT-3′.
MTT assay
U87 and U251 glioma cells were trypsinized,
resuspended, seeded into a 96-well plate at a concentration of
2,000 cells/well, and incubated at 37°C. The number of viable cells
was measured at daily intervals. At each time point, 10 µl of 5
mg/ml MTT (DingGuo Biotechnology, Co., Ltd., Beijing, China) was
added, and incubation was continued for 4 h. Then, the medium was
removed carefully and 100 µl dimethyl sulfoxide was added at the
end of the incubation. The absorbance was measured at 490 nm on a
spectrophotometer.
Colony formation assay
U87 and U251 glioma cells were seeded into 6-well
plates. The medium was replaced at regular time intervals. After 14
days of culture at 37°C, the natural colonies were washed with
phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde
for 30 min at room temperature. The colonies were then stained with
methylene blue for 10 min, washed with water and air-dried. The
total number of colonies with >50 cells was counted under
fluorescence microscopy.
Scratch wound assay
U87 and U251 glioma cells were infected with
sh-LINK-A or sh-control. Wounds were created in adherent cells
using a 20 µl pipette tip, 48 h after infection. The cells were
then washed 3 times with PBS to remove any free-floating cells and
debris. Medium without serum was added, and the cells were
incubated under normal conditions. Wound healing was observed after
24 h under light microscopy. Representative scrape lines were
photographed using digital microscopy after the culture inserts
were removed. Each experiment was repeated in triplicate.
Invasion assays
Cells (5×105) were seeded on the top side
of a polycarbonate Transwell filter coated with Matrigel (for
Transwell matrix penetration assay) in the upper chamber of the
QCM™ 24-Well Cell Invasion Assay (Cell Biolabs, Inc., San Diego,
CA, USA). For the invasion assay, cells were suspended in medium
without serum, and medium supplemented with serum was used as a
chemoattractant in the lower chamber. The cells were incubated at
37°C for 24 h. The non-invasive cells in the top chambers were
removed with cotton swabs. The invaded cells on the lower membrane
surface were fixed with methanol and stained with crystal violet.
Cells were counted visually in 5 random fields under a light
microscope (10X objective lens). In addition, invaded cells were
dissociated, lysed and quantified at 570 nm using a
spectrophotometer.
Western blotting
U87 and U251 glioma cells were lysed with RIPA lysis
buffer (Beyotime, Beijing, China). Whole extracts were prepared,
and protein concentrations were determined using the BCA protein
assay kit (Boster, Wuhan, China). Whole-cell extracts (20 µg) were
then fractionated by electrophoresis through 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were
run at 120 V for 2 h before transfer onto a polyvinylidene
difluoride (PVDF) membrane (Millipore Corporation Billerica, MA,
USA). After blocking against non-specific protein binding,
nitrocellulose blots were incubated for 1 h with primary antibodies
diluted in TBS/Tween-20 (0.075% Tween-20) containing 3% MARVEL.
An-LDH-A (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was
diluted at 1:500. Following incubation with the primary antibody,
blots were washed 3 times in TBS/Tween-20 before incubation for 1 h
with goat anti-mouse horseradish peroxidase-conjugated antibody at
a 1:10,000 dilution in TBS/Tween-20 containing 5% milk. After
extensive washing in TBS/Tween-20, the blots were rinsed with
distilled water and proteins were detected using the enhanced
chemiluminescence system. Proteins were visualized with a
chemiluminescent (ECL) kit (ECL Plus; Thermo Fisher Scientific,
Inc., Waltham, MA, USA).
Measurement of glucose consumption and
lactate production
LDHA-expressing vector or the emplty vector was
transfected into U87 and U251 glioma cells. Cell culture media were
collected 48 h after the transfection. Lactate production and
glucose uptake were measured using a lactate assay kit (Sigma, St.
Louis, MO, USA) and Amplex Red Glucose/Glucose Oxidase Assay kit
(Invitrogen), respectively. The results were normalized according
to total cellular protein amounts.
Statistical analysis
All data are expressed as mean ± SD of 3 independent
experiments, in which each assay was performed in triplicate. Data
were analyzed with SPSS 16.0 software. Evaluation of the data was
performed by Students t-test (two-sided) and one-way ANOVA.
P<0.05 was considered statistically significant.
Results
LINK-A is upregulated in glioma
cells
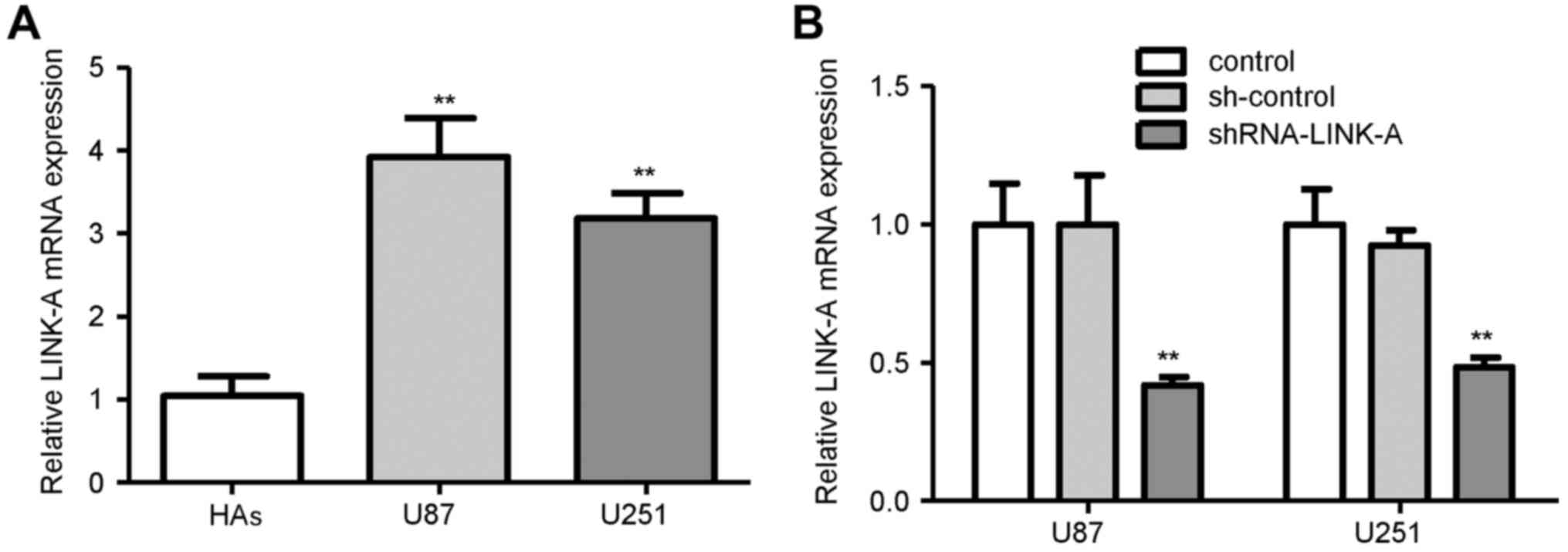
The expression levels of LINK-A in glioma cell lines
(U87 and U251) and normal HAs were examined. As shown in Fig. 1A, the mRNA expression of LINK-A was
higher in the glioma cell lines than that in the HAs, suggesting
that LINK-A upregulation may play important roles in human
glioma.
LINK-A promotes cell growth in glioma
cells
To further investigate the function of LINK-A in
glioma cells, a lentivirus carrying a specific shRNA against LINK-A
(shRNA-LINK-A) to knockdown its expression was infected into U87
and U251 glioma cells. U87 and U251 cells with non-target shRNA
(sh-control) served as the control. As shown in Fig. 1B, knockdown of LINK-A significantly
decreased LINK-A mRNA expression in the U87 glioma cells compared
with that in the sh-control group. Similar results were found in
the U251 glioma cells (Fig. 1B).
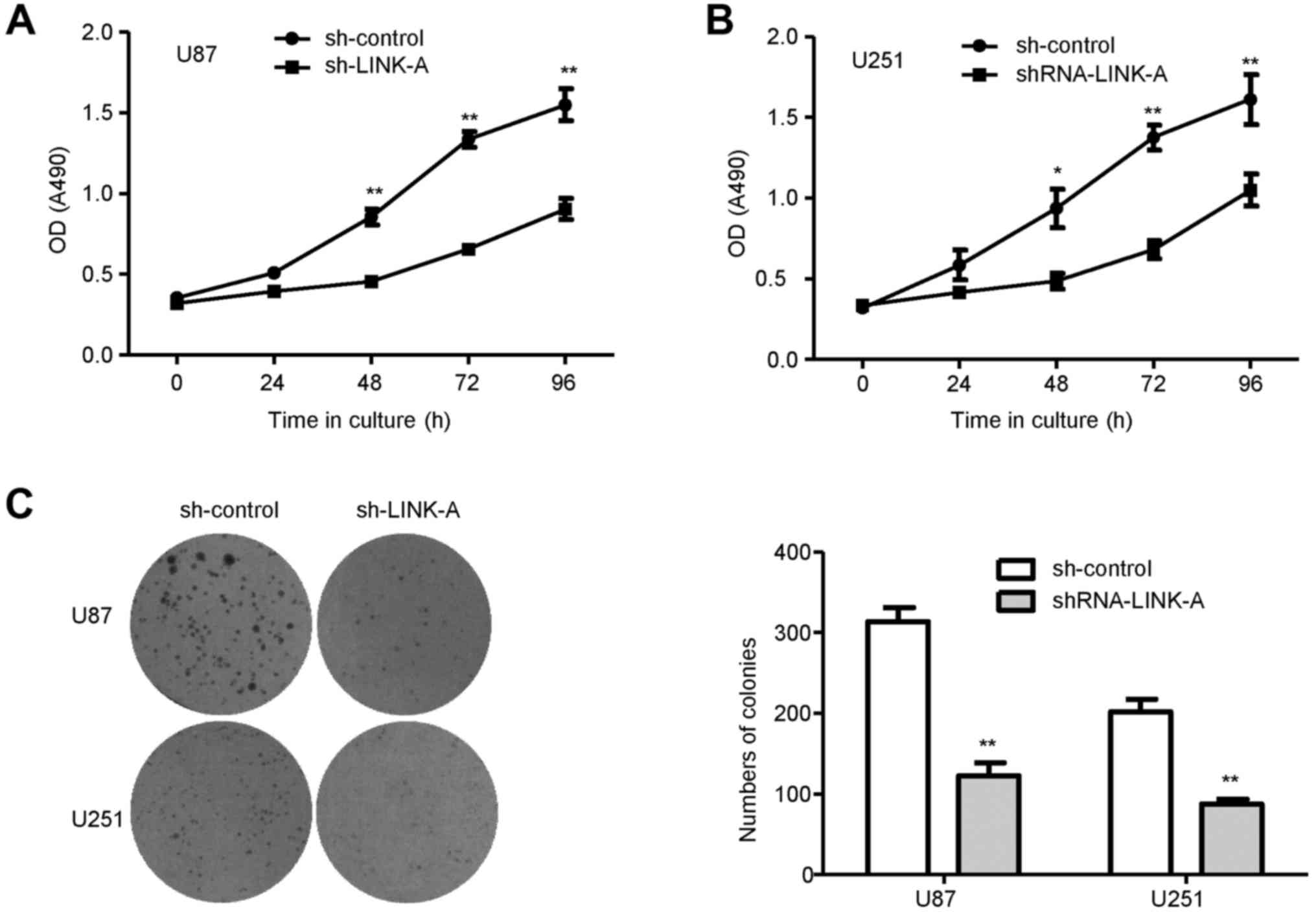
Then, we performed MTT assays to detect the effects of LINK-A on
glioma cell proliferation. As shown in Fig. 2A and B, U87 and U251 glioma cells
transfected with shRNA-LINK-A exhibited a lower proliferative rate,
in comparison with that noted in the respective sh-control groups.
To examine whether LINK-A has an influence on the colony-forming
capacity of glioma cells, a colony formation assay was performed in
the U87 and U251 glioma cells. The numbers of colonies formed in
the shRNA-LINK-A groups were significantly decreased when compared
with the numbers in the sh-control groups (Fig. 2C), suggesting that the reduced
expression of LINK-A significantly inhibited colony formation in
the glioma cells.
LINK-A promotes glioma cell migration
and invasion
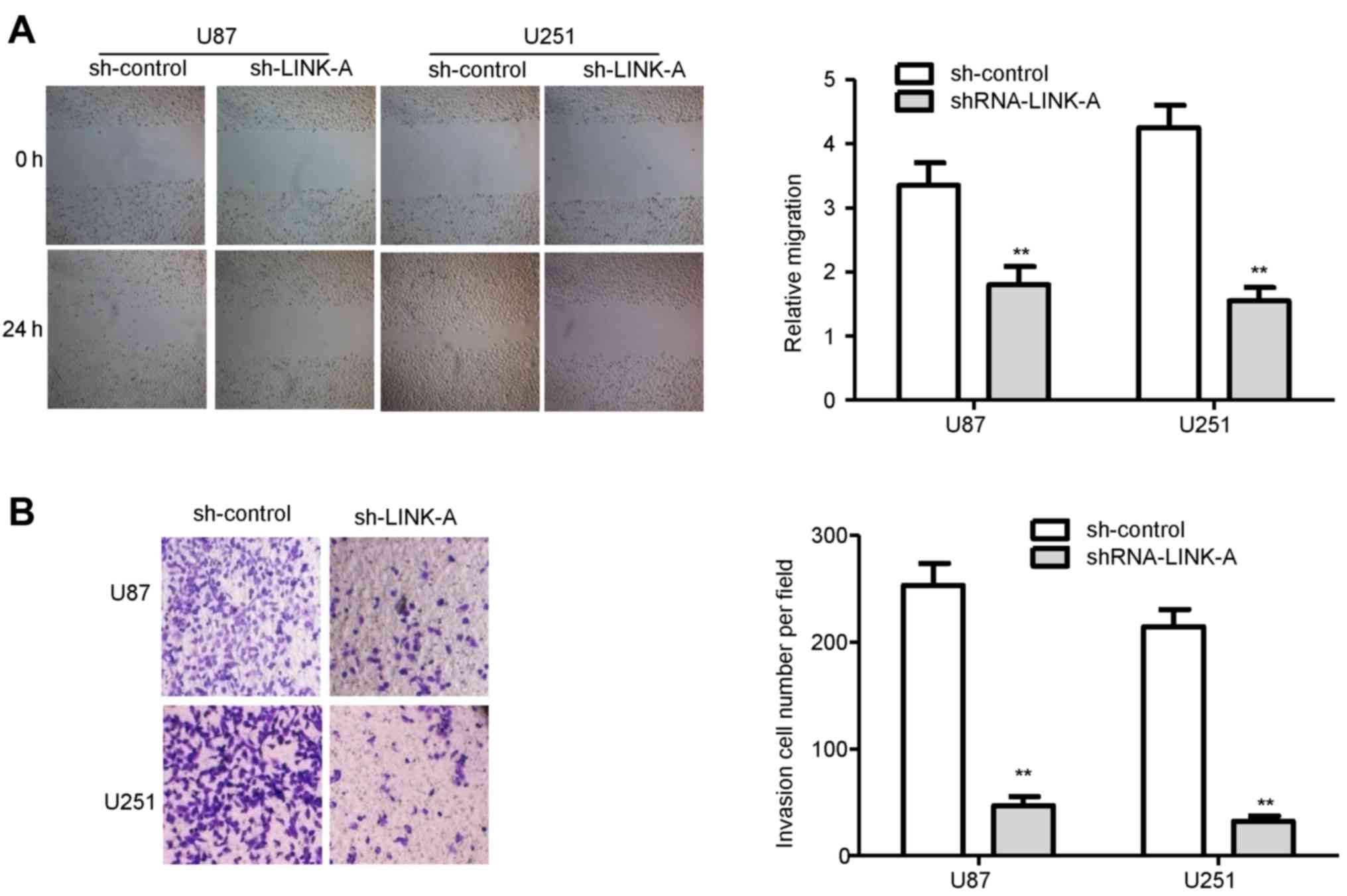
To ascertain whether LINK-A is involved in the
migration and invasion of glioma cells, we performed wound healing
and invasion assays. The migration assays showed that the migratory
rate of the U87 and U251 glioma cells in the shRNA-LINK-A groups
was significantly reduced in comparison with the sh-control groups
(Fig. 3A). Similarly, Transwell
invasion assays confirmed that LINK-A knockdown reduced the
invasion ability of the U87 and U251 glioma cells (Fig. 3B).
Suppression of LDH-A by LINK-A
knockdown
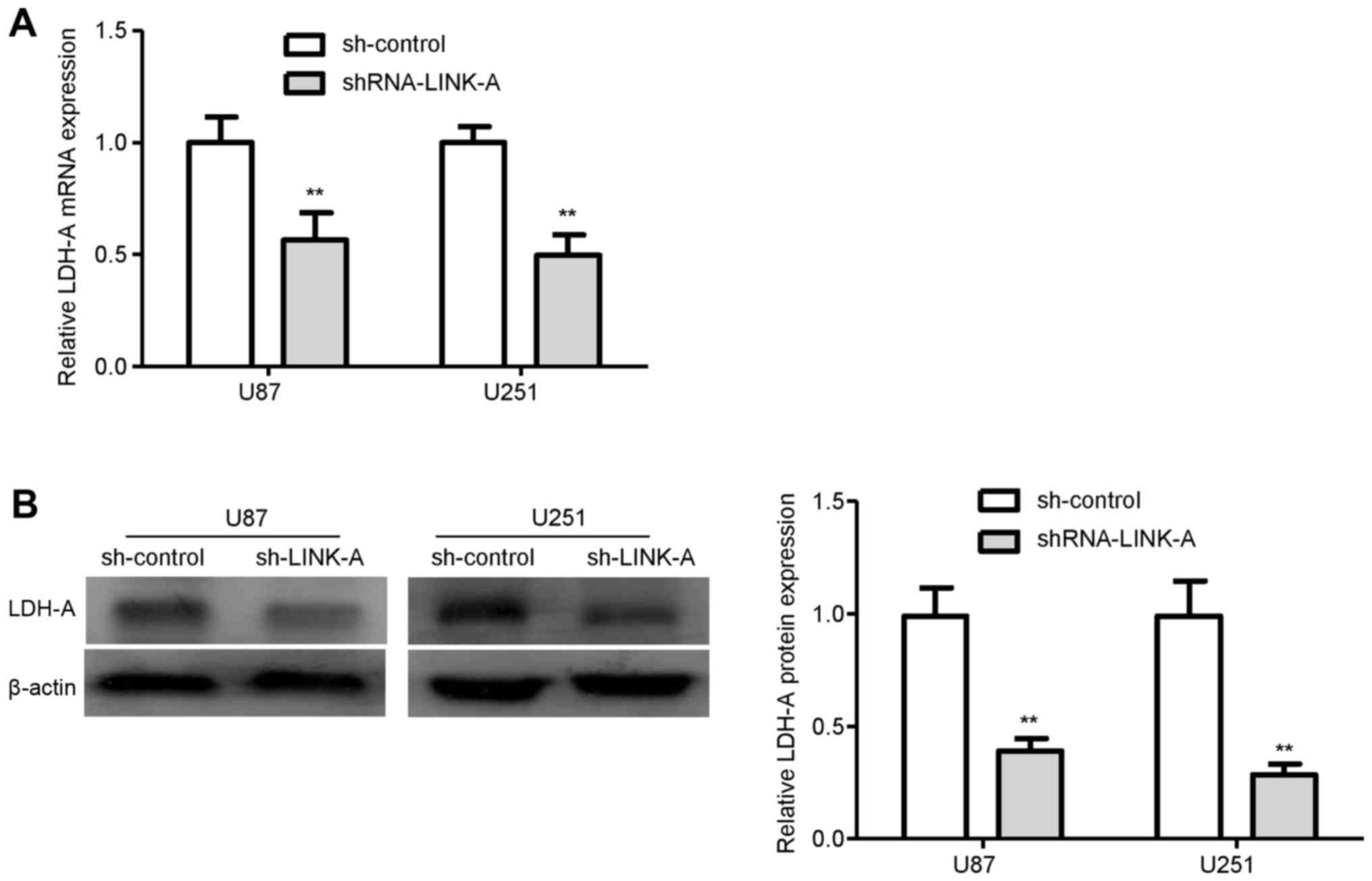
To explore the molecular mechanism by which LINK-A
exerts biological function in glioma cells, we performed qRT-PCR
analysis to investigate the effects of LINK-A knockdown on LDH-A,
which is frequent aberrantly activated in cancer (20). qRT-PCR analysis showed that the
levels of LDH-A mRNA expression were markedly reduced in the
sh-LINK-A-infected U87 and U251 glioma cells compared with levels
in the sh-control groups (Fig. 4A).
Similarly, knockdown of LINK-A significantly reduced the protein
expression of LDH-A in both the U87 and U251 glioma cells (Fig. 4B). These results indicated that
LDH-A may be involved in the LINK-A-induced proliferation and
invasion of glioma cells.
LDH-A promotes glycolysis and cell
proliferation
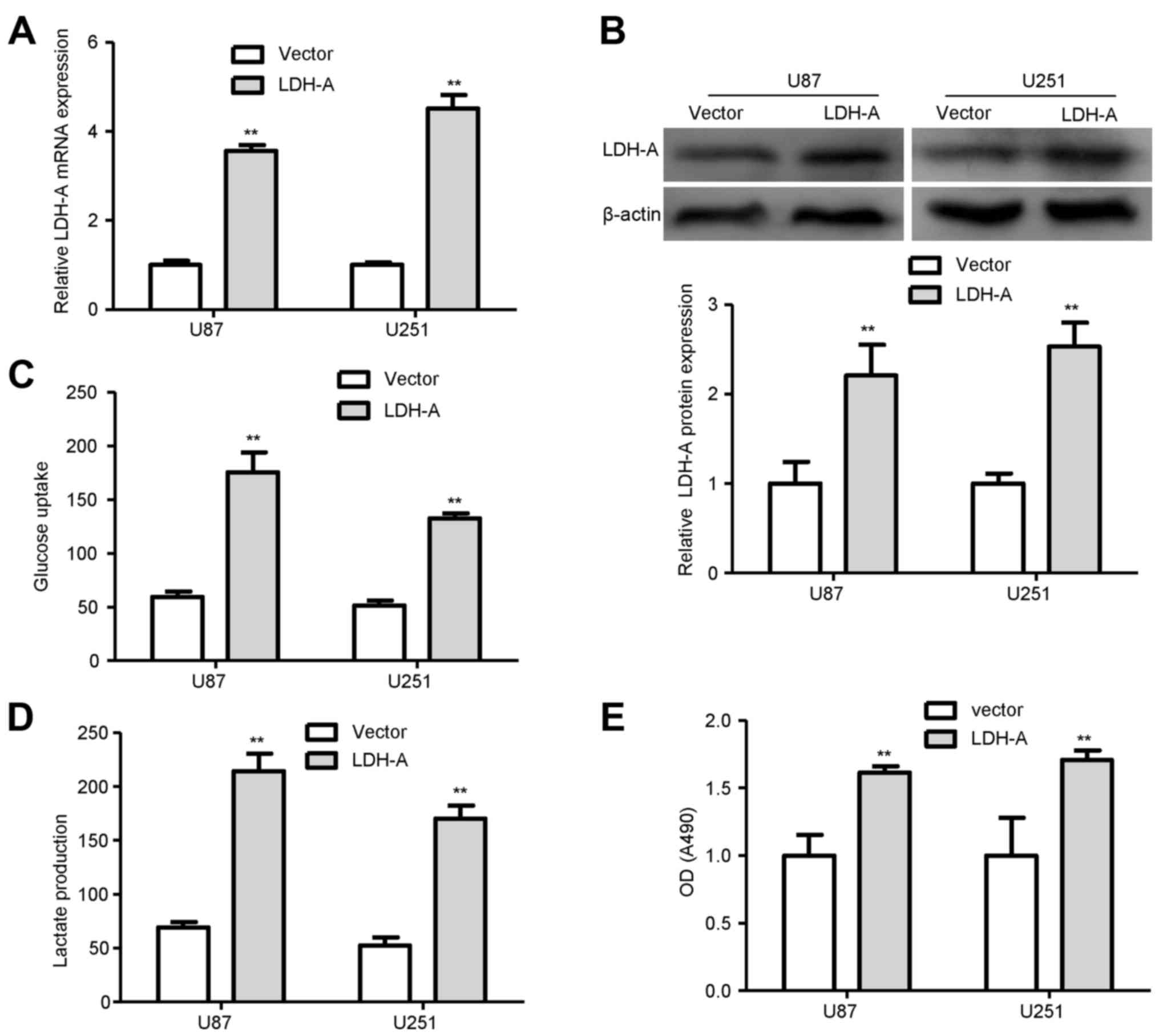
To assess the biological effects of LDH-A in glioma,
LDHA-expressing vector or the empty vector was transfected into U87
and U251 glioma cells, respectively. qRT-PCR and western blot
analysis demonstrated that the transfection was successful
(Fig. 5A and B). Next, we examined
the differences in metabolic parameters and we found that increased
expression of LDH-A largely influenced aerobic glycolysis in the
glioma cells, e.g., increased glucose uptake and lactate production
(Fig. 5C and D). To confirm the
role of LDH-A in glioma cells, we performed a proliferation assay
in glioma cells. We found that overexpression of LDH-A in the U87
and U251 glioma cells significantly promoted cell proliferation
compared with that in the empty vector groups (Fig. 5E).
LDH-A mediates the tumor-suppressive
effects of sh-LINK-A in glioma cells
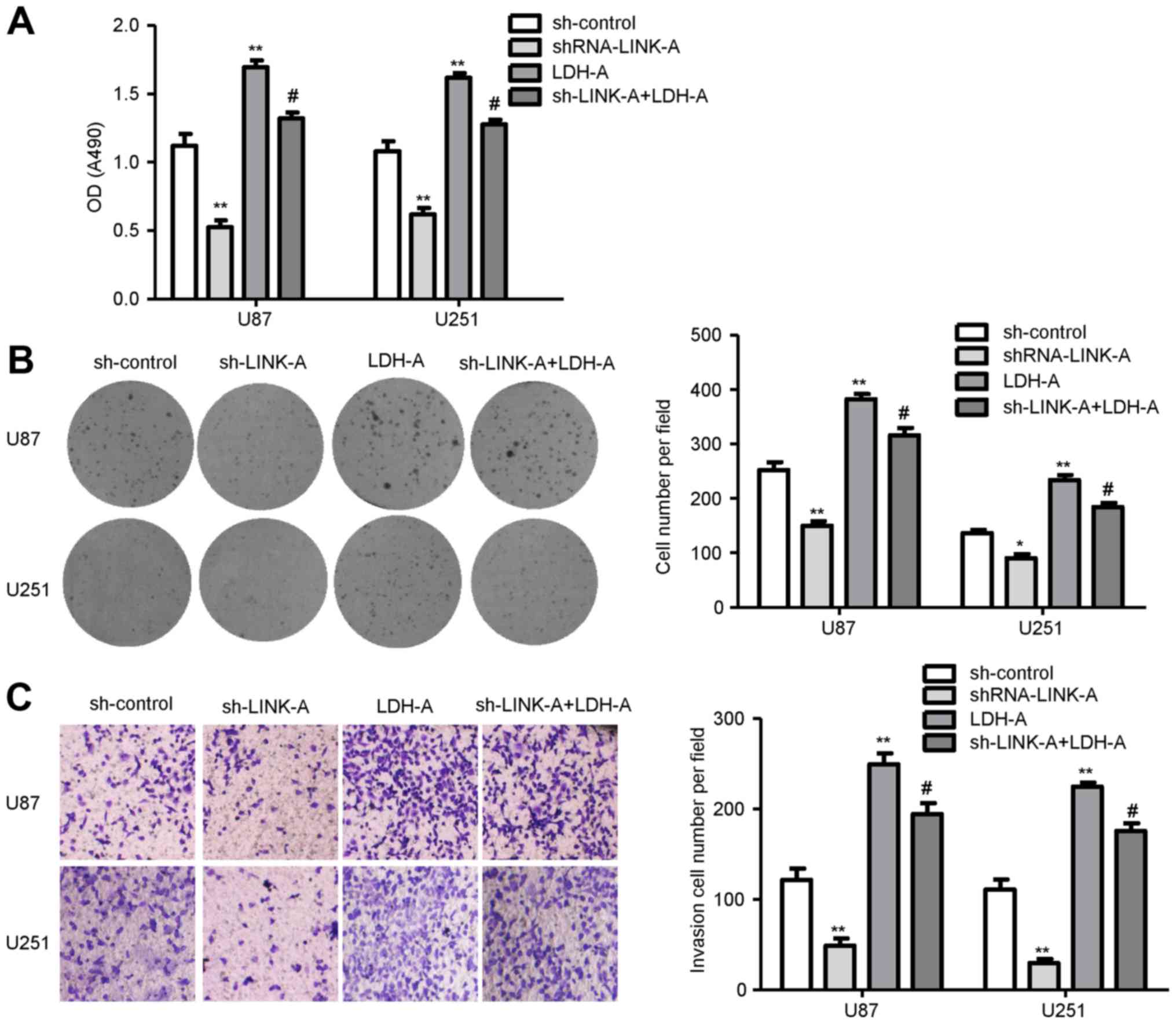
To clarify whether the tumor-suppressive effects of
shRNA-LINK-A were mediated by LDH-A, we transfected the LDH-A
plasmid into the U87 and U251 glioma cells infected by
shRNA-LINK-A. In rescue studies, cell proliferation assay results
showed that shRNA-LINK-A inhibited glioma cell proliferation and
LDH-A promoted glioma cell proliferation. Co-transfection of
shRNA-LINK-A and the LDH-A plasmid showed that LDH-A rescued the
decrease in glioma cell proliferation by shRNA-LINK-A (Fig. 6A). Colony formation assay was used
to further assess the proliferation ability. As shown in Fig. 6B, overexpression of LDH-A rescued
U87 and U251 glioma cell cloning capability inhibited by
shRNA-LINK-A. Moreover, overexpression of LDH-A reversed the
invasive ability in the LINK-A knockdown glioma cells (Fig. 6C). Therefore, these results indicate
that the tumor oncogene function of LINK-A is via LDH-A in glioma
cells.
Discussion
Genome-wide surveys have revealed that ~90% of the
genome is actively transcribed into non-coding RNAs (ncRNAs), while
<2% of the genome sequences encode proteins (21). Although ncRNAs were initially argued
to be spurious transcriptional noise, recent evidence suggests that
the transcriptional noise of the genome may play a major biological
role in cellular development and human diseases (22,23).
The newly discovered lncRNAs, identified as one type of ncRNAs, are
poorly conserved and capable to regulate gene expression at various
levels (24,25). Generally, lncRNAs have been involved
in gene-regulatory roles, such as chromosome dosage-compensation,
imprinting, epigenetic regulation, cell cycle control, nuclear and
cytoplasmic trafficking, transcription, translation, splicing and
cell differentiation (23,26,27).
Recently, it has been found that lncRNAs affect many cellular
processes in tumor cells, such as cell cycle, proliferation,
migration and invasion (28,29).
During recent years, an accumulated number of
studies have focused on the functional role of lncRNAs in
tumorigenesis. Zhang et al reported that HOTAIR is a cell
cycle-related lncRNA in human glioma, and its expression is closely
related to glioma staging and poor prognosis (30). In addition, MALAT1 expression was
lower than that in normal brain tissues, whereas overexpression of
MALAT1 caused significant reduction in cell proliferation and
invasion in vitro, and tumorigenicity in both subcutaneous
and intracranial human glioma xenograft models. Furthermore,
MALAT1-mediated tumor suppression in glioma cells may be via the
attenuation of ERK/MAPK-mediated growth and MMP2-mediated
invasiveness (31).
Long intergenic non-coding RNA for kinase activation
(LINK-A), known as LOC339535, is a highly prognostic lncRNA in
triple-negative breast cancer, which mediates HIF1α
phosphorylation, and then causes HIF1α stabilization and activation
of HIF1α transcriptional programs (19). In the present study, we found that
LINK-A was significantly upregulated in glioma cells. This result
prompted us to speculate that downregulation of LINK-A may be
essential for glioma cells, and its knockdown may suppress tumor
growth of glioma. Via successful cell infection with shRNA-LINK-A,
and the detection of glioma cell proliferation, migration and
invasion, it was demonstrated that knockdown of LINK-A suppressed
the growth of glioma cells. In addition, knockdown of LINK-A and
the related assays were also performed to confirm the promoting
effect of LINK-A on migration and invasion of glioma cells.
Lactate dehydrogenase A (LDH-A) is thought to be a
major molecular mediator of the Warburg effect and to play a
critical role in the metabolism of tumor cells (32–34).
LDH-A increases the efficiency of the LDH complex, allowing the
rapid flux via glycolysis that is responsible for the energy needs
of rapidly proliferating cells (35). It has been reported that elevated
levels of LDH-A are a hallmark of many tumors, including glioma,
and is associated with the clinicopathological features and
survival outcomes of patients (36–38).
Inhibition of LDH-A typically results in accelerated oxygen
consumption, reduced cell malignant transformation and markedly
delayed tumor formation, indicating the underlying role of LDH-A in
tumor initiation or maintenance (20,39).
Recently, Lin et al reported that knockdown
of LINK-A in triple-negative breast cancer cells markedly reduced
the expression of LDH-A, and impaired glycolysis, suggesting that
LDH-A may represent an important downstream effector of LINK-A
(19). Therefore, we speculated
that LDH-A was probably also regulated by LINK-A in glioma cells.
Thus, we determined the levels of LDH-A in LINK-A-knockdown glioma
cells. As a result, we found that LINK-A knockdown in glioma cells
downregulated the LDH-A expression. Moreover, LDH-A facilitated
glucose uptake, lactate production and markedly enhanced cell
proliferation in glioma cells. These findings suggest that LDH-A
may promote glioma malignant potential via the glycolysis
pathway.
To investigate whether LDH-A is involved in the
inhibition of cell proliferation and invasion regulated by LINK-A,
shRNA-LINK-A was infected into glioma cells where LDH-A was
overexpressed by transfection with pcDNA-LDH-A. LDH-A
overexpression reversed the inhibitory effect mediated by LINK-A
knockdown. Therefore, we conclude that LDH-A is involved in the
proliferation and invasion of glioma cells influenced by
LINK-A.
In conclusion, we found that knockdown of LINK-A
inhibited glioma cell proliferation and invasion. Moreover, the
involvement of LDH-A in the proliferation and invasion of glioma
cells was mediated by LINK-A. Therefore, LINK-A may serve as an
oncogenic lncRNA that promotes proliferation and invasion of glioma
cells through LDH-A.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81470112).
Glossary
Abbreviations
Abbreviations:
|
GBM
|
glioblastoma
|
|
lncRNAs
|
long non-coding RNAs
|
|
LINK-A
|
long intergenic non-coding RNA for
kinase activation
|
|
HA
|
normal human astrocyte
|
|
RNAi
|
RNA interference
|
|
ncRNAs
|
non-coding RNAs
|
|
LDH-A
|
lactate dehydrogenase A
|
References
|
1
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol.
15:(Suppl 2). ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei J, Gabrusiewicz K and Heimberger A:
The controversial role of microglia in malignant gliomas. Clin Dev
Immunol. 2013:2852462013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: The avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gupta K and Salunke P: Molecular markers
of glioma: An update on recent progress and perspectives. J Cancer
Res Clin Oncol. 138:1971–1981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hochberg FH, Atai NA, Gonda D, Hughes MS,
Mawejje B, Balaj L and Carter RS: Glioma diagnostics and
biomarkers: An ongoing challenge in the field of medicine and
science. Expert Rev Mol Diagn. 14:439–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen LL and Carmichael GG: Long noncoding
RNAs in mammalian cells: What, where, and why? Wiley Interdiscip
Rev RNA. 1:2–21. 2010.PubMed/NCBI
|
|
8
|
Jeon Y, Sarma K and Lee JT: New and
Xisting regulatory mechanisms of X chromosome inactivation. Curr
Opin Genet Dev. 22:62–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Quek XC, Thomson DW, Maag JL, Bartonicek
N, Signal B, Clark MB, Gloss BS and Dinger ME: lncRNAdb v2.0:
Expanding the reference database for functional long noncoding
RNAs. Nucleic Acids Res. 43:D168–D173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bian EB, Ma CC, He XJ, Wang C, Zong G,
Wang HL and Zhao B: Epigenetic modification of miR-141 regulates
SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget.
7:30610–30625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J
and Xue YX: Long non-coding RNA CASC2 suppresses malignancy in
human gliomas by miR-21. Cell Signal. 27:275–282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Wang Y, Li J, Zhang Y, Yin H and
Han B: CRNDE, a long-noncoding RNA, promotes glioma cell growth and
invasion through mTOR signaling. Cancer Lett. 367:122–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao X, Wang P, Liu J, Zheng J, Liu Y,
Chen J and Xue Y: Gas5 exerts tumor-suppressive functions in human
glioma cells by targeting miR-222. Mol Ther. 23:1899–1911. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Bian EB, He XJ, Ma CC, Zong G, Wang
HL and Zhao B: Epigenetic repression of long non-coding RNA MEG3
mediated by DNMT1 represses the p53 pathway in gliomas. Int J
Oncol. 48:723–733. 2016.PubMed/NCBI
|
|
19
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Le A, Cooper CR, Gouw AM, Dinavahi R,
Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL and Dang
CV: Inhibition of lactate dehydrogenase A induces oxidative stress
and inhibits tumor progression. Proc Natl Acad Sci USA.
107:2037–2042. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote
cancer metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang K, Sun X, Zhou X, Han L, Chen L, Shi
Z, Zhang A, Ye M, Wang Q, Liu C, et al: Long non-coding RNA HOTAIR
promotes glioblastoma cell cycle progression in an EZH2 dependent
manner. Oncotarget. 6:537–546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X,
Sun T, Xie X, Zhou Y and Du Z: Tumor-suppressive function of long
noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and
inactivation of ERK/MAPK signaling. Cell Death Dis. 7:e21232016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J,
Zhu Z, Gao Y and Xie K: A novel KLF4/LDHA signaling pathway
regulates aerobic glycolysis in and progression of pancreatic
cancer. Clin Cancer Res. 20:4370–4380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Su D, Zhao L, Zhang D, Xu J, Wan
J, Fan S and Chen M: Different effects of LDH-A inhibition by
oxamate in non-small cell lung cancer cells. Oncotarget.
5:11886–11896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang
P, Xu YH, Dong B, Xiong Y, Lei QY, et al: Lysine-5 acetylation
negatively regulates lactate dehydrogenase A and is decreased in
pancreatic cancer. Cancer Cell. 23:464–476. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chesnelong C, Chaumeil MM, Blough MD,
Al-Najjar M, Stechishin OD, Chan JA, Pieper RO, Ronen SM, Weiss S,
Luchman HA, et al: Lactate dehydrogenase A silencing in IDH mutant
gliomas. Neuro-oncol. 16:686–695. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cai Z, Zhao JS, Li JJ, Peng DN, Wang XY,
Chen TL, Qiu YP, Chen PP, Li WJ, Xu LY, et al: A combined
proteomics and metabolomics profiling of gastric cardia cancer
reveals characteristic dysregulations in glucose metabolism. Mol
Cell Proteomics. 9:2617–2628. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Crane CA, Austgen K, Haberthur K, Hofmann
C, Moyes KW, Avanesyan L, Fong L, Campbell MJ, Cooper S, Oakes SA,
et al: Immune evasion mediated by tumor-derived lactate
dehydrogenase induction of NKG2D ligands on myeloid cells in
glioblastoma patients. Proc Natl Acad Sci USA. 111:12823–12828.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim J, Han J, Jang Y, Kim SJ, Lee MJ, Ryu
MJ, Kweon GR and Heo JY: High-capacity glycolytic and mitochondrial
oxidative metabolisms mediate the growth ability of glioblastoma.
Int J Oncol. 47:1009–1016. 2015.PubMed/NCBI
|
|
39
|
Fantin VR, St-Pierre J and Leder P:
Attenuation of LDH-A expression uncovers a link between glycolysis,
mitochondrial physiology, and tumor maintenance. Cancer Cell.
9:425–434. 2006. View Article : Google Scholar : PubMed/NCBI
|