Introduction
Lung cancer is one of the most common cancers in the
world (1). Non-small cell lung
cancer (NSCLC), which mainly contains lung adenocarcinoma (ADC) and
squamous cell carcinoma (SCC), is the most frequent type
(accounting for ~80%) of lung cancer (2–4). The
prognosis of NSCLC patients remains poor with a 5-year survival
rate being less than 15% (5,6).
Therefore, searching for a useful prognostic biomarker is crucial
to identify the patients with high risk of recurrence and prolong
the survival of NSCLC patients.
LIM homeobox (LH) members are one of the most
important subfamilies of LIM-homeodomain proteins. There are at
least 12 LH genes in mammalian genomes. Studies have shown that
these genes are critical for the development of specialized cells
in various tissues (7–12). Although many reports have identified
important roles on organism development, little is known about
functions of these proteins in disease. Recent studies have shown
that some LH genes play roles in various cancers, including Lhx1,
Lhx2 and Lhx4 (13–18). LHX3, located at 9q34.3, is involved
in the development of spinal cord motor neurons and pituitary
(19–22). Only recently, LHX3 methylation
pattern has been shown to be associated with breast cancer
(23). However, the expression,
prognostic significance and functional role of LHX3 remain unclear
in cancer.
In the present study, we identified LHX3 as a
differentially expression gene in NSCLC tumor and non-tumor tissues
by RT-PCR, RT-qPCR and immunohistochemistry. Next, the prognostic
significance and functional roles of LHX3 were explored. This study
indicates that LHX3 is an important early-stage and
radiosensitivity prognostic factor and acts as a novel oncogene in
ADC.
Materials and methods
Patient samples and cell lines
A total of 180 lung adenocarcinoma patients
(undergone surgical resection between 2005 and 2009) were obtained
from the Yanan Hospital in Kunming, China. The clinicopathological
information includes sex, age, lymph node status, tumor size,
histological grade, tumor location, clinical stage, radiotherapy
and overall survival (OS) rates of 5–10 years. This study was
approved by the ethics committee of the Yanan Hospital Affiliated
to Kunming Medical University, and all experiments were carried out
in accordance with the approved guidelines of Kunming Medical
University. Informed consent was signed by all the patients.
The NSCLC cell lines (A549, H460 and SPC-A1) were
obtained from the Cell Bank of the Chinese Academy of Science
(Shanghai, China), cultured in F12K (Sigma-Aldrich, St. Louis, MO,
USA) and RPMI-1640 (HyClone Laboratories, Inc., Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone
Laboratories).
Total RNA isolation
Total RNAs were extracted from frozen tissues.
Approximately 3.0 µg of total RNAs was treated with DNase I to
eliminate genomic DNA contamination. The treated RNAs were then
reverse-transcribed to cDNAs.
RT-PCR and RT-qPCR
Series of PCRs with different cycles were performed
and appropriate cycles were chosen for RT-PCR. Human β-actin was
used as endogenous control. The primers were: (5′-3′): LHX3-F, AAG
TGT CTC AAG TGC AGC GA and LHX3-R, GTG GTA CAC GAA GTC CTG GG;
β-actin-F, TTC TAC AAT GAG CTG CGT GTG and β-actin-R, GGG GTG TTG
AAG GTC TCA AA. RT-qPCR was performed using the iQ5 Real-Time
Detection system (Bio-Rad Laboratories, Hercules, CA, USA) and
GoTaq® qPCR Master Mix (Promega, Madison, WI, USA). The
expression was calculated by the equation 2−ΔΔCT. The
primers are (5′-3′): LHX3-F1, ACG GAC CCA GTT CTG ACC TA and
LHX3-R1, TGG TCT ACC TCA TCC AGC CA; β-actin-F2, TGA CGT GGA CAT
CCG CAA AG and β-actin-R2, CTG GAA GGT GGA CAG CGA GG. All assays
were performed in triplicate.
Tissue microarray (TMA)
To construct the TMA slides, two cores were taken
from cancerous and adjacent non-cancerous tissue (within a distance
of 25 mm). The adjacent non-cancerous tissues were stained with
hematoxylin-eosin (H&E) and compared with normal tissues. The
TMAs were then constructed as previously described (24).
Immunohistochemical analysis
IHC was performed using LHX3 antibody (1:200; Abcam)
as previously described (25).
Tumor cell staining was quantified and classified into 5 grades:
<10% positive cells for 0; 10–25% for 1; 26–50% for 2; 51–75%
for 3 and ≥76% for 4. Staining intensity was graded as negative
(0), weak (1), moderate (2) and strong (3). The expression levels were defined by
product of grade and intensity for staining.
Construction of LHX3 overexpression
vector
Full-length human LHX3 was cloned, verified by
sequencing and subcloned into the pIRES2-EGFP vector (Invitrogen,
Carlsbad, CA, USA). Cancer cells were transfected using
Lipofectamine 2000 (Invitrogen).
Western blot analysis
Protein was run on 12% SDS-PAGE, transferred to PVDF
membrane (Millipore, Billerica, MA, USA) and incubated with primary
LHX3 antibody (1:1,500; Abcam). The proteins on the membrane were
detected by chemiluminescence (Pierce, Rockford, IL, USA). The same
membrane was also incubated with β-actin antibody (1:2,000;
Sigma-Aldrich).
Cell viability assay
Transfected cells were plated in 96-well plate at
4,000 cells/well. Cell viability was evaluated by MTS (Promega) at
1–5 days as previously described (26). The experiments were carried out in
triplicate.
Flow cytometry assay
Cancer cell apoptosis was determined by Annexin V
and APC/7-AAD dual staining Detection kit (Nanjing KeyGen Biotech,
Co., Ltd., Nanjing, China) at 48 h after transfection according to
the instructions. The apoptosis cells were analyzed by the
FACSCalibur and FlowJo software (Tree Star, Inc., San Carlos, CA,
USA).
Invasion assay
Transwell assays were performed in 12-well plates
with Matrigel (8 µm; Corning, Inc., Corning, NY, USA). The
transfected cells were suspended in the upper well of the chamber
with serum-free medium. The lower well contained media supplemented
with 10% FBS. The cells of the lower chamber were stained with 0.1%
crystal violet and counted. The experiments were performed in
triplicate.
Clinical microarray database
analysis
A Kaplan-Meier Plotter (http://kmplot.com/analysis/index.php) containing the
gene expression data and survival information of lung patients was
used (27). The probe was
221670_s_at. The patients were grouped according to the auto
selection of best cut-off. The P<0.05 was considered to be
statistically significant.
Statistical analysis
Statistical analyses were performed using the SPSS
17.0 software (SPSS, Inc., Chicago, IL, USA). The difference in
categorical variables was analyzed by the Chi-square or the
Linear-by-Linear test. Kaplan-Meier method and log-rank test were
used to calculate and evaluate the OS. Cox regression was used for
multivariate analysis of prognostic predictors. The high or low
expression was categorized by median score. The P<0.05 was taken
as statistically significant.
Results
Increased LHX3 expression is observed
in NSCLC tissues
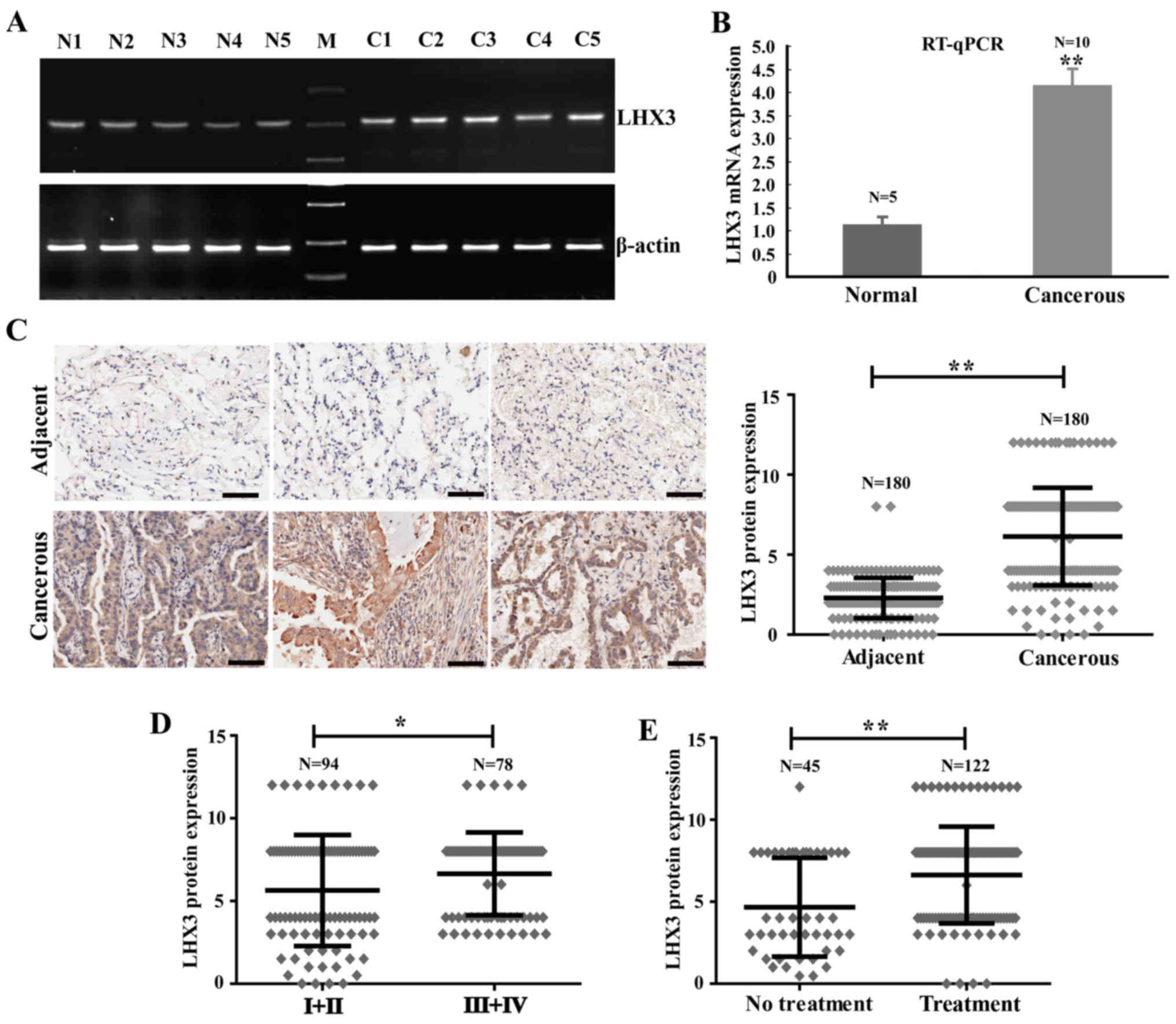
Our previous study revealed that LHX3 might be a
preferentially expressed gene in NSCLC tissues. To further
investigate the LHX3 expression pattern, the expression of LHX3 was
determined in normal and tumor tissues by RT-PCR and RT-qPCR. The
results indicated that LHX3 expression was elevated in tumor
tissues compared to normal tissues (Fig. 1A). The relative mean expression
level of LHX3 was 4.12±0.39 in tumor tissues remarkably higher than
1.10±0.19 in normal tissues (Fig.
1B). To confirm the altered expression of LHX3 at protein
level, we performed immunohistochemistry (IHC) for LHX3 on tissue
microarray (TMA) containing 180 lung adenocarcinoma and
corresponding para-carcinoma tissues. From the staining, LHX3
protein was also increased in carcinoma tissues compared to
para-carcinoma tissues (Fig. 1C).
The positive staining of tumor cells was then quantified using the
scoring system for intensity and positive staining percentage. The
mean protein expression level of LHX3 was 6.13±3.06 in cancer
tissues sharply higher than 2.29±1.27 in para-carcinoma tissues
(P<0.0001; Fig. 1C). In
addition, LHX3 was expressed in both the nucleus and cytoplasm of
tumor cells. These results revealed that LHX3 expression is
increased in tumor tissues.
LHX3 is associated with clinical stage
and radiotherapy of NSCLC patients
Based on the quantified expression above, we
classified LHX3 expression into high and low groups (the scores
greater than or less than the mean expression were obtained in two
groups: low and high). After investigating association between LHX3
expression and clinicopathological features, LHX3 expression was
found to be associated with clinical stage (n=172, P=0.032) and
radiotherapy (n=167, P=0.022) of the NSCLC patients (Table I). However, LHX3 expression was not
associated with age (P=0.359), histological grade (P=0.865), sex
(P=0.550), tumor size (P=0.368), tumor location (P=0.171) or lymph
node status (P=0.361) of the patients (Table I). To further confirm the
association between LHX3 expression and clinical stage or
radiotherapy, we then analyzed the changes of LHX3 expression in
the patients at early stages (stage I–II, n=94) or advanced stages
(stage III–IV, n=78) and the patients with (n=122) or without
(n=45) radiotherapy treatment. The data indicated that LHX3
expression is much higher in the patients at advanced stages than
in the patients at early stages (P=0.0304; Fig. 1D) and LHX3 expression is clearly
increased in the patients with radiotherapy treatment (P=0.0002;
Fig. 1E).
 | Table I.Correlations of LHX3 with
clinicopathological features of patients (n=180). |
Table I.
Correlations of LHX3 with
clinicopathological features of patients (n=180).
|
|
| LHX3 expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Total | High (n=96) | Low (n=84) | P-value |
|---|
| Age (years) |
|
|
| 0.359 |
|
<60 | 70 | 34 | 36 |
|
|
≥60 | 110 | 62 | 48 |
|
| Histological
grade |
|
|
| 0.865 |
| 1 | 22 | 12 | 10 |
|
| 2 | 116 | 62 | 54 |
|
| 3 | 42 | 22 | 20 |
|
| Sex |
|
|
| 0.550 |
|
Male | 98 | 50 | 48 |
|
|
Female | 82 | 46 | 36 |
|
| Clinical stage |
|
|
| 0.032 |
|
I+II | 92 | 42 | 50 |
|
|
III+IV | 80 | 50 | 30 |
|
| Tumor size
(cm) |
|
|
| 0.368 |
|
<4 | 80 | 46 | 34 |
|
| ≥4 | 100 | 50 | 50 |
|
| Tumor location |
|
|
| 0.171 |
|
Right | 102 | 60 | 42 |
|
|
Left | 76 | 36 | 40 |
|
| Lymph node
status |
|
|
| 0.361 |
|
Negative | 78 | 38 | 40 |
|
|
Positive | 96 | 54 | 42 |
|
| Radiotherapy |
|
|
| 0.022 |
|
Yes | 122 | 72 | 50 |
|
| No | 45 | 17 | 28 |
|
LHX3 expression is associated with
poor OS of NSCLC patients
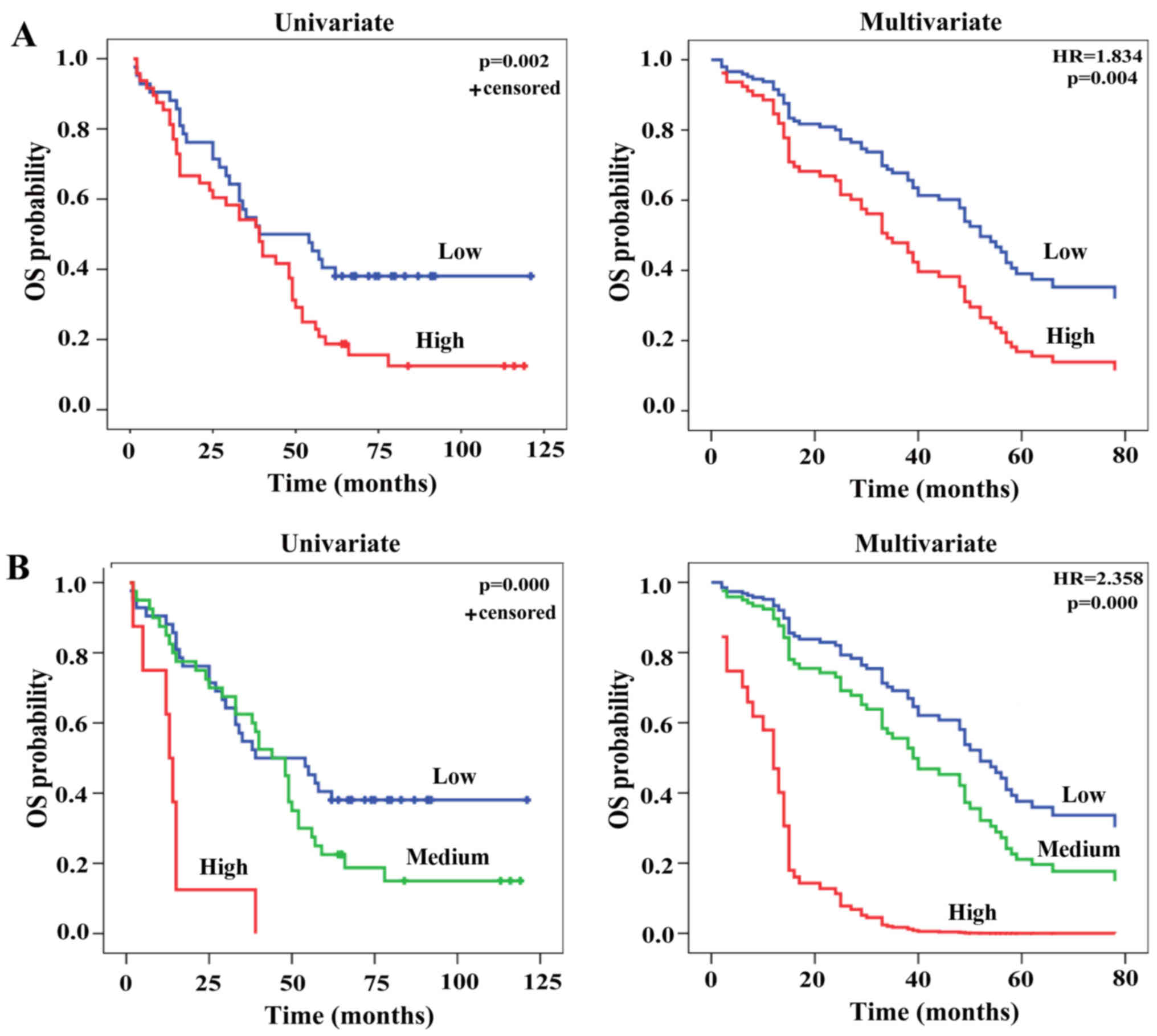
LHX3 expression was then analyzed with respect to
survival data of the patients. Kaplan-Meier analysis revealed a
poor OS in the patients with high LHX3 expression compared to that
with low LHX3 expression (P=0.002; Fig.
2A). To correct for bias, LHX3 expression as well as other
parameters (sex, age, histological grade, tumor size, tumor
location, lymph node, clinical stage and radiotherapy) were
analyzed by multivariate Cox-regression analysis. In addition to
clinical stage [hazard ratio (HR)=1.545, P=0.001], LHX3 expression
is an independent prognostic factor (HR, 1.834, P=0.004) for OS of
the patients (Fig. 2A and Table II). To confirm these results, the
survival analyses were also performed by Kaplan-Meier and
Cox-regression methods base on three groups of LHX3 expression:
low, intermediate and high. The results observed were consistent
with the ones based on two groups (HR, 2.358, P=0.000; Fig. 2B). To further validate the
correlation between the LHX3 expression and the survival of
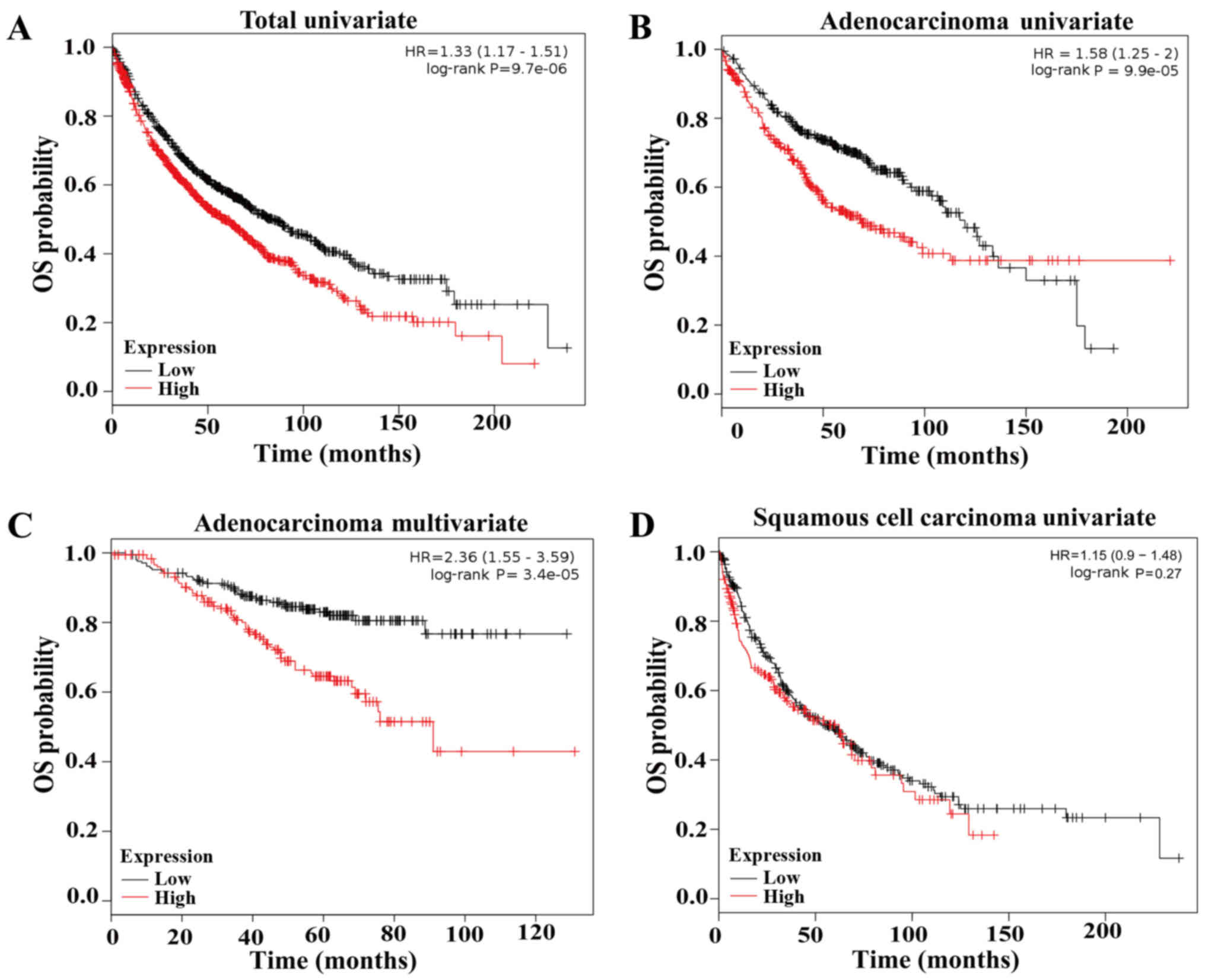
patients, the contribution of LHX3 to survival of patients was also
examined in a clinical microarray database (http://kmplot.com/analysis/index, 27). The analyses
revealed that LHX3 expression still predicts poor OS of patients
(Fig. 3A). These findings show that
LHX3 is an unfavorable and independent prognostic factor of NSCLC
patients.
 | Table II.Multivariate analysis of prognostic
factors in NSCLC patients (n=180). |
Table II.
Multivariate analysis of prognostic
factors in NSCLC patients (n=180).
| Expression | Variables | HR | 95% CI | P-value |
|---|
| The LHX3
expression | LHX3
expression | 1.834 | 1.209–2.780 | 0.004 |
|
| Gender | 1.352 | 0.893–2.048 | 0.154 |
|
| Age | 1.001 | 0.979–1.023 | 0.930 |
|
| Histological
grade | 1.101 | 0.781–1.554 | 0.582 |
|
| Tumor size | 1.090 | 0.980–1.212 | 0.110 |
|
| Tumor location | 1.122 | 0.738–1.705 | 0.591 |
|
| Lymph node
status | 1.030 | 0.985–1.078 | 0.190 |
|
| Clinical stage | 1.545 | 1.195–1.996 | 0.001 |
|
| Radiotherapy | 1.454 | 0.906–2.334 | 0.121 |
LHX3 expression is associated with
poor OS outcomes in ADC patients
We then investigated the correlations between the
LHX3 expression and the survival of ADC and SCC patients,
respectively. In ADC patients, survival analysis indicated that the
patients with low LHX3 expression had a significantly prolonged OS
compared to those with high LHX3 expression (HR=2.36, multivariate
P=0.000/univariate P=0.000; Fig. 3B and
C). In SCC patients, survival analysis demonstrated that LHX3
expression was not associated with OS of the patients (P=0.27;
Fig. 3D). These results reveal that
LHX3 is an unfavorable and independent prognostic factor of ADC
patients, but is not associated with OS of SCC patients.
LHX3 is an early-stage prognostic
factor of ADC patients
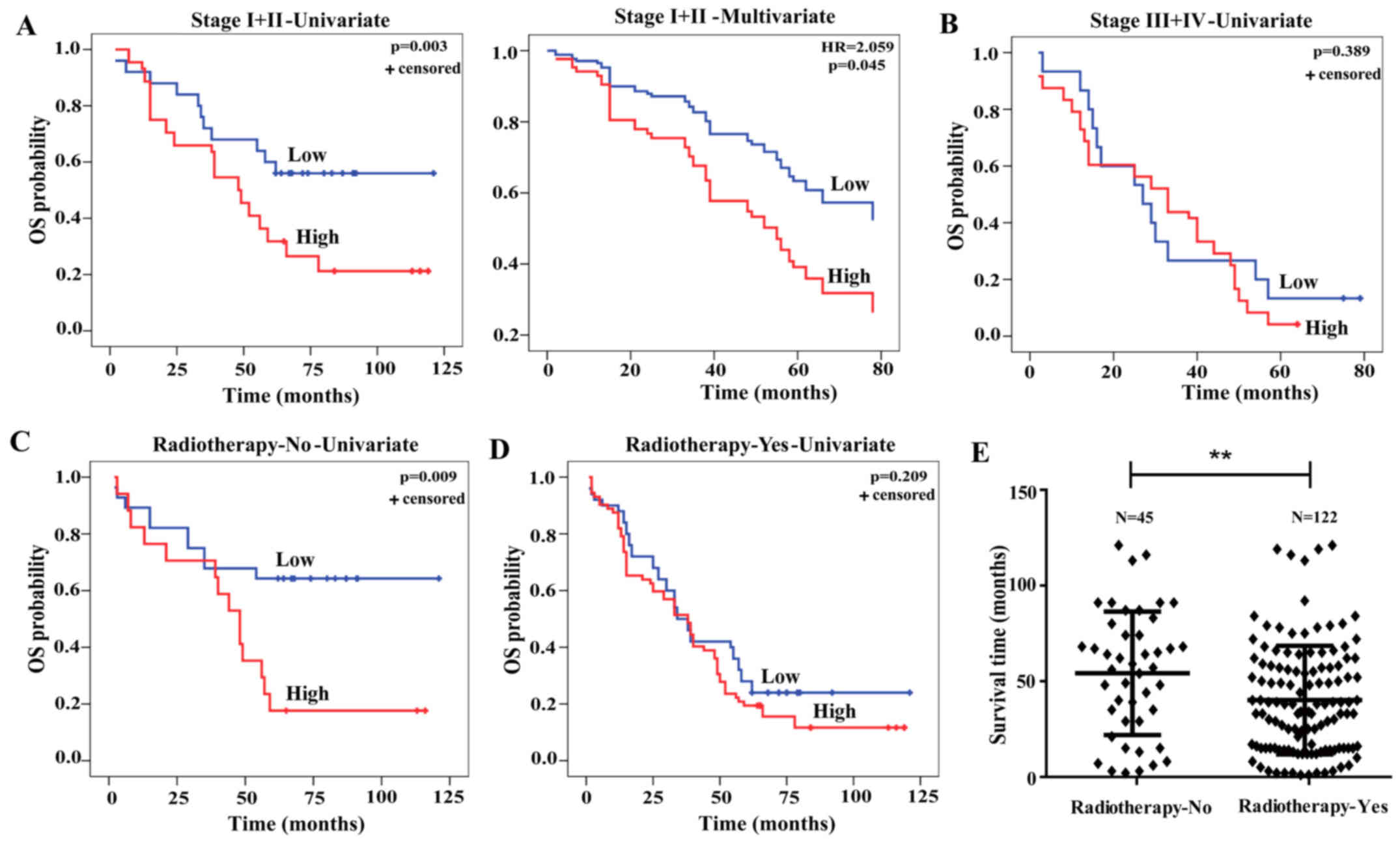
The associations between LHX3 expression and
survival of ADC patients at early stages (stage I–II, n=94) or
advanced stages (stage III–IV, n=78) were further evaluated.
Kaplan-Meier and Cox-regression analyses indicated that LHX3
expression was obviously associated with the OS (P=0.003) and
represented an independent prognostic factor of ADC patients at
early stage (HR, 2.059, P=0.045; Fig.
4A), but not associated with OS of the ones at advanced stage
(P=0.389; Fig. 4B). These data
indicate that LHX3 is an early stage prognostic biomarker of ADC
patients.
LHX3 is a radiosensitivity prognostic
biomarker of ADC patients
We then ascertained whether LHX3 expression
predicted poor OS of ADC patients treated with or without
radiotherapy. From the results, we found a significant effect of
LHX3 expression on poor OS of the patients without radiotherapy
treatment (P=0.009; Fig. 4C), while
LHX3 expression was not associated with OS of the patients with
radiotherapy treatment (P=0.209; Fig.
4D). To further validate whether radiotherapy treatment has an
effect on ADC patient survival, we analyzed the survival time of
the patient treated with or without radiotherapy. The results
showed that the patient without radiotherapy (54.2 months) had a
significantly prolonged survival compared to those with
radiotherapy (40.15 months, P=0.0069; Fig. 4E). These results demonstrate that
LHX3 is a radiosensitivity prognostic biomarker of ADC
patients.
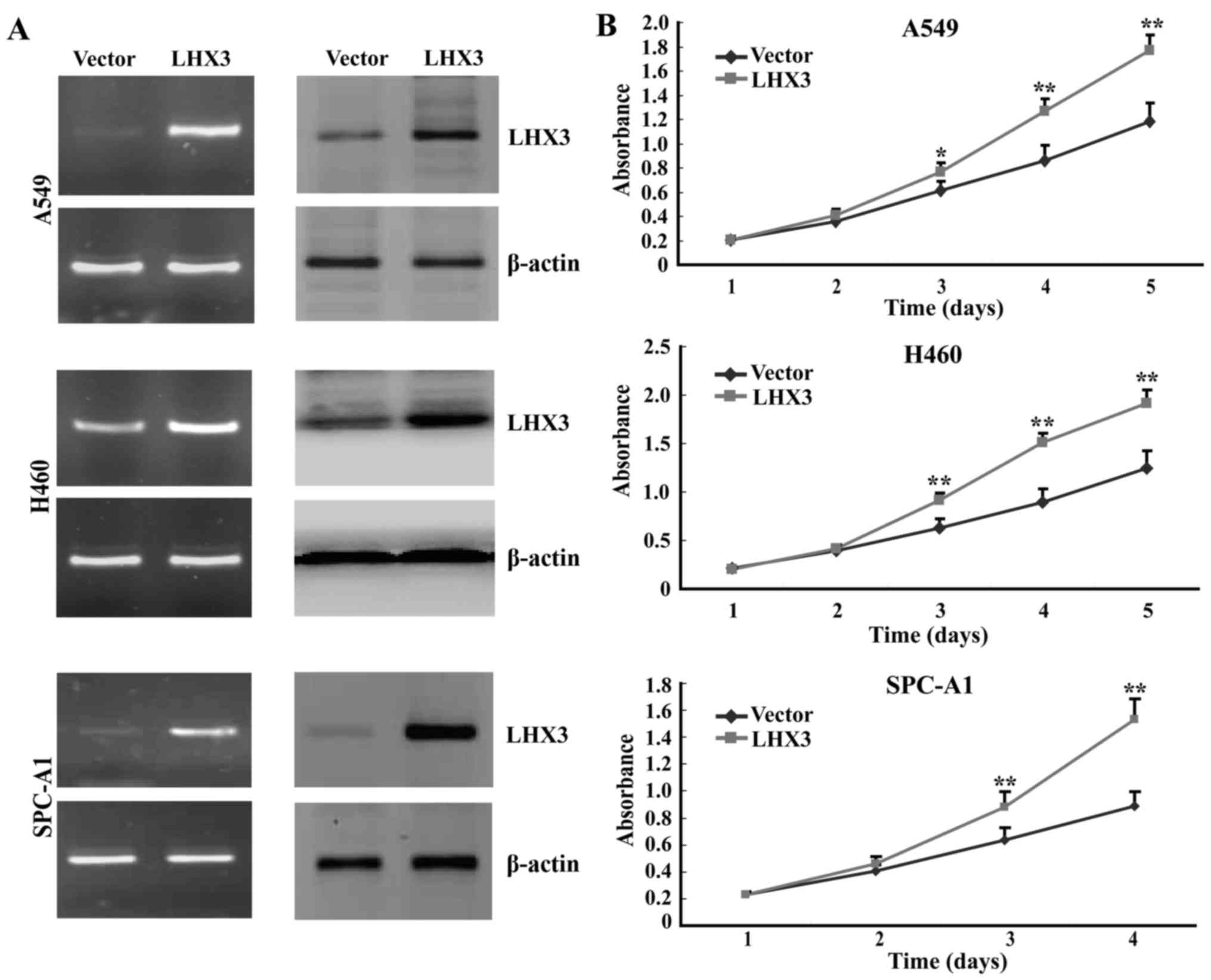
LHX3 promotes cancer cell growth and
proliferation
To evaluate the possible roles of LHX3, we examined
cancer cell growth and proliferation by MTS assay with exogenous
expression of LHX3 in A549 (human adenocarcinoma cell line), H460
(the human large cell lung cancer cell line) and SPC-A1 (human
adenocarcinoma cell line) cells. The results of MTS assay indicated
that ectopic expression of LHX3 significantly promoted cancer cell
growth and proliferation compared with the empty vector control
(Fig. 5). These data indicate that
LHX3 remarkably activates cancer cell proliferation and growth.
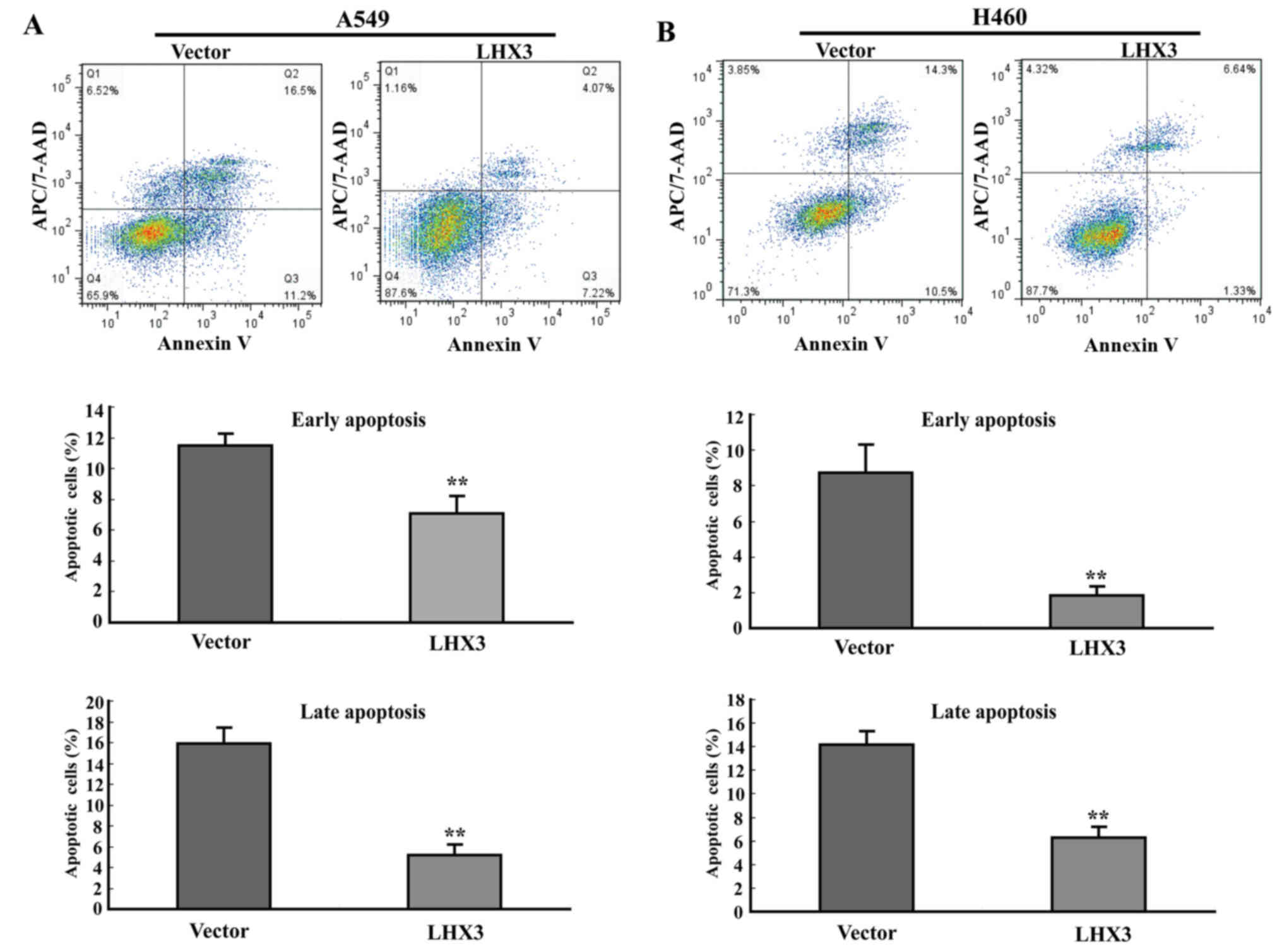
LHX3 inhibits cancer cell
apoptosis
To determine whether the promotion of cancer cell
proliferation of LHX3 related to cell apoptosis, we conducted
Annexin V-APC/7AAD (amino-actinomycin D) double staining the
transfected cancer cells followed by flow cytometric analysis. The
experimental data showed that LHX3 overexpression resulted in a
significant decrease of early apoptotic cells and late apoptotic
cells in A549 and H460 (Fig. 6).
These results demonstrate that LHX3 markedly inhibits cancer cell
apoptosis.
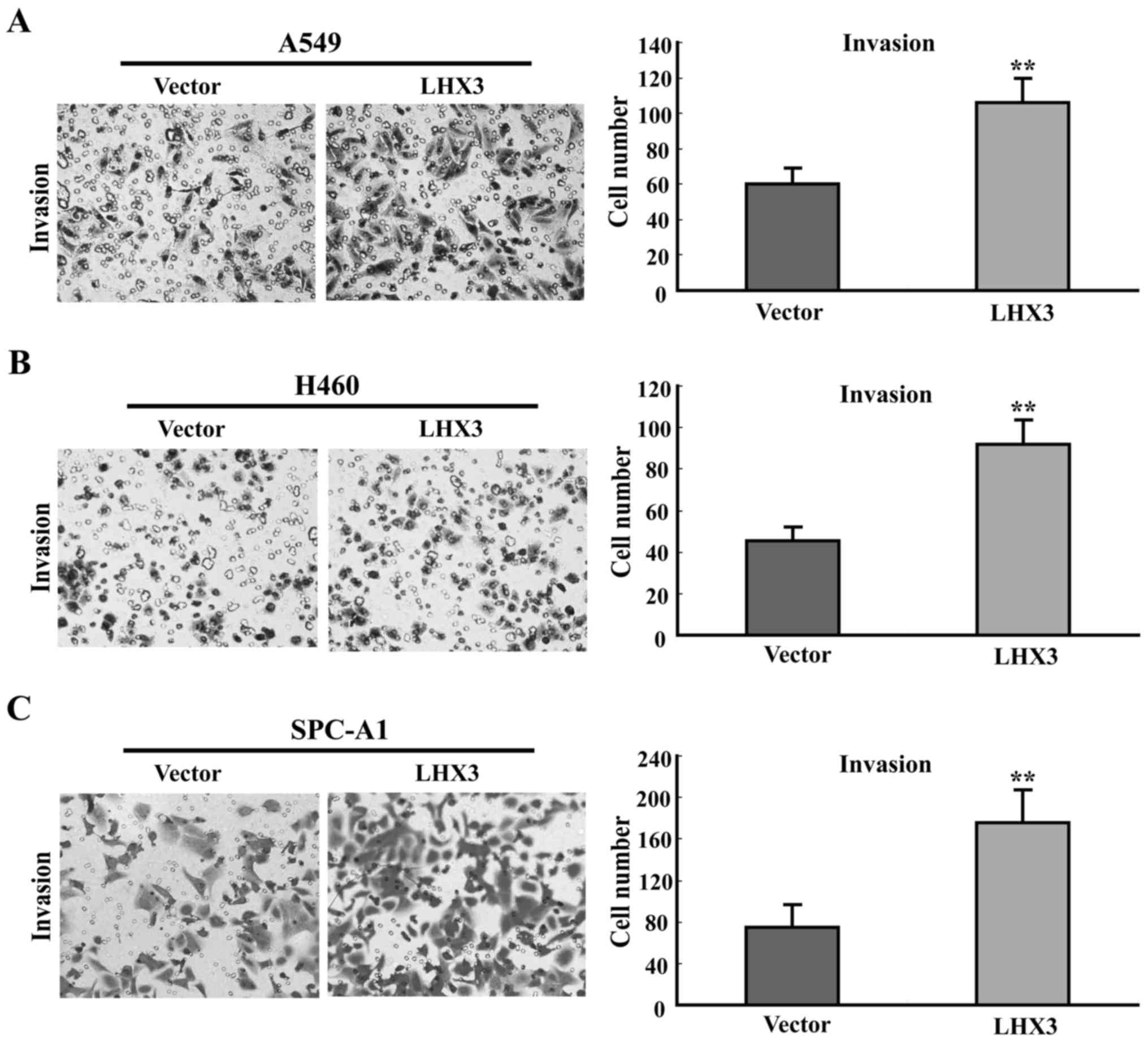
LHX3 stimulates cancer cell
invasion
To investigate the effects of LHX3 on cancer cell
invasion, Transwell assays with Matrigel were performed. A
significant increase of cancer cell invasion was found in
LHX3-transfected cells compared with empty vector-transfected cells
(Fig. 7). These data suggest that
LHX3 markedly stimulates cancer cell invasion.
Discussion
In the present study, the expression, the clinical
relevance and the functional roles of LHX3 were investigated in
NSCLC for the first time. Our data reveal that LHX3 is generally
increased in cancer tissues; LHX3 is obviously associated with
clinical stage and radiotherapy of NSCLC patients, and is an
unfavorable independent prognostic factor of ADC patients;
furthermore, LHX3 is an early stage and radiosensitivity prognostic
factor in ADC patients; functionally, LHX3 promotes cancer cell
proliferation and invasion, whereas inhibits cancer cell
apoptosis.
Previous studies have shown that some LH genes are
involved in various cancers progression (13–18).
However, little is known about their clinical relevance. In the
present study, to determine the clinical relevance of LHX3,
relational analysis and survival analysis were performed.
Relational analysis indicates that LHX3 is obviously associated
with clinical stage and radiotherapy of patients. To confirm the
association between the LHX3 expression and the clinical stage or
radiotherapy of patients, the expression patterns were performed.
LHX3 expression is higher in the advanced stage patients than that
in the early stages patients, and similarly LHX3 expression is
remarkably increased in the patients with radiotherapy. These
results suggest a close association between LHX3 and tumor
metastasis or radiotherapy treatment.
To further evaluate the prognostic significance of
LHX3, Kaplan-Meier and Cox-regression analysis were performed. The
patients with low LHX3 expression have prolonged OS compared to the
ones with high LHX3 expression, and LHX3 is an independent
prognostic factor for OS in NSCLC patients. Notably, LHX3 is an
unfavorable and independent prognostic factor in ADC patients, but
is not associated with survival of SCC patients. Noticeably, LHX3
is an early stage and radiosensitivity prognostic factor in ADC
patients and the radiotherapy treatment significantly shorten the
survival of ADC patients suggesting radiotherapy is an unfavorable
therapeutic strategy for ADC patients. Considering the effect of
radiotherapy on LHX3 expression, we speculate that radiotherapy as
an unfavorable factor for patient survival may be mediated by
upregulation of LHX3 expression.
Previous evidence has indicated that LHX3 plays an
important role in spinal cord motor neurons and pituitary
development (19–22). In a recent study, the DNA
methylation of LHX3 has been reported to be associated with breast
cancer (23). However, no studies
on its function in cancer were reported until now. In the present
study, LHX3 expression was evaluated in 180 NSCLC patients, and
increased LHX3 expression was found in tumor tissues compared to
non-tumor tissues. This result suggests that LHX3 may play roles in
tumorigenesis of NSCLC. To determine the roles of LHX3,
gain-of-function assays were investigated. The functional analysis
show that LHX3 significantly promotes cancer cell proliferation and
invasion, whereas inhibits cancer cell apoptosis. These findings
indicate that LHX3 functions as a potential oncogene in cancer.
Previous studies have reported that several other
LHX genes, such as LHX4, LHX6 and LHX9, are implicated in
tumorigenesis, and act as tumor suppressors in different types of
cancers (16,18,28,29).
LHX3 belongs to the same LHX family, and it is plausible to assume
that LHX3 may function in a similar way. However, LHX3 was found as
an oncogene in the present study. After systematic retrieval of
literature, genes of the same family with different (tumor
suppressive or oncogenic) roles are a common phenomenon in cancer
development, such as the well known SOX and HOX family gene
(30–32). We then used The UCSC Cancer Genomics
Browser (https://genome-cancer.soe.ucsc.edu/proj/site/hg
Heatmap/) to analyze the gene expression in cancer and normal
lung tissues. The results showed that decreased LHX6 expression was
observed in lung cancer tissues, while increased LHX3 expression
was observed in lung cancer tissues compare with normal lung
tissues in the same cohort consistent with our results (data not
shown). According to these data, the conclusion that LHX6 is a
tumor suppressor whereas LHX3 is an oncogene in lung cancer is
reasonable. Furthermore, we then evaluated the functional role of
LHX3 in another lung cancer cell line (SPC-A1), and the results
revealed that LHX3 also promotes cancer cell proliferation and
invasion, which is consistent with the results of A549 and H460
cells. All the data suggest that LHX3 has an oncogenic role in lung
cancer.
In conclusion, LHX3 is a potential early stage and
radiosensitivity biomarker for prognosis evaluation of patient and
acts as a candidate oncogene in ADCs. However, further researches
are needed to explore the molecular mechanism of LHX3 in
cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81502551). The authors
thank all the patients involved in this study.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boyle P and Ferlay J: Cancer incidence and
mortality in Europe, 2004. Ann Oncol. 16:481–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ridge CA, McErlean AM and Ginsberg MS:
Epidemiology of lung cancer. Semin Intervent Radiol. 30:93–98.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pao W and Chmielecki J: Rational,
biologically based treatment of EGFR-mutant non-small-cell lung
cancer. Nat Rev Cancer. 10:760–774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen YT, Kobayashi A, Kwan KM, Johnson RL
and Behringer RR: Gene expression profiles in developing nephrons
using Lim1 metanephric mesenchyme-specific conditional mutant mice.
BMC Nephrol. 7:12006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shawlot W and Behringer RR: Requirement
for Lim1 in head-organizer function. Nature. 374:425–430. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Richter K, do Pinto OP, Hägglund AC,
Wahlin A and Carlsson L: Lhx2 expression in hematopoietic
progenitor/stem cells in vivo causes a chronic myeloproliferative
disorder and altered globin expression. Haematologica.
88:1336–1347. 2003.PubMed/NCBI
|
|
10
|
Raetzman LT, Ward R and Camper SA: Lhx4
and Prop1 are required for cell survival and expansion of the
pituitary primordia. Development. 129:4229–4239. 2002.PubMed/NCBI
|
|
11
|
Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee
E, Huang S, Taira M and Westphal H: Control of hippocampal
morphogenesis and neuronal differentiation by the LIM homeobox gene
Lhx5. Science. 284:1155–1158. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grigoriou M, Tucker AS, Sharpe PT and
Pachnis V: Expression and regulation of Lhx6 and Lhx7, a novel
subfamily of LIM homeodomain encoding genes, suggests a role in
mammalian head development. Development. 125:2063–2074.
1998.PubMed/NCBI
|
|
13
|
Dormoy V, Béraud C, Lindner V, Thomas L,
Coquard C, Barthelmebs M, Jacqmin D, Lang H and Massfelder T:
LIM-class homeobox gene Lim1, a novel oncogene in human renal cell
carcinoma. Oncogene. 30:1753–1763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gorantla B, Asuthkar S, Rao JS, Patel J
and Gondi CS: Suppression of the uPAR-uPA system retards
angiogenesis, invasion, and in vivo tumor development in pancreatic
cancer cells. Mol Cancer Res. 9:377–389. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim MS, Lee J, Oh T, Moon Y, Chang E, Seo
KS, Hoehn BD, An S and Lee JH: Genome-wide identification of OTP
gene as a novel methylation marker of breast cancer. Oncol Rep.
27:1681–1688. 2012.PubMed/NCBI
|
|
16
|
Hung TM, Hu RH, Ho CM, Chiu YL, Lee JL,
Jeng YM, Shih DT and Lee PH: Downregulation of alpha-fetoprotein
expression by LHX4: A critical role in hepatocarcinogenesis.
Carcinogenesis. 32:1815–1823. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Estécio MR, Youssef EM, Rahal P, Fukuyama
EE, Góis-Filho JF, Maniglia JV, Goloni-Bertollo EM, Issa JP and
Tajara EH: LHX6 is a sensitive methylation marker in head and neck
carcinomas. Oncogene. 25:5018–5026. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jung S, Jeong D, Kim J, Yi L, Koo K, Lee
J, Kim CJ, Kim CH, An S, Yang Y, et al: Epigenetic regulation of
the potential tumor suppressor gene, hLHX6.1, in human cervical
cancer. Int J Oncol. 38:859–869. 2011.PubMed/NCBI
|
|
19
|
Sharma K, Sheng HZ, Lettieri K, Li H,
Karavanov A, Potter S, Westphal H and Pfaff SL: LIM homeodomain
factors Lhx3 and Lhx4 assign subtype identities for motor neurons.
Cell. 95:817–828. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sheng HZ, Zhadanov AB, Mosinger B Jr,
Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA and
Westphal H: Specification of pituitary cell lineages by the LIM
homeobox gene Lhx3. Science. 272:1004–1007. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sheng HZ, Moriyama K, Yamashita T, Li H,
Potter SS, Mahon KA and Westphal H: Multistep control of pituitary
organogenesis. Science. 278:1809–1812. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thaler JP, Lee SK, Jurata LW, Gill GN and
Pfaff SL: LIM factor Lhx3 contributes to the specification of motor
neuron and interneuron identity through cell-type-specific
protein-protein interactions. Cell. 110:237–249. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dietrich D, Lesche R, Tetzner R, Krispin
M, Dietrich J, Haedicke W, Schuster M and Kristiansen G: Analysis
of DNA methylation of multiple genes in microdissected cells from
formalin-fixed and paraffin-embedded tissues. J Histochem Cytochem.
57:477–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY,
Xiao YS, Xu Y, Li YW and Tang ZY: Intratumoral balance of
regulatory and cytotoxic T cells is associated with prognosis of
hepatocellular carcinoma after resection. J Clin Oncol.
25:2586–2593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han F, Dong Y, Liu W, Ma X, Shi R, Chen H,
Cui Z, Ao L, Zhang H, Cao J, et al: Epigenetic regulation of sox30
is associated with testis development in mice. PLoS One.
9:e972032014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han F, Liu W, Jiang X, Shi X, Yin L, Ao L,
Cui Z, Li Y, Huang C, Cao J, et al: SOX30, a novel epigenetic
silenced tumor suppressor, promotes tumor cell apoptosis by
transcriptional activating p53 in lung cancer. Oncogene.
34:4391–4402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu WB, Jiang X, Han F, Li YH, Chen HQ,
Liu Y, Cao J and Liu JY: LHX6 acts as a novel potential tumour
suppressor with epigenetic inactivation in lung cancer. Cell Death
Dis. 4:e8822013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vladimirova V, Mikeska T, Waha A,
Soerensen N, Xu J, Reynolds PC and Pietsch T: Aberrant methylation
and reduced expression of LHX9 in malignant gliomas of childhood.
Neoplasia. 11:700–711. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thu KL, Becker-Santos DD, Radulovich N,
Pikor LA, Lam WL and Tsao MS: SOX15 and other SOX family members
are important mediators of tumorigenesis in multiple cancer types.
Oncoscience. 1:326–335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thu KL, Radulovich N, Becker-Santos DD,
Pikor LA, Pusic A, Lockwood WW, Lam WL and Tsao MS: SOX15 is a
candidate tumor suppressor in pancreatic cancer with a potential
role in Wnt/β-catenin signaling. Oncogene. 33:279–288. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, Dang Y, Liu J and Ouyang X: The
function of homeobox genes and lncRNAs in cancer. Oncol Lett.
12:1635–1641. 2016.PubMed/NCBI
|