Introduction
Cervical cancer (CC) is one of the most common
cancers and the third leading cause of cancer-related deaths in
women worldwide (1,2). Despite notable advances in surgery,
radiotherapy and chemotherapy, even the novel molecular-targeted
therapy, the long-term survival of advanced CC patients remains
poor due to the high rates of tumor recurrence and distal
metastasis (3–5). To elucidate the development and
progression of CC, it is crucial to identify the molecular etiology
and molecular mechanisms underlying the progression and metastasis
in CC and thus improve therapeutic strategies and prognosis.
MicroRNAs (miRNAs) are a class of small, non-coding
RNAs that negatively regulate protein expression by binding to the
3′-untranslated region (3′-UTR) of target mRNAs and function as
post-transcriptional regulators of gene expression (6,7).
Accumulating studies have confirmed that aberrant expression of
miRNAs is involved in various types of cancer, including CC, and
participate in diverse biological processes such as cell growth,
invasion, differentiation and metastasis (8–12).
Recently, miR-106a, which is derived from the precursor
miR-106a-363 on chromosome Xq26.2, was revealed to play a critical
role in the progression of cancers (13–17). A
previous study revealed that miR-106a functions as a regulator of
cell invasion, proliferation and drug sensitivity (18). miR-106a functioned as an oncogene in
human gastric cancer and contributed to proliferation and
metastasis in vitro and in vivo and a high expression
of miR-106a was associated with the poor survival rate of gastric
cancer patients (19,20). Upregulated expression of miR-106a by
DNA hypomethylation played an oncogenic role in hepatocellular
carcinoma by targeting TP53INP1 and CDKN1A (21). However, the expression of miR-106a
was downregulated in glioma, renal carcinoma and osteosarcoma
cancer (22–24). miR-106a inhibited glioma cell growth
by targeting E2F1 independent of p53 status (25). miR-106a inhibited the proliferation
of renal carcinoma cells by targeting IRS-2 (24). Moreover, miR-106a suppressed the
proliferation, migration and invasion of osteosarcoma cells by
targeting HMGA2 (22). Thus, the
functional significance of miR-106a in cancer initiation,
development and progression appears to be cancer-type specific.
However, the functional role and the underlying molecular mechanism
by which miR-106a regulates the development and progression of CC
have not been elucidated.
In this study, we investigated the expression and
biological function of miR-106a in CC progression. We demonstrated
that the expression of miR-106a was upregulated in both CC tissues
and cell lines. Increased expression of miR-106a was associated
with adverse prognostic characteristics and poor 5-year survival of
CC patients. MiR-106a promoted CC migration and invasion in
vitro by gain- and loss-of-function experiments by directly
targeting tissue inhibitor of metalloproteinase (TIMP)2.
Collectively, these data confirmed the underlying mechanism by
which miR-106a promoted migration and invasion of CC and identified
miR-106a as a novel prognostic biomarker for CC patients.
Materials and methods
Clinical samples and cell culture
CC tissues and corresponding adjacent normal tissues
were obtained from 135 patients who received routine surgery at the
Affiliated Tumor Hospital of Xinjiang Medical University from
January 2007 to December 2010. None of the patients had received
any therapy before surgery. All tissues were stored at −80°C until
RNA extraction. Informed consent was obtained from all the patients
and this study was approved by the ethics committee of the local
institutional review boards in accordance with the Declaration of
Helsinki.
Four CC cell lines (C33A, HeLa, CaSki, SiHa) and a
normal human cervical epithelial cell line (H8) were obtained from
the Institute of Biochemistry and Cell Biology (Chinese Academy of
Sciences, Shanghai, China) and were cultured in Dulbecco's modified
Eagle's medium (DMEM; Hyclone, Logan, UT, USA) containing 10% fetal
bovine serum (FBS; Gibco, Grand Island, NY, USA) with 1%
penicillin/streptomycin (Sigma, St. Louis, MO, USA) at 37°C with 5%
CO2.
Quantitative real-time reverse
transcription polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
used to extract RNA from tissues and cell lines according to the
manufacturer's instructions. RNA was reverse-transcribed using a
TaqMan human miRNA assay kit (Applied Biosystems, Foster City, CA,
USA) and cDNA was then amplified with a SYBR® Premix Ex
Taq™ II (Perfect Real-Time) kit (Takara Bio, Inc., Shiga, Japan)
and analyzed with an Applied Biosystems 7900 real-time PCR system.
Hsa-miR-106a primer (HmiRQP0027), snRNA U6 qPCR primer (HmiRQP9001)
and GAPDH (HQP006940) were purchased from Genecopoeia (Guangzhou,
China). The primers for TIMP2 were as follows: sense,
5′-AGCACCACCCAGAAGAAGAG-3′; and antisense,
5′-GTGACCCAGTCCATCCAGAG-3′ and were synthesized by GenePharma
(Shanghai, China).
Western blot analysis
Whole tissue and cell proteins were collected in
RIPA buffer (Beyotime, China) and the protein concentration was
determined using a BCA protein assay kit (Thermo Scientific,
Rockford, IL, USA). An equal amount of protein was separated by 10%
SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica,
MA, USA). The membranes were blocked with 5% non-fat milk in TBST
and then incubated with primary antibodies: TIMP2, matrix
metalloproteinase (MMP)2, MMP9 and GAPDH (1:1,000; Cell Signaling
Technology, Inc.) at 4°C overnight. After being washed by TBST, the
membranes were incubated with an appropriate peroxidase-conjugated
secondary antibody for 2 h at room temperature (ZSGB-Bio, China).
The protein bands were visualized using an enhanced
chemiluminescence kit (Amersham, Little Chalfont, UK).
Immunohistochemical analysis
(IHC)
IHC was performed on 4-µm thickness of
paraformaldehyde-fixed paraffin sections. MMP2 and MMP9 (1:100;
Cell Signaling Technology, Inc., Danvers, MA, USA) antibodies were
used in immunohistochemistry with the streptavidin
peroxidase-conjugated (SP-IHC) method. The staining results for the
protein were semiquantitatively calculated by multiplying the
staining intensity and the percentage of positive cells. The
staining intensity was expressed as four grades: 0= none; 1= weak;
2= moderate; and 3= strong. The percentage of positive cells was
expressed as the following grades: 0, <5%; 1, 6–25%; 2, 26–50%;
3, 51–75%; and 4, >75%.
Cell transfection
The cells were seeded in 6-well plates and
transfected with vectors using Lipofectamine 2000 reagent
(Invitrogen Life Technologies) in accordance with the
manufacturer's protocol. miRNA expression vectors, including
miR-106a expression vector (HmiR0392), the control vector
(CmiR0001-MR04), the miR-106a inhibitor (HmiR-AN0027) and the
negative control (CmiR-AN0001-AM04) were obtained from Genecopoeia.
The TIMP2 expression plasmid and specific siRNA were producted by
GenePharma.
Transwell migration and invasion
assays
Cell migration and invasion assays were assessed by
8-µM pore-sized Transwell inserts (Nalge Nunc International Corp.,
Naperville, IL, USA). Cells (2.5×104) in 200 µl of
serum-free DMEM medium and were loaded into the top chamber, and
750 µl of DMEM medium with 10% FBS were placed in the bottom
chamber. For the invasion, the upper chambers were pre-coated with
Matrigel (BD Biosciences). After 24 h of incubation, the cells were
fixed in 4% paraformaldehyde and stained with 0.3% crystal violet.
The cells remaining in the top layer were swabbed carefully and the
bottom cells were counted under a microscope.
Luciferase reporter assay
Luciferase activity was determined by the
Dual-Luciferase assay (Promega, Madison, WI, USA) according to the
manufacturer's protocol. The wild-type 3′-UTR of TIMP2 containing
the putative miR-106a target site was amplified and cloned into the
pGL3-control luciferase reporter vector (Promega). The mutant
constructs were generated using a QuickChange Site-Directed
Mutagenesis kit (Stratagene, La Jolla, CA, USA). The experiment was
performed in duplicate in three independent experiments.
Statistical analysis
Data are presented as the mean ± SD from at least
three independent replicates. SPSS software, 16.0 (SPSS., Inc,
Chicago, IL, USA) and Graphpad Prism 6.0 (GraphPad Software, Inc.,
La Jolla, CA, USA) were used for a two-tailed Students t-test,
Pearson's correlation analysis, Kaplan-Meier method and the
log-rank test to evaluate the statistical significance. Differences
were defined as P<0.05.
Results
miR-106a is significantly upregulated
in CC tissues and cell lines
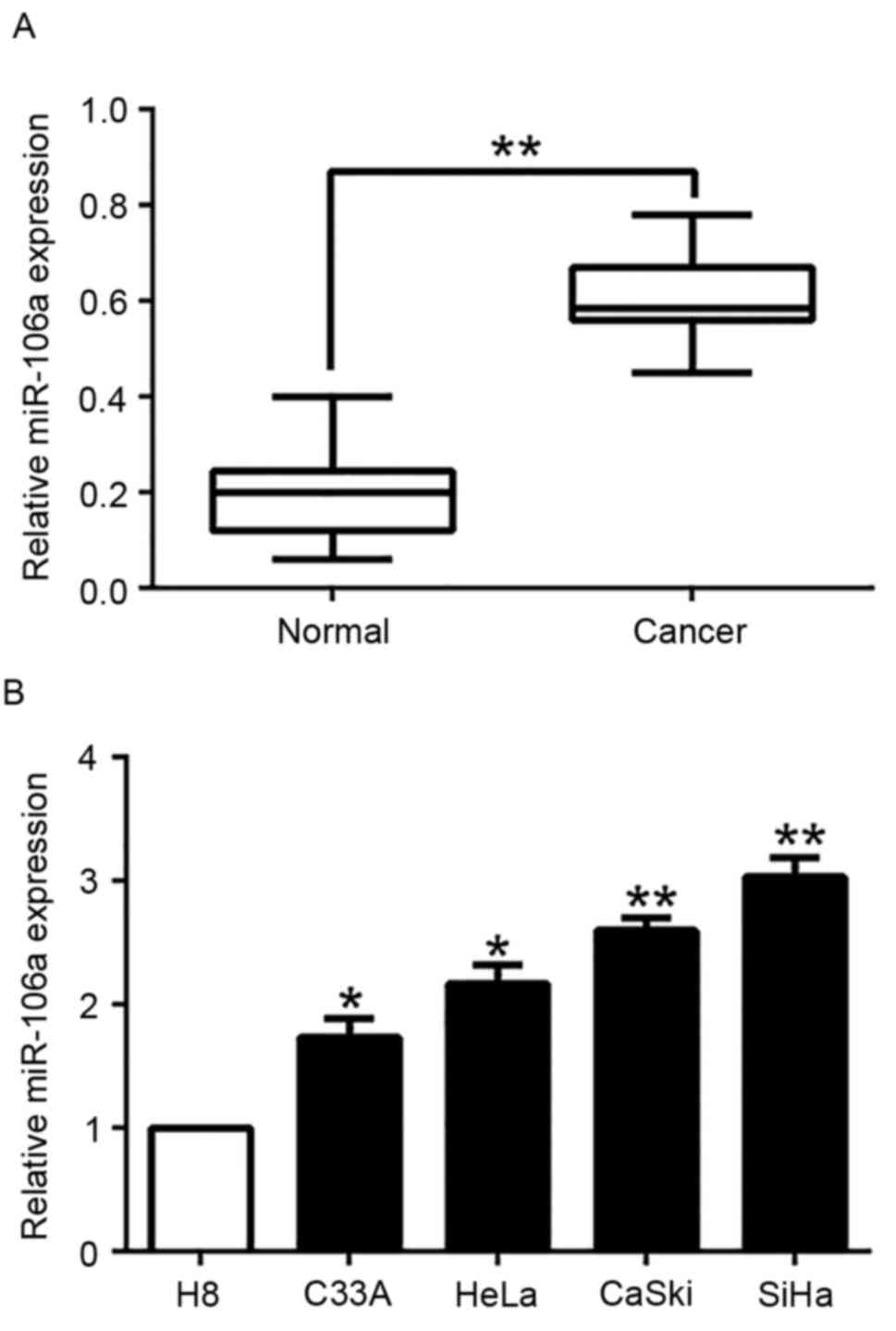
To explore the expression of miR-106a in CC tissues
and cell lines, qRT-PCR was performed. We found that miR-106a
expression was significantly higher in human CC tissues than that
of adjacent normal cervical tissues (P<0.05, Fig. 1A). Furthermore, we analyzed miR-106a
expression in a panel of CC cell lines and normal human cervical
epithelial cell line (H8). Consistently, our data revealed that
miR-106a was significantly upregulated in the CC cell lines
compared to the H8 cells (P<0.05, Fig. 1B). These results indicated that the
increased expression of miR-106a is potentially correlated with the
development and progression of CC.
Correlation between the expression of
miR-106a and the clinical characteristics in CC samples
To assess its clinical relevance in CC samples, we
defined the high and low expression of miR-106a according to the
median expression level. As shown in Table I, high miR-106a expression was
significantly associated with lymph node metastasis (LNM, P=0.007),
lymphovascular space invasion (LVSI, P=0.001) and advanced FIGO
stage (P=0.038). Hence, these findings indicated that increased
miR-106a was involved in the development and progression of CC.
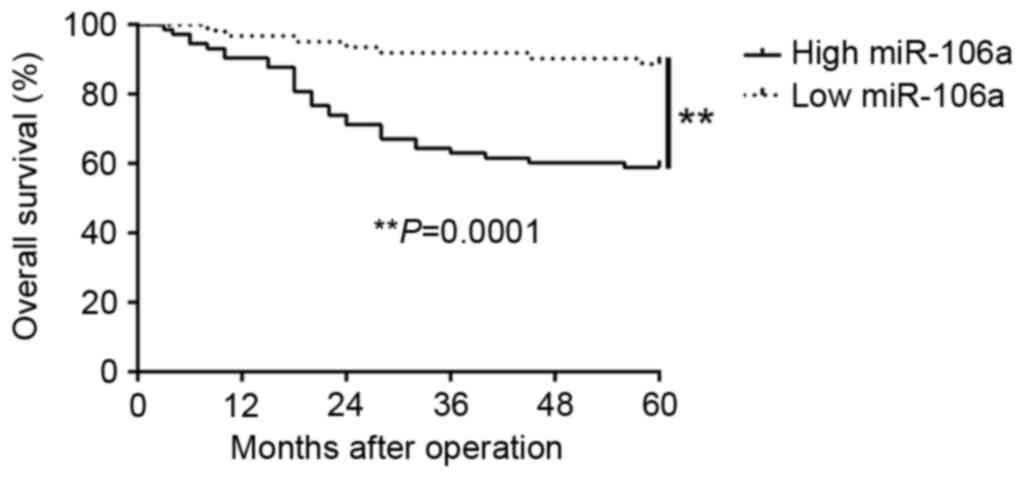
Moreover, survival analysis revealed that upregulation of miR-106a
was markedly correlated with shorter overall survival (P=0.0001,
Fig. 2) in CC patients.
Furthermore, miR-106a expression was an independent prognostic
factor for predicting the overall survival in CC patients (P=0.002,
0.009, respectively, Table II).
These results revealed that miR-106a could serve as a potential
prognostic biomarker in CC patients.
 | Table I.Clinical correlation of miR-106a
expression in CC (n=135). |
Table I.
Clinical correlation of miR-106a
expression in CC (n=135).
|
|
| Expression level |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Cases (n) |
miR-106ahigh (n=73) |
miR-106alow (n=62) | P-value
(ap<0.05) |
|---|
| Age (years) |
|
|
| 0.642 |
| <45 | 33 | 19 | 14 |
|
| ≥45 | 102 | 54 | 48 |
|
| FIGO stage |
|
|
| 0.038a |
| I | 107 | 53 | 54 |
|
| II | 28 | 20 | 8 |
|
| Tumor size
(cm) |
|
|
| 0.680 |
|
<4 | 109 | 58 | 51 |
|
| ≥4 | 26 | 15 | 11 |
|
| LNM |
|
|
| 0.007a |
|
Negative | 117 | 58 | 59 |
|
|
Positive | 18 | 15 | 3 |
|
| LVSI |
|
|
| 0.001a |
|
Negative | 112 | 53 | 59 |
|
|
Positive | 23 | 20 | 3 |
|
| Vaginal
invasion |
|
|
| 0.992 |
|
Negative | 111 | 60 | 51 |
|
|
Positive | 24 | 13 | 11 |
|
| Histology |
|
|
| 0.625 |
|
Squamous | 120 | 64 | 56 |
|
|
Adenocarcinoma | 15 | 9 | 6 |
|
| Parametrial
extension |
|
|
| 0.570 |
|
Negative | 122 | 65 | 57 |
|
|
Positive | 13 | 8 | 5 |
|
 | Table II.Univariate and multivariate analyses
of the 5-year overall survival of 135 CC patients. |
Table II.
Univariate and multivariate analyses
of the 5-year overall survival of 135 CC patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-106a
expression | 4.832 | 1.852–12.257 | 0.002a | 3.982 | 1.382–10.237 | 0.009a |
| FIGO stage | 1.543 | 1.083–5.985 | 0.032a | 1.123 | 1.108–3.668 | 0.038a |
| LNM | 3.192 | 1.426–7.678 | 0.007a | 1.747 | 1.212–4.649 | 0.018a |
| LVSI | 3.312 | 1.382–7.681 | 0.014a | 1.223 | 1.019–5.348 | 0.023a |
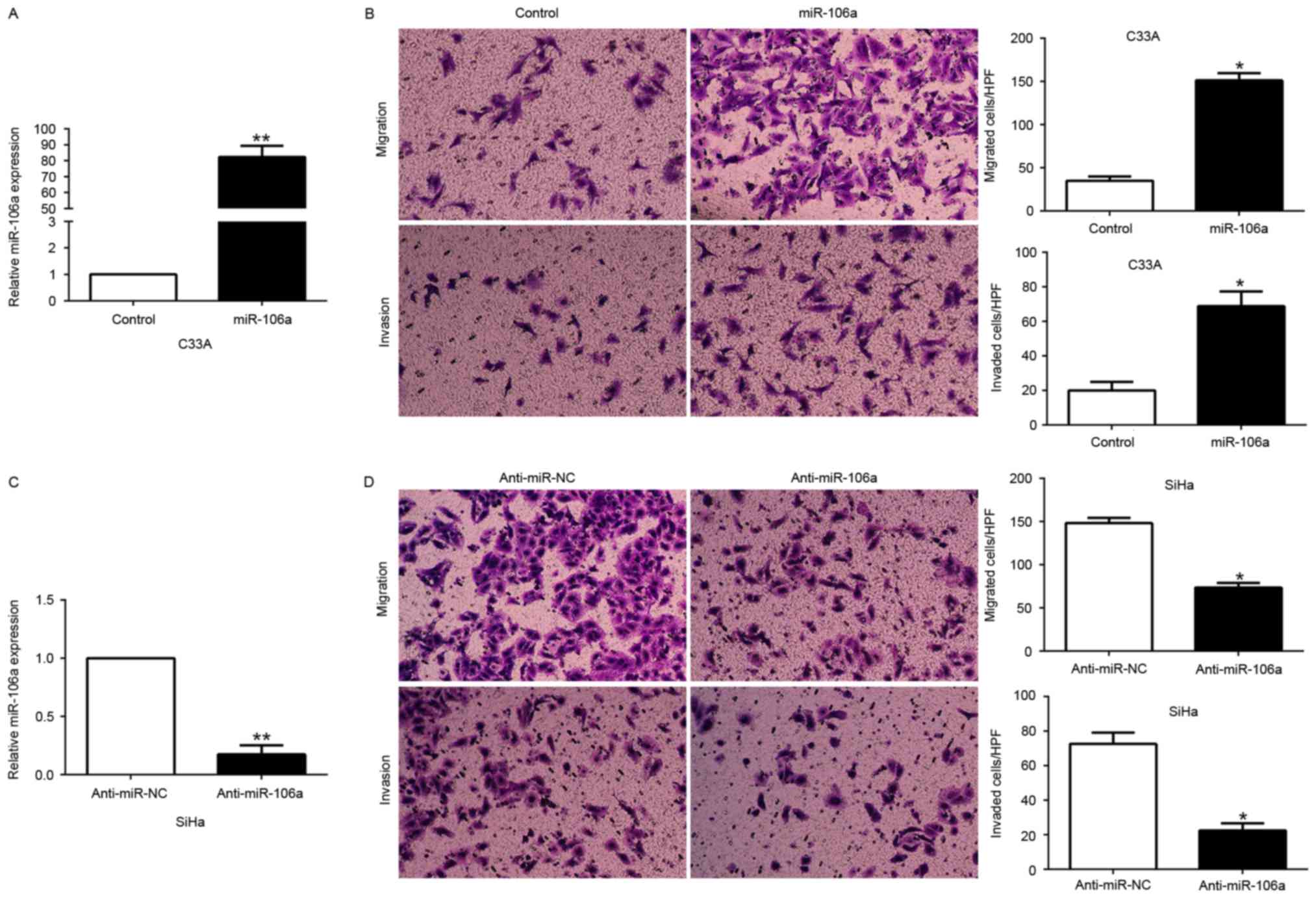
miR-106a promotes CC cell migration
and invasion in vitro
To further illustrate the biological function of
miR-106a in CC cells, we transfected the miR-106a expression vector
into C33A cells or the anti-miR-106a vector into SiHa cells which
contained different endogenous miR-106a levels. As assessed by
qRT-PCR, we confirmed that miR-106a effectively increased miR-106a
in the C33A cells (P<0.05, Fig.
3A) or decreased miR-106a in the SiHa cells (P<0.05,
Fig. 3C). Transwell migration and
invasion assays were performed to evaluate the effects of miR-106a
on the migration and invasion of the C33A and SiHa cells. We found
that miR-106a overexpression significantly promoted the migration
and invasion of the C33A cells (P<0.05, Fig. 3B), whereas miR-106a knockdown
markedly inhibited the number of migrated and invaded SiHa cells
(P<0.05, Fig. 3D). These data
revealed that miR-106a promoted CC cell migration and invasion
in vitro.
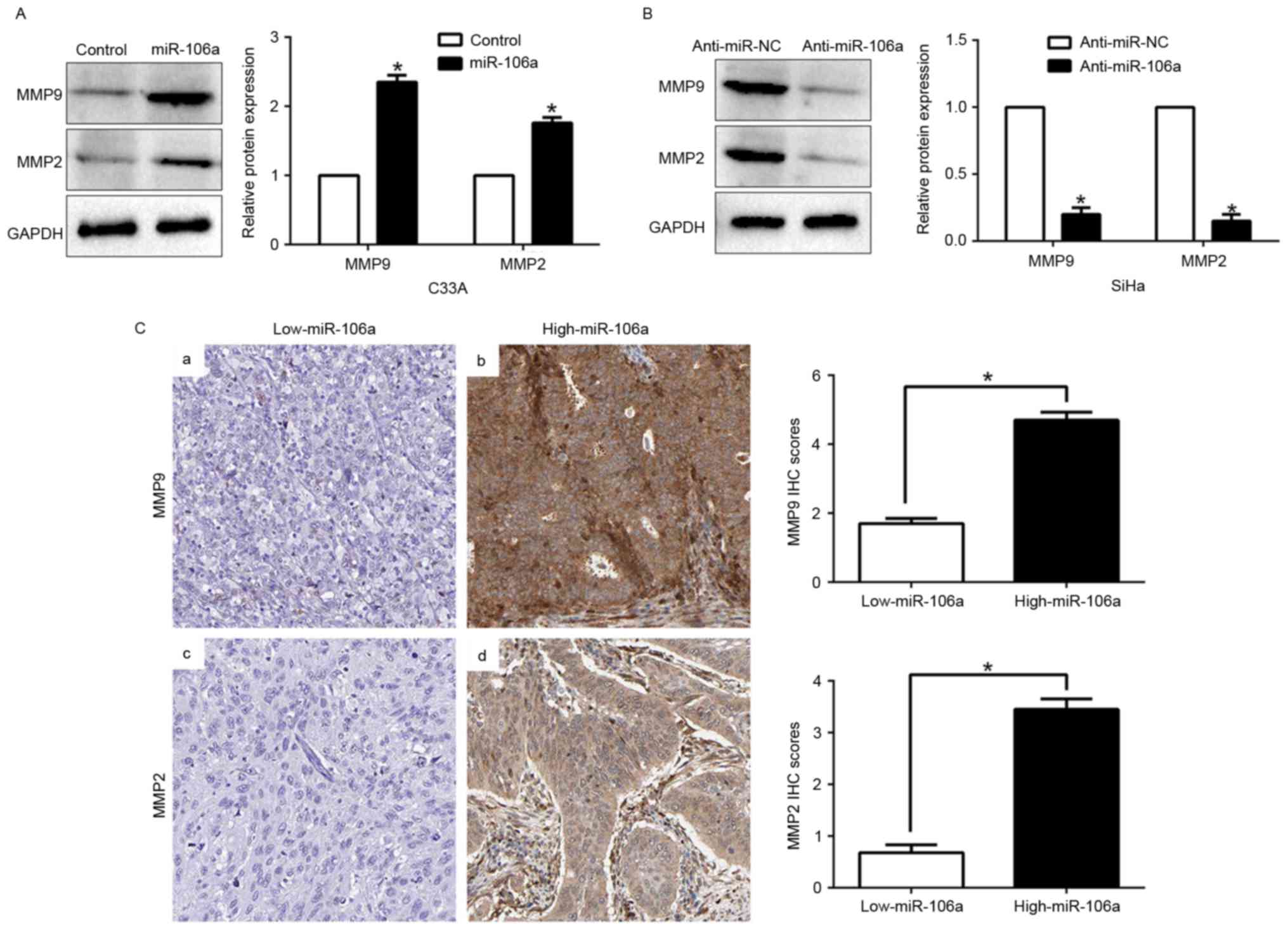
miR-106a modulates the expression of
MMPs
Previous studies confirmed that MMP2 and MMP9 are
important members of the MMP family, which play critical roles in
cancer migration and invasion, including CC. Thus, we investigated
whether miR-106a could regulate invasion-related genes. Our data
revealed that miR-106a overexpression significantly upregulated the
expression of MMP2 and MMP9 (P<0.05, Fig. 4A), however, miR-106a knockdown
markedly downregulated the expression of MMPs (P<0.05, Fig. 4B). In addition, we further explored
the correlation between miR-106a expression and MMPs in CC tissues.
We found that the expression of MMP2 and MMP9 in the high
miR-106a-expression group was higher than that in the low
miR-106a-expression group (P<0.05, Fig. 4C). Collectively, these results
revealed that miR-106a is vital for CC cell invasion by modulating
the expression of MMPs.
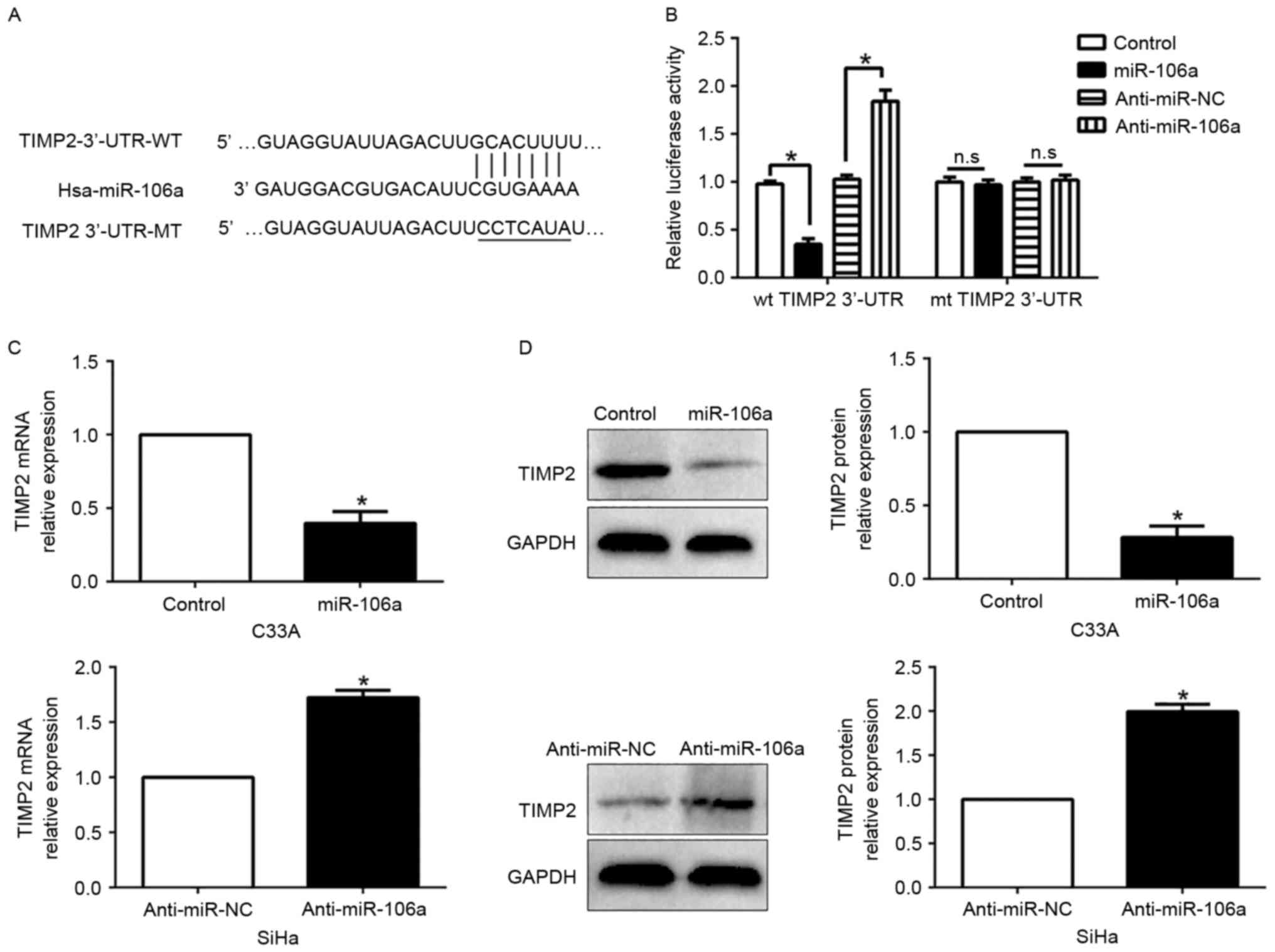
TIMP2 is a direct target of miR-106a
in CC cells
To investigate the mechanism of miR-106a in CC cell
migration and invasion, we used algorithm TargetScan to search for
candidate targets of miR-106a and found that the 3′-UTR of TIMP2
matched the ‘seed sequence’ of miR-106a (Fig. 5A). To ascertain whether TIMP2 was a
direct target of miR-106a in CC cells, we carried out a luciferase
reporter assay to confirm that miR-106a could bind to the 3′-UTR of
TIMP2. The reporter assays revealed that the increased miR-106a
markedly inhibited the luciferase activity of the wild-type (wt)
TIMP2 3′-UTR (P<0.05, Fig. 5B)
while it had no influence on that of the mutant (mt) TIMP2 3′-UTR
(P>0.05, Fig. 5B). Conversely,
the miR-106a decrease increased the luciferase activity of the wt
TIMP2 3′-UTR (P<0.05, Fig. 5B)
but did not affect the luciferase activity of the mt TIMP2 3′-UTR
constructs. Furthermore, miR-106a overexpression markedly
suppressed the mRNA and protein expression of TIMP2 in the C33A
cells (P<0.05, respectively, Fig. 5C
and D). By contrast, the expression of TIMP2 mRNA and protein
were significantly increased by the inhibition of miR-106a in the
SiHa cells (P<0.05, respectively, Fig. 5C and D).
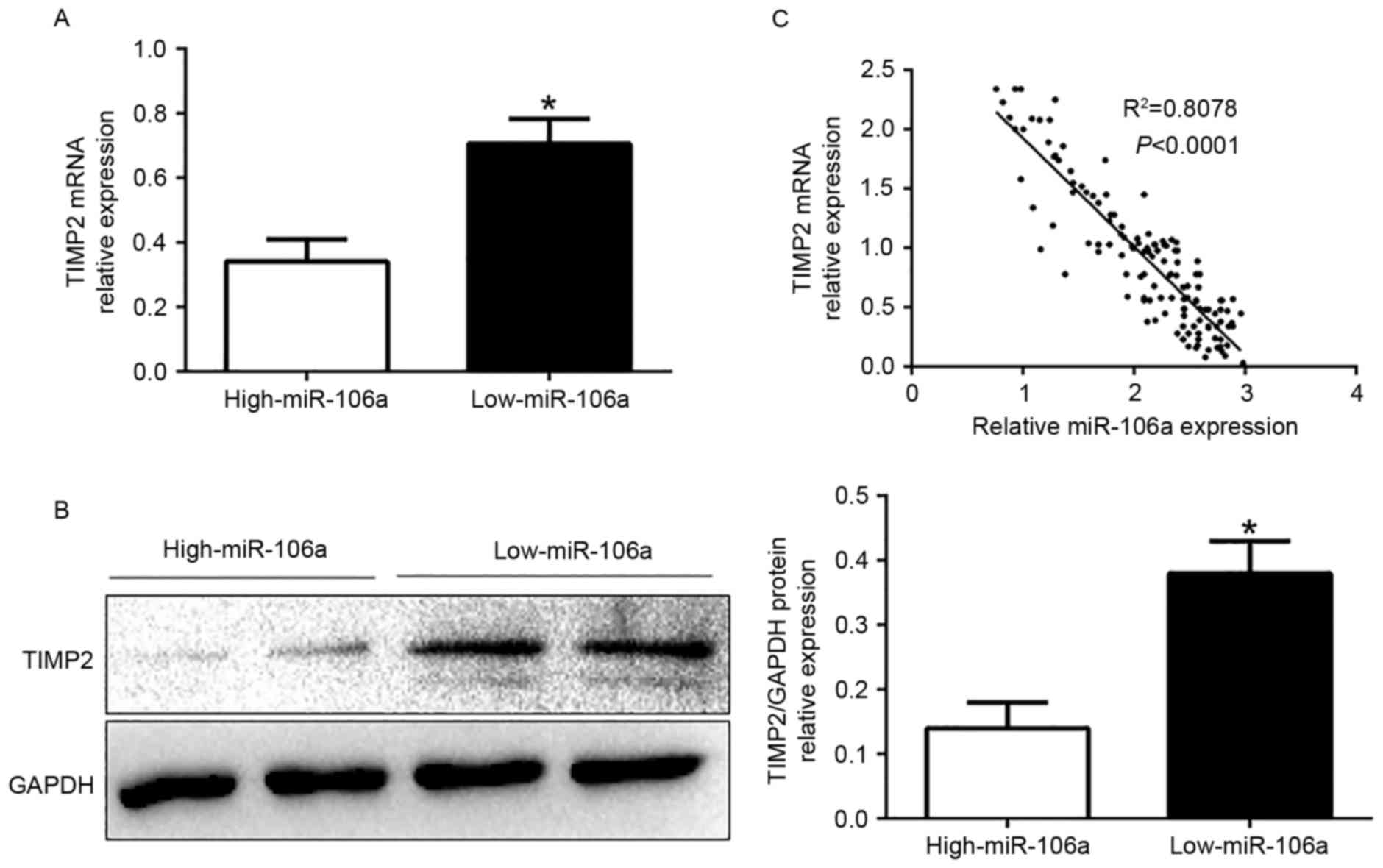
miR-106a expression is inversely
correlated with TIMP2 in CC tissues
To further investigate the relationship between
miR-106a and TIMP2 in vivo, we examined the mRNA and protein
expression in diverse miR-106a expression groups. The results
revealed that TIMP2 mRNA and protein levels were significantly
lower in the high miR-106a-expression group than that in the low
miR-106a-expression group in CC (P<0.05, Fig. 6A and B). In addition, we
demonstrated that the mRNA level of TIMP2 in the CC tissues was
inversely correlated with miR-106a expression (R2=0.8078,
P<0.0001, Fig. 6C). In
conclusion, these data revealed that TIMP2 was a direct downstream
target of miR-106a in CC.
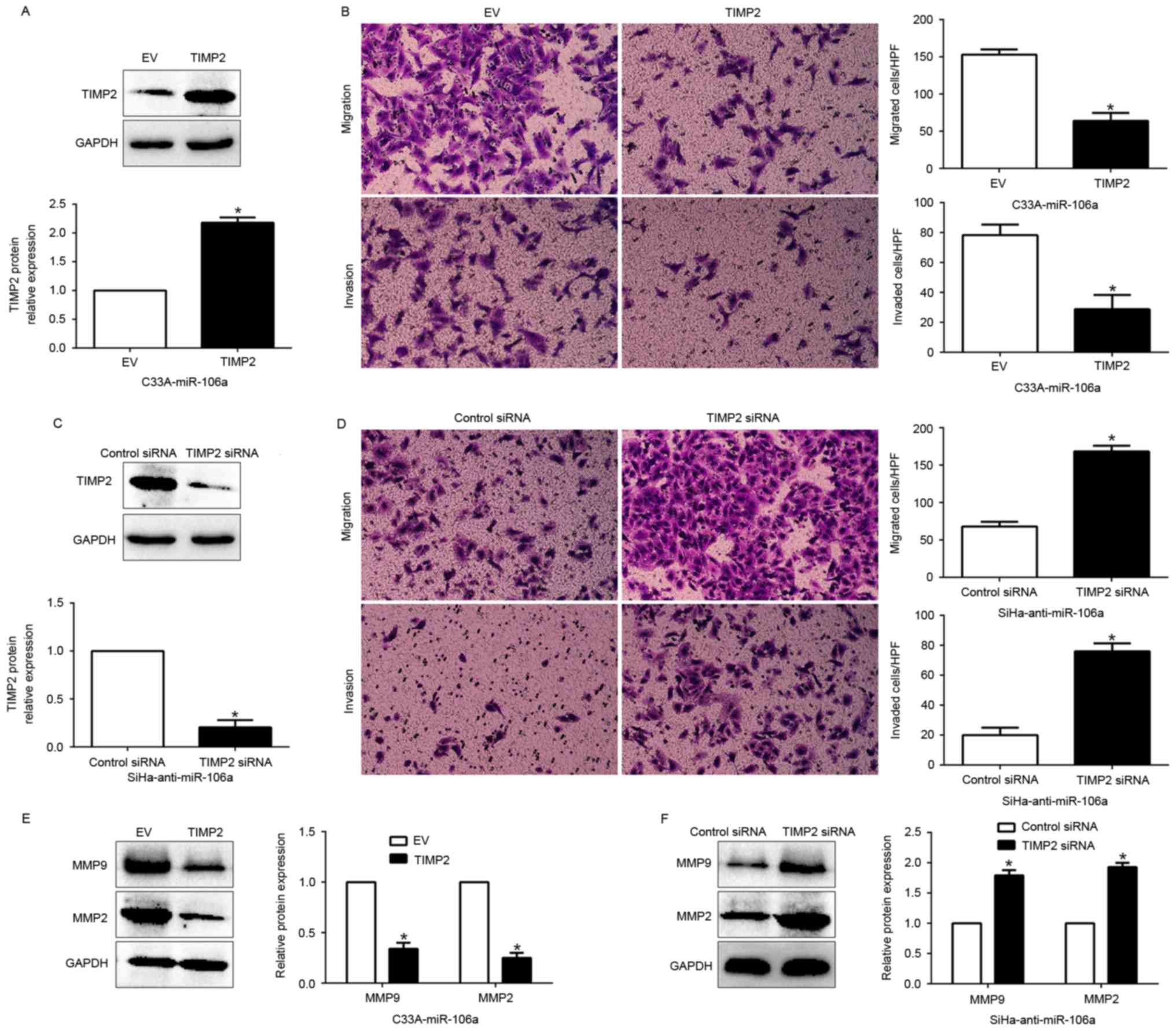
Functional role of TIMP2 in
miR-106a-mediated cell migration, invasion and invasion-related MMP
expression
Having confirmed TIMP2 as a target of miR-106a, we
next explored the biological function of TIMP2 in miR-106a-mediated
cell migration, invasion and increase of MMPs. We restored TIMP2
expression in the C33A-miR-106a cells by transfecting them with
TIMP2 expression plasmid (P<0.05, Fig. 7A). Furthermore, TIMP2 overexpression
inhibited the cell migration, and invasion (P<0.05,
respectively, Fig. 7B) and
suppressed the expression of MMPs (P<0.05, Fig. 7E). Conversely, TIMP2 knockdown by a
specific siRNA in the miR-106a-suppressive SiHa cells (P<0.05,
Fig. 7C) significantly increased
cell migration and invasion (P<0.05, respectively, Fig. 7D) and promoted the expression of
MMPs (P<0.05, Fig. 7F). These
data demonstrated that TIMP2 is a downstream mediator in the
function of miR-106a in CC.
Discussion
Accumulating evidence has demonstrated that aberrant
expression of miRNAs plays pivotal roles in the development and
progression of cancer by inhibiting the expression of their target
genes and potentially serve as biomarkers for the prediction and
prognosis in diverse cancers, including CC (26–28).
In previous studies, Zhu et al demonstrated that miR-106a
targets TIMP2 to regulate the invasion and metastasis of gastric
cancer (15). The phenomenon was
also observed in pancreatic cancer (16). These data strongly support our
present data in regards to CC. Moreover, miR-106a was frequently
upregulated in gastric cancer and inhibited the extrinsic apoptotic
pathway by targeting FAS (13). In
addition, miR-106a targeted autophagy and enhanced sensitivity of
lung cancer cells to Src inhibitors (29). However, conversely, miR-106a
inhibited the proliferation and migration of astrocytoma cells and
promoted apoptosis by targeting FASTK (23). Moreover, miR-106a suppressed the
proliferation, migration, and invasion of osteosarcoma cells by
targeting HMGA2 (22). These data
indicated that the expression level and biological effect of
miR-106a was dependent on the type of cancer.
In present study, we found that miR-106a was
significantly upregulated in CC tissue samples and cell lines,
which was consistent with previous microarray analyses data
(30). Increased miR-106a
expression was prominently associated with malignant
clinicopathological features of CC patients, including lymph node
metastasis, lymphovascular space invasion and advanced FIGO stage.
Moreover, we found that the high miR-106a-expression group had a
poorer 5-year overall survival for CC patients than the low
miR-106a-expression group. Multivariate Cox regression analysis
revealed that miR-106a was an independent prognostic factor for
predicting the survival of CC patients. In conclusion, these
results suggest that miR-106a is critical for the prognosis outcome
of CC patients. What is more, gain- and loss-of-function
experiments demonstrated that miR-106a promoted cell migration,
invasion and the expression of invasion-related genes, at least
partially by targeting the TIMP2-mediated MMP signaling pathway.
Furthermore, miR-106a was inversely correlated with TIMP2
expression, which was downregulated in CC tissues. In addition,
miR-106a could negatively modulate TIMP2 accumulation and regulate
MMP2 and MMP9 expression, which play an important role in the
metastasis of CC. Collectively, these results demonstrated that
miR-106a functions as oncogene in the migration, invasion and the
expression of MMPs by directly inhibiting the TIMP2 pathway.
The degradation of the extracellular matrix (ECM) is
crucial for the invasion and metastasis of CC. MMPs, especially
MMP2 and MMP9, are able to degrade ECM to promote endothelial cell
migration and trigger an angiogenic switch by releasing VEGF from
ECM (31). TIMPs are specific
inhibitors of MMP2 and MMP9, which were decreased in CC (32,33).
Our results revealed that TIMP2 abolished the stimulatory effect of
miR-106a on CC cells. Collectively, these data demonstrated that
the promotive effect of miR-106a was mediated by targeting TIMP2
thus inhibiting MMP expression in CC.
In conclusion, we demonstrated that miR-106a was
upregulated in CC tissues and cell lines, and its increased
expression was associated with malignant clinicopathological
features. Furthermore, we confirmed that miR-106a promoted cell
migration, invasion and the expression of MMPs by inhibiting TIMP2.
These results suggest that miR-106a is a potential
metastasis-associated tumor oncogene in CC. Collectively, the
dysregulation of miR-106a may play an important role in tumor
metastasis and may be a novel prognostic factor and potential
therapeutic target for CC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dueñas-Gonzalez A, Cetina L, Mariscal I
and de la Garza J: Modern management of locally advanced cervical
carcinoma. Cancer Treat Rev. 29:389–399. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Glick SB, Clarke AR, Blanchard A and
Whitaker AK: Cervical cancer screening, diagnosis and treatment
interventions for racial and ethnic minorities: A systematic
review. J Gen Intern Med. 27:1016–1032. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sakuragi N: Up-to-date management of lymph
node metastasis and the role of tailored lymphadenectomy in
cervical cancer. Int J Clin Oncol. 12:165–175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Su Y, Xiong J, Hu J, Wei X, Zhang X and
Rao L: MicroRNA-140-5p targets insulin like growth factor 2 mRNA
binding protein 1 (IGF2BP1) to suppress cervical cancer growth and
metastasis. Oncotarget. 7:68397–68411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yi Y, Li H, Lv Q, Wu K and Zhang W, Zhang
J, Zhu D, Liu Q and Zhang W: miR-202 inhibits the progression of
human cervical cancer through inhibition of cyclin D1. Oncotarget.
7:72067–72075. 2016.PubMed/NCBI
|
|
10
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting
p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin Y, Peng D, Shen Y, Xu M, Liang Y, Xiao
B and Lu J: MicroRNA-376c inhibits cell proliferation and invasion
in osteosarcoma by targeting to transforming growth factor-alpha.
DNA Cell Biol. 32:302–309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C,
Quan L, Bai J and Xu N: miR-106a is frequently upregulated in
gastric cancer and inhibits the extrinsic apoptotic pathway by
targeting FAS. Mol Carcinog. 52:634–646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao B, Guo J, Miao Y, Jiang Z, Huan R,
Zhang Y, Li D and Zhong J: Detection of miR-106a in gastric
carcinoma and its clinical significance. Clin Chim Acta.
400:97–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu M, Zhang N, He S, Lui Y, Lu G and Zhao
L: MicroRNA-106a targets TIMP2 to regulate invasion and metastasis
of gastric cancer. FEBS Lett. 588:600–607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li P, Xu Q, Zhang D, Li X, Han L, Lei J,
Duan W, Ma Q, Wu Z and Wang Z: Upregulated miR-106a plays an
oncogenic role in pancreatic cancer. FEBS Lett. 588:705–712. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang R, Li Y, Hou Y, Yang Q, Chen S, Wang
X, Wang Z, Yang Y, Chen C, Wang Z, et al: The
PDGF-D/miR-106a/Twist1 pathway orchestrates epithelial-mesenchymal
transition in gemcitabine resistance hepatoma cells. Oncotarget.
6:7000–7010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen L, Zhang F, Sheng XG, Zhang SQ, Chen
YT and Liu BW: MicroRNA-106a regulates phosphatase and tensin
homologue expression and promotes the proliferation and invasion of
ovarian cancer cells. Oncol Rep. 36:2135–2141. 2016.PubMed/NCBI
|
|
19
|
Zhu M, Zhang N, He S, Yan R and Zhang J:
MicroRNA-106a functions as an oncogene in human gastric cancer and
contributes to proliferation and metastasis in vitro and in vivo.
Clin Exp Metastasis. 33:509–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hou X, Zhang M and Qiao H: Diagnostic
significance of miR-106a in gastric cancer. Int J Clin Exp Pathol.
8:13096–13101. 2015.PubMed/NCBI
|
|
21
|
Yuan R, Zhi Q, Zhao H, Han Y, Gao L, Wang
B, Kou Z, Guo Z, He S, Xue X, et al: Upregulated expression of
miR-106a by DNA hypomethylation plays an oncogenic role in
hepatocellular carcinoma. Tumour Biol. 36:3093–3100. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He QY, Wang GC, Zhang H, Tong DK, Ding C,
Liu K, Ji F, Zhu X and Yang S: miR-106a-5p suppresses the
proliferation, migration, and invasion of osteosarcoma cells by
targeting HMGA2. DNA Cell Biol. 35:506–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhi F, Zhou G, Shao N, Xia X, Shi Y, Wang
Q, Zhang Y, Wang R, Xue L, Wang S, et al: miR-106a-5p inhibits the
proliferation and migration of astrocytoma cells and promotes
apoptosis by targeting FASTK. PLoS One. 8:e723902013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Y, Zhang H, He X, Song H, Qiang Y, Li
Y, Gao J and Wang Z: miR-106a* inhibits the proliferation of renal
carcinoma cells by targeting IRS-2. Tumour Biol. 36:8389–8398.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang G, Zhang R, Chen X, Mu Y, Ai J, Shi
C, Liu Y, Shi C, Sun L, Rainov NG, et al: miR-106a inhibits glioma
cell growth by targeting E2F1 independent of p53 status. J Mol Med
(Berl). 89:1037–1050. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deng Y, Xiong Y and Liu Y: miR-376c
inhibits cervical cancer cell proliferation and invasion by
targeting BMI1. Int J Exp Pathol. 97:257–265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deng B, Zhang Y, Zhang S, Wen F, Miao Y
and Guo K: MicroRNA-142-3p inhibits cell proliferation and invasion
of cervical cancer cells by targeting FZD7. Tumour Biol.
36:8065–8073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu S, Zhang P, Chen Z, Liu M, Li X and
Tang H: MicroRNA-7 downregulates XIAP expression to suppress cell
growth and promote apoptosis in cervical cancer cells. FEBS Lett.
587:2247–2253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rothschild SI, Gautschi O, Batliner J,
Gugger M, Fey MF and Tschan MP: MicroRNA-106a targets autophagy and
enhances sensitivity of lung cancer cells to Src inhibitors. Lung
Cancer. 107:73–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Servín-González LS, Granados-López AJ and
López JA: Families of microRNAs expressed in clusters regulate cell
signaling in cervical cancer. Int J Mol Sci. 16:12773–12790. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nagase H and Woessner JF Jr: Matrix
metalloproteinases. J Biol Chem. 274:21491–21494. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cardeal LB, Boccardo E, Termini L,
Rabachini T, Andreoli MA, di Loreto C, Filho A Longatto, Villa LL
and Maria-Engler SS: HPV16 oncoproteins induce MMPs/RECK-TIMP-2
imbalance in primary keratinocytes: Possible implications in
cervical carcinogenesis. PLoS One. 7:e335852012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Braicu EI, Fotopoulou C, Chekerov R,
Richter R, Blohmer J, Kümmel S, Stamatian F, Yalcinkaya I, Mentze
M, Lichtenegger W, et al: Role of serum concentration of VEGFR1 and
TIMP2 on clinical outcome in primary cervical cancer: Results of a
companion protocol of the randomized, NOGGO-AGO phase III adjuvant
trial of simultaneous cisplatin-based radiochemotherapy vs.
carboplatin and paclitaxel containing sequential radiotherapy.
Cytokine. 61:755–758. 2013. View Article : Google Scholar : PubMed/NCBI
|