Introduction
Colorectal cancer (CRC) ranks as the third most
common malignancy throughout the world, with high incidence and
mortality rates (1). Although
advanced treatment including resection, chemotherapy, as well as
radiation are commonly used to improve the outcomes of patients
with CRC, the 5- and 10-year survival rates for CRC patients remain
unsatisfactory, and are ~65 and 58%, respectively (2,3).
Further clarification of the underlying molecular mechanisms for
CRC development and progression are urgently needed (4). Therefore, better understanding of the
precise molecular mechanism contributing to the initiation and
progression of CRC may provide new diagnostic approaches or
therapeutic strategies for patients with CRC.
Long non-coding RNAs (lncRNAs) are types of RNAs
which are over 200 nucleotides in length without protein-coding
capabilities (5–7). lncRNAs, frequently located in the
nucleus, are present with lower abundance, and act in a more
disease-specific or tissue-specific manner with poorer interspecies
conservation in comparison with message RNAs (mRNAs) (8). Growing evidence demonstrates that
lncRNAs are involved in a large range of biological progresses
including proliferation, metastasis, differentiation, inflammation,
angiogenesis, and metabolism (9–14).
Alteration of lncRNAs has been demonstrated to be involved in the
onset and development of various cancers. For example, upregulation
of lncRNA DSCAM-AS1 in breast cancer mediates tumor progression
through direct interaction with hnRNPL (15). LncSox4, highly expressed in
hepatocellular carcinoma (HCC) and liver tumor-initiating cells
(TICs), is required for TIC self-renewal and tumor initiation via
recruitment of Stat3 to the promoter of SOX4 (16). Highly expressed lncRNA SNHG20
facilitates cellular proliferation and predicts poor prognosis for
patients with HCC (17). LINC00673,
a tumor suppressor, mitigates SRC-ERK oncogenic signaling through
reinforcement of the interaction of PTPN11 and PRPF19, and promotes
PTPN11 degradation in the proteasome pathway (18). However, the function of numerous
lncRNAs remain largely unclear.
Small nucleolar RNA host gene 3 (SNHG3; GenBank
accession no. NR_036473.1), located on 1q35.3, is a newly
identified lncRNA. Zhang et al found SNHG3 was highly
expressed in HCC and significantly associated with malignant status
and poor prognosis in HCC (19).
However, its functions and the underlying mechanisms in CRC remain
to be explored. Herein, we seek to determine the expression level
and function of SNHG3, and further explore the potential molecular
mechanism of SNHG3 in colorectal carcinogenesis. We found that
SNHG3 was highly expressed in CRC tissues and CRC cell lines.
Moreover, overexpression of SNHG3 promoted CRC cell proliferation,
whereas silencing of SNHG3 impaired cellular proliferation ability.
In addition, SNHG3 acted as a competing endogenous RNA (ceRNA) to
sponge miR-182-5p, thus leading to the release of c-Myc from
miR-182-5p and the regulation of the expression of c-Myc.
Materials and methods
Cell culture
Human CRC cell lines HT29, HCT116, SW480, and LoVo
were purchased from the American Type Culture Collection (ATCC;
Maryland, MD, USA). The normal colonic epithelial cell line NCM460
was obtained from INCELL (San Antonio, TX, USA). Colorectal cell
lines were cultured in RPMI-1640 medium, L-15 or Dulbeccos modified
Eagles medium (DMEM; Gibco, Carlsbad, USA) supplemented with 10%
fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml
streptomycin. All cell lines were incubated in a humidified
incubator containing 5% CO2 at 37°C.
RNA extraction, reverse transcription,
and qRT-PCR
The total RNA of the CRC cell lines was extracted
using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA).
After quantitating the amount of total RNA, 500 ng RNA from each
sample was converted to cDNA using a Reverse Transcription kit
(Takara Biotechnology, Dalian, China). Real-time PCR was performed
using the SYBR-Green PCR kit purchased from Takara Biotechnology,
and the data collection was carried out on an ABI Prism 7500
Sequence detection system (Applied Biosystems, Foster City, CA,
USA). Primer sequences used in this study were synthesized by
Invitrogen (Shanghai, China) and the sequences were as follows:
SNHG3 forward, 5′-TTCAAGCGATTCTCGTGCC-3′ and reverse,
5′-AAGATTGTCAAACCCTCCCTGT-3′; c-Myc forward,
5′-TACAACACCCGAGCAAGGAC-3′ and reverse, 5′-GAGGCTGCTGGTTTTCCACT-3′;
CCNB1 forward, 5′-CAACTTGAGGAAGAGCAAGCA-3′ and reverse,
5′-AGCATCTTCTTGGGCACACA-3′; CCND2 forward,
5′-CAGCTGTCACTCCTCATGACT-3′ and reverse,
5′-TTGAGACAATCCACGTCTGTGTT-3′; CDK4 forward,
5′-TACAACACCCGAGCAAGGAC-3′ and reverse, 5′-GAGGCTGCTGGTTTTCCACT-3′;
E2F1 forward, 5′-GAGGAGACCGTAGGTGGGAT-3′ and reverse,
5′-GGACAACAGCGGTTCTTGC-3′; GAPDH forward,
5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse,
5′-ATCCGTTGACTCCGACCTTCAC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′ and miR-183-5p forward,
5′-ACACTCCAGCTGGGTTTGGCAATGGTAGAACT-3′ and reverse,
5′-TGGTGTCGTGGAGTCG-3′. Fold change was calculated using the
2−ΔΔCt method. U6 and GAPDH were used as internal
controls.
CCK-8 assay
The cell proliferation ability was assessed using
Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) following the
manufacturer's instructions. Cancer cells (2×103) were
seeded into 96-well plates 12 h before transfection. Ten
microliters of CCK-8 reagent was added into each well of the
96-well plates every 24 h, mixed gently and cultured at 37°C for 2
h. Finally, the absorption was assessed at a wavelength of 450
nm.
Small interfering RNA, miRNA mimics,
and inhibitor of transfection
The small interfering RNAs (siRNAs) specifically
targeting SNHG3, miR-182-5p mimics and miR-182-5p inhibitor were
commercially designed and synthesized by Shanghai GenePharma
(Shanghai, China). Cells were transfected with siRNAs, miR-182-5p
mimics, or miR-182-5p inhibitor with a cell density of 60% using
Lipofectamine 3000 (Invitrogen, USA). The cells were then harvested
for qRT-PCR, western blot analyses, or for further studies 48 h
after transfection.
Lentivirus production and construction
of stable cell lines
A lentivirus harboring full length SNHG3 was
constructed by Shanghai Jikai (Shanghai, China). SW480 cells were
infected with a lentivirus containing full length SNHG3 using the
recombinant lentivirus-transducing units plus 8 mg/ml Polybrene
obtained from Jikai. Three days later, the cells transfected with
the lentivirus were subjected to FACS analysis for GFP to obtain
cells stably overexpressing SNHG3.
Luciferase reporter assay
Wild-type c-Myc 3′-UTR SNHG3 sequences and mutant
c-Myc 3′-UTR SNHG3 sequences were cloned and inserted into
psiCHEK2.0 vector (Promega, Madison, WI, USA) to construct the
psiCHEK-c-Myc-wt, psiCHEK-c-Myc-mut, psiCHEK-SNHG3-wt, and
psiCHEK-SNHG3-mut plasmids, respectively. SW480 cells were
co-transfected with 1,000 ng of luciferase construct vectors along
with miR-182-5p mimics, miR-182-5p inhibitor, or a plasmid
harboring SNHG3. Luciferase activities were assessed using the
dual-luciferase reporter assay system 48 h after transfection.
RIP assay
RIP assay was performed according to the
instructions of the Magna RIP RNA-Binding Protein
Immunoprecipitation kit (Millipore, Bedford, MA, USA). SW480 cells
transfected with MS2-SNHG3-wt and MS2-SNHG3-mut were lysed in lysis
buffer containing a protease inhibitor cocktail and an RNase
inhibitor. Magnetic beads were incubated with anti-GFP antibody or
anti-rabbit IgG for 30–60 min at room temperature. The lysates were
immunoprecipitated with beads at 4°C overnight. The RNA-protein
complex was eluted from the beads. Purified RNA from the
RNA-protein complex was analyzed using qRT-PCR.
Animal studies
The animal experiments were approved by the
Institutional Animal Care and Use Committee of the Huizhou
Municipal Central Hospital. Male BALA/c nude mice (4–6 weeks old)
were used to detect the effect of SNHG3 on the tumor growth in
vivo. Briefly, 1×107 SW480 cells with SNHG3
overexpression (right side) or control vector (left side) were
injected into the bilateral flanks of nude mice. Six weeks later,
the tumors were removed and subjected to further experiments.
Western blot analysis
Western blot analysis was used to assess the
expression level of c-Myc, as previously described (20).
Expression datasets
To explore the expression level of SNHG3 and
miR-182-5p in CRC tissues, we analyzed the data from the Cancer
Genome Atlas project (TCGA; https://tcga-data.nci.nih.gov/tcga/) and Gene
Expression Omnibus (GEO, accession number GSE54632, platform:
GPL8786, [miRNA-1_0] Affymetrix miRNA Array), respectively. In
general, a total of 456 CRC patients with mRNA expression data from
TCGA were enrolled in the current study. Among these patients, 40
patients had SNHG3 expression data from both CRC tumor tissues and
non-tumor tissues, while miRNA expression data from 5 patients in
the GSE54632 dataset were used to compare the expression level of
miR-182-5p in CRC tumor tissues and non-tumor tissues.
Gene Set Enrichment Analysis
(GSEA)
GSEA is a method used to analyze and interpret
microarray data through biological technology, which has been
previously described (21). The
data is analyzed in terms based on their differential enrichment in
a predefined co-expression or biochemical pathway (gene set) in a
previous experiment. If the majority of a gene set had high
expression accompanied with a high risk score, this gene set would
have a positive enrichment score and would be termed ‘enriched’.
GSEA was carried out by the JAVA program (http://www.broadinstitute.org/gsea) using GSEA version
2.0. Data from TCGA dataset was subjected to GSEA analysis.
Parameters used in the current study were as follows: 1,000 random
sample permutations were used to calculate the p-value. An FDR
<25% and a nominal p-value <0.05 were considered as
significant.
Statistical analysis
Statistical analysis was performed by SPSS 20.0
(Abbott Laboratories, North Chicago, IL, USA). All data were
expressed as the mean ± standard error of mean (SEM) from at least
three independent experiments. The t-test was used to analyze data
between two groups and a parametric generalized linear model with
random effects was used for cell proliferation analysis. All
statistical tests were two-sided and a P<0.05 was regarded as
statistically significant.
Results
Upregulation of SNHG3 is correlated
with poor prognosis in patients with CRC
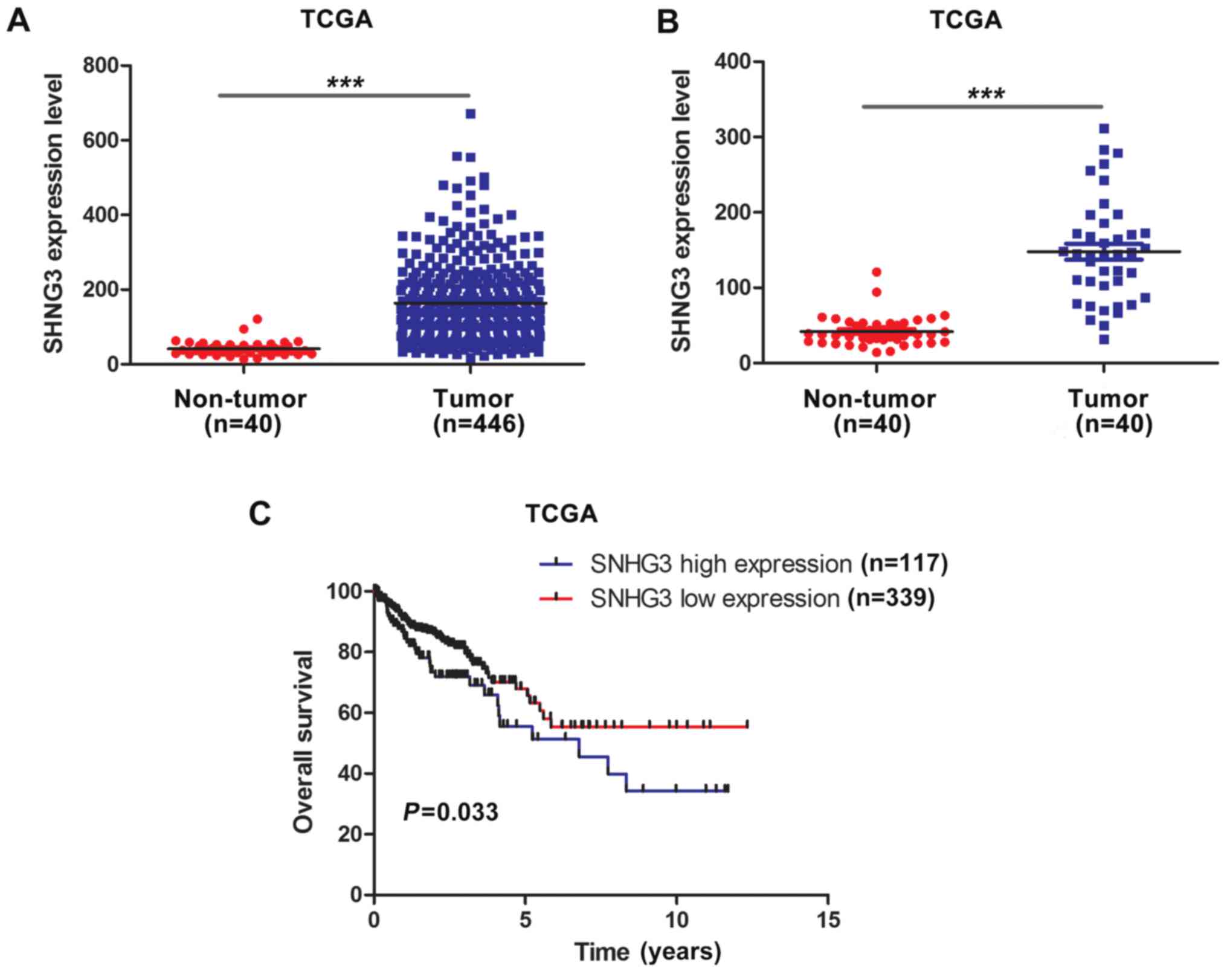
To determine the expression of SNHG3 in CRC, we
analyzed the data of 456 colon adenocarcinoma patients from TCGA
dataset. As shown in Fig. 1A, the
expression of SNHG3 was significantly overexpressed in CRC tumor
tissues in comparison with non-tumor tissues (P<0.001). To
eliminate the possibility that the difference of expression of
SNHG3 between the CRC tumor tissues and non-tumor tissues was
caused by the imbalance of the sample size, we reanalyzed the
expression of SNHG3 in 40 paired CRC tumor tissues and adjacent
non-tumor tissues. The results confirmed that SNHG3 was markedly
upregulated in CRC (Fig. 1B,
P<0.001). Notably, Kaplan-Meier analysis and log-rank test
demonstrated that patients with overexpression of SNHG3 had poorer
overall survival time than those with low expression of SNHG3
(Fig. 1C, P=0.033).
SNHG3 promotes CRC cell
proliferation
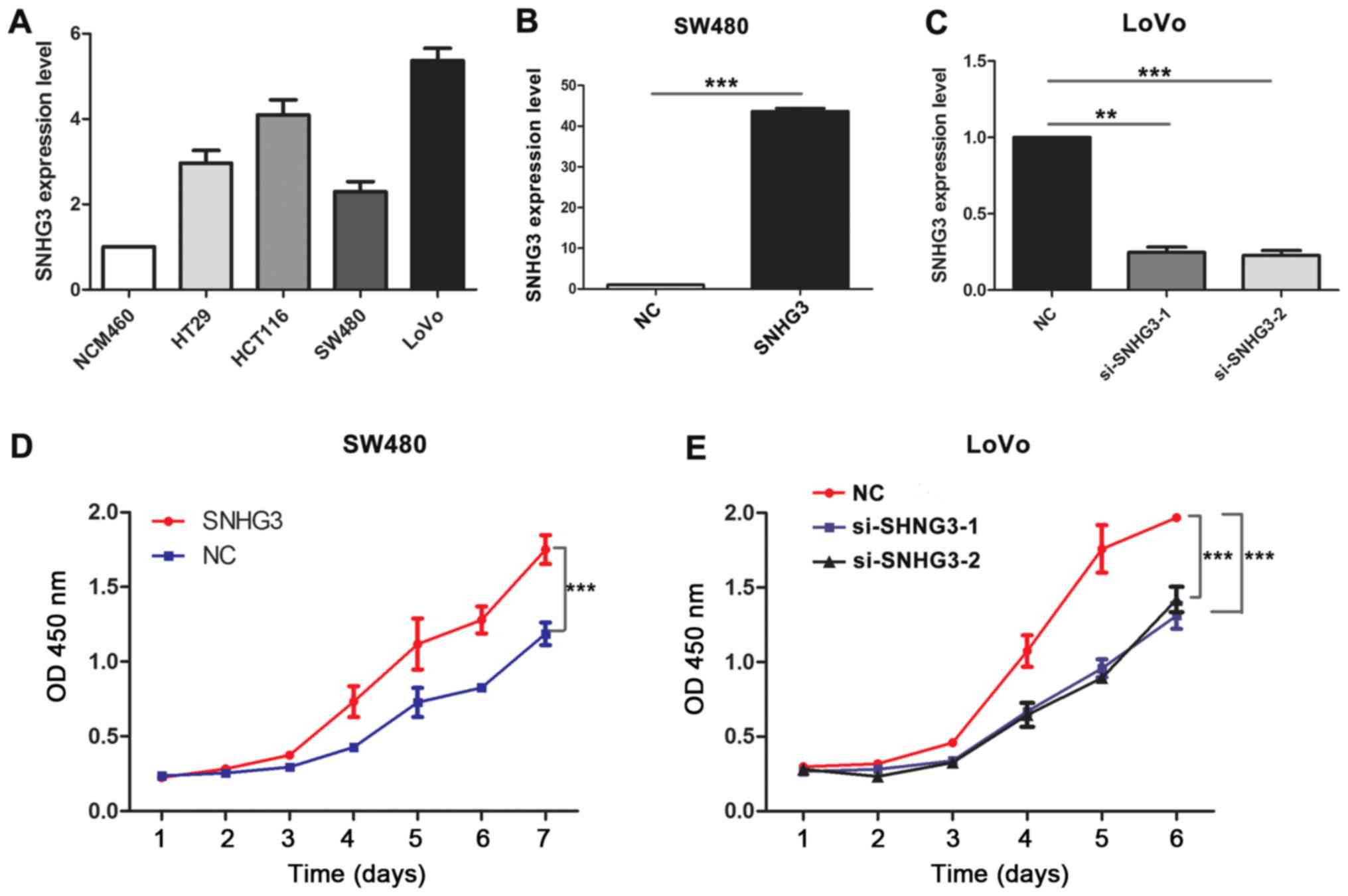
To explore the biological functions of SNHG3 in CRC,
we first analyzed the expression of SNHG3 in CRC cell lines using
qRT-PCR. The results revealed that the expression of SNHG3 was
significantly upregulated in the CRC cell lines compared with the
normal colonic epithelial cells (NCM460, Fig. 2A). Moreover, the expression level of
SNHG3 was the highest in the CRC cell line with the more malignant
phenotype (LoVo), whereas the expression of SNHG3 was the lowest in
the SW480 cells. These results revealed that SNHG3 may be
associated with the progression of CRC. Lentiviral vectors
harboring SNHG3 were introduced into SW480 cells, whereas the
endogenous SNHG3 was knocked down using siRNAs in LoVo cells. As
shown in Fig. 2B, the expression of
SNHG3 was markedly increased in SW480 cells transfected with
lentiviral vectors harboring SNHG3 (P<0.0001). Meanwhile, siRNAs
specifically targeting SNHG3 decreased the expression of SNHG3 in
LoVo cells (Fig. 2C, P<0.001).
CCK-8 assays revealed that overexpression of SNHG3 visibly
increased the proliferation abilities of SW480 cells (Fig. 2D, P<0.001). Consistently,
silencing of SNHG3 in LoVo cells significantly impaired the
cellular proliferation abilities of the LoVo cells (Fig. 2E, P<0.001).
Overexpression of SNHG3 correlates
with c-Myc and its target genes
To investigate the biological pathway and
progression involved in CRC pathogenesis through SNHG3, GSEA
analysis was performed on the CRC tumor samples in TCGA database.
An FDR <25% and a nominal p-value <0.05 were considered as
significant. GSEA analysis indicated that high expression of SNHG3
was associated with c-Myc and its target genes including cell
cycle-related genes like CCNB1, CCND2, CDK4, and E2F1 (Fig. 3A), which suggested that SNHG3 may
regulate CRC cell proliferation through the c-Myc pathway. To find
out whether SNHG3 regulated c-Myc and its target genes, expression
of c-Myc and its target genes were evaluated after overexpression
or silencing of SNHG3 in SW480 and LoVo cells, respectively.
Notably, upregulation of SNHG3 significantly increased the
expression of c-Myc and its target genes related to the cell cycle
including CCNB1, CCND2, CDK4 and E2F1 (Fig. 3B). Consistently, knockdown of SNHG3
obviously decreased the expression of c-Myc and its target genes
(Fig. 3C). We further explored the
correlation between SNHG3 and the mRNA levels of c-Myc and its
target genes in 456 CRC tumor tissues from TCGA dataset. Positive
correlations between SNHG3 and c-Myc, CCNB1, CCND2, CDK4, and E2F1
were found in CRC tumor tissues (Fig.
3D).
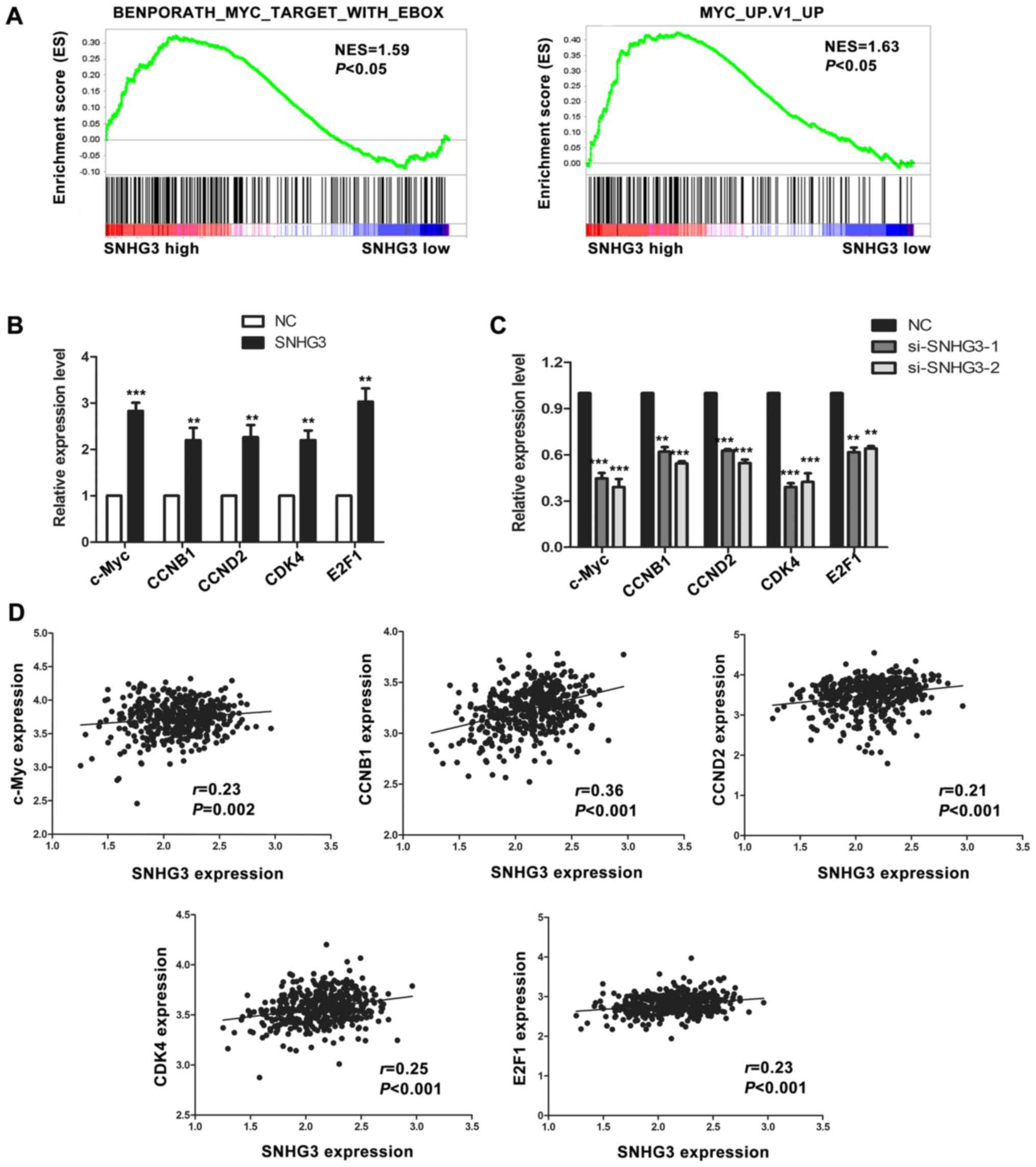 | Figure 3.SNHG3 is correlated with c-Myc and its
target genes. (A) GSEA plot indicated a significant correlation
between high expression of SNHG3 and c-Myc as well as its target
genes. (B) Overexpression of SNHG3 increased the mRNA expression
level of c-Myc and its target genes related to the cell cycle
(CCNB1, CCND2, CDK4, E2F1) in SW480 cells as detected by qRT-PCR
(***P<0.001, **P<0.01). (C) Silencing of SNHG3 decreased the
mRNA expression of c-Myc and its targets in LoVo cells by qRT-PCR
(***P<0.001, **P<0.01). (D) Correlation between SNHG3 and
c-Myc, CCNB1, CCND2, CDK4, and E2F1 in CRC samples from TCGA
database. SNHG3, small nucleolar RNA host gene 3; CRC, colorectal
cancer; GSEA, Gene Set Enrichment Analysis. |
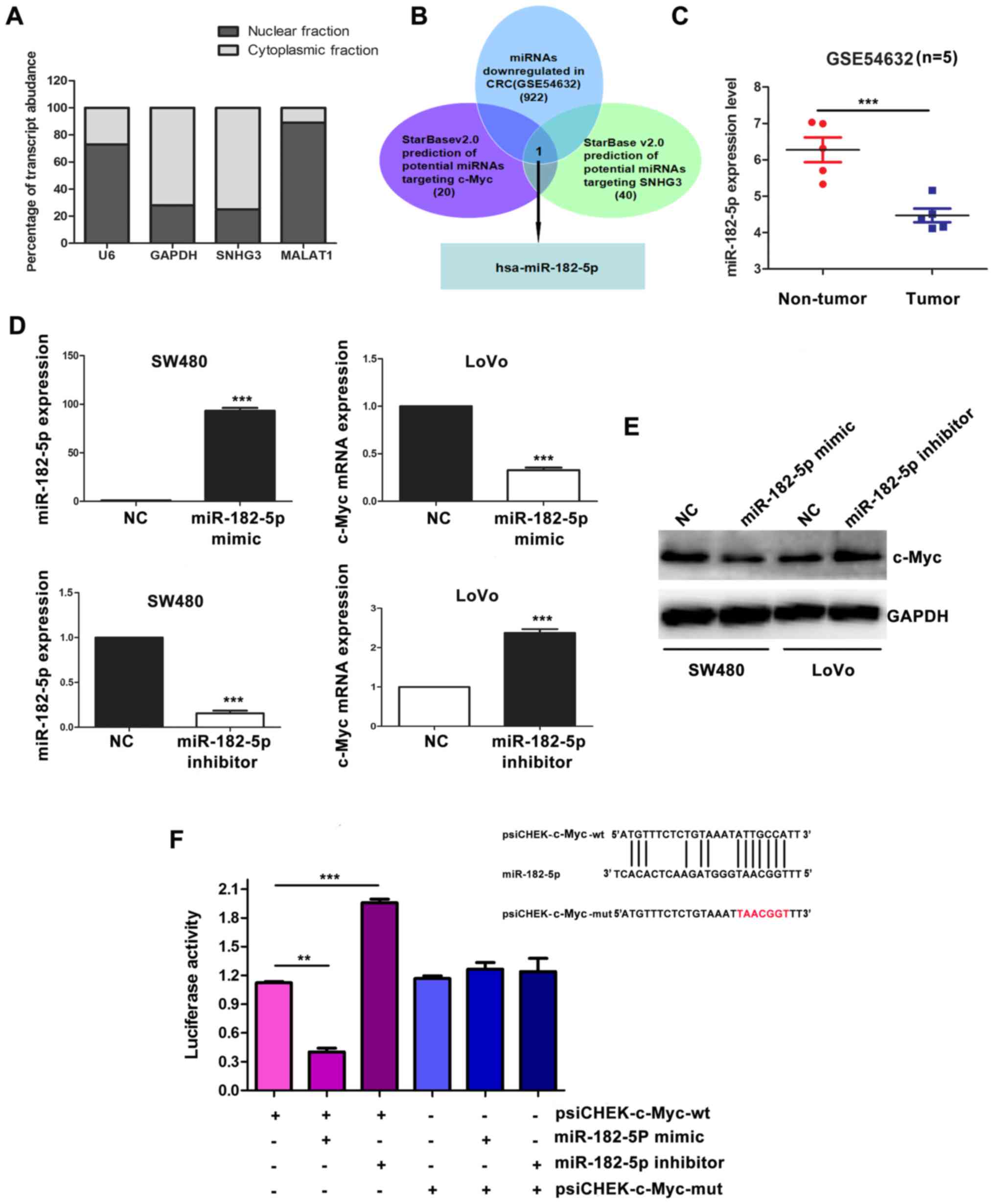
c-Myc is the target of miR-182-5p
It has been reported that the activities of lncRNAs
depend on their subcellular localization (22). Thus to gain further insight into the
mechanism by which SNHG3 promoted the malignant phenotypes of CRC
cells, subcellular distribution of SNHG3 was detected using
cytoplasmic and nuclear RNA fractions from CRC cells. We found that
similar to GAPDH, SNHG3 was mainly located in the cytoplasm
(Fig. 4A). Growing evidence
suggests that cytoplasmic distribution of lncRNAs may function
through interaction with mRNAs and proteins (23). Recently, lncRNA has been reported to
act as a ceRNA via sponging miRNA and releasing the target mRNA
from specific miRNA (24). It was
important to determine whether SNHG3 upregulated c-Myc in a ceRNA
dependent manner, thus bioinformatic analysis was performed to
investigate the potential miRNAs that can target both SNHG3 and
c-Myc. Using starBase2.0, 20 potential miRNAs were predicted to
target c-Myc, and 40 miRNAs were predicted to target SNHG3. After
overlapping these 60 miRNAs, we found 4 potential miRNAs
(miR-186-5p, miR-135b-5p, miR-135a-5p, miR-182-5p) which can target
both SNHG3 and c-Myc. To further narrow the range, we overlapped
these 4 miRNAs with 922 miRNAs downregulated in CRC from the
GSE54632 database. Finally, miR-182-5p was the only candidate
obtained through this process. MiR-182-5p was significantly
downregulated in CRC (Fig. 4C,
P<0.001). To further investigate whether c-Myc is the target of
miR-182-5p, miR-182-5p mimics and miR-182-5p inbihitor were
introduced into SW480 and LoVo cells, respectively (Fig. 4D). qRT-PCR and western blot assays
revealed that the miR-182-5p mimic decreased c-Myc both in mRNA and
protein levels (Fig. 4D and E).
Conversely, inhibition of miR-182-5p increased the expression of
c-Myc (Fig. D and E). In addition,
plasmids harboring wild-type c-Myc 3′-UTR (psiCHEK-c-Myc-wt) and
mutant c-Myc 3′-UTR (psiCHEK-c-Myc-mut) were transiently introduced
into SW480 cells. miR-182-5p mimic decreased the luciferase
activity of the psiCHEK-c-Myc-wt luciferase reporter, while
inhibition of miR-182-5p increased the psiCHEK-c-Myc-wt luciferase
reporter activity. However, neither miR-182-5p mimic nor miR-182-5p
inhibition had an effect on the psiCHEK-c-Myc-mut luciferase
reporter activity. These results indicated c-Myc was the direct
target of miR-182-5p.
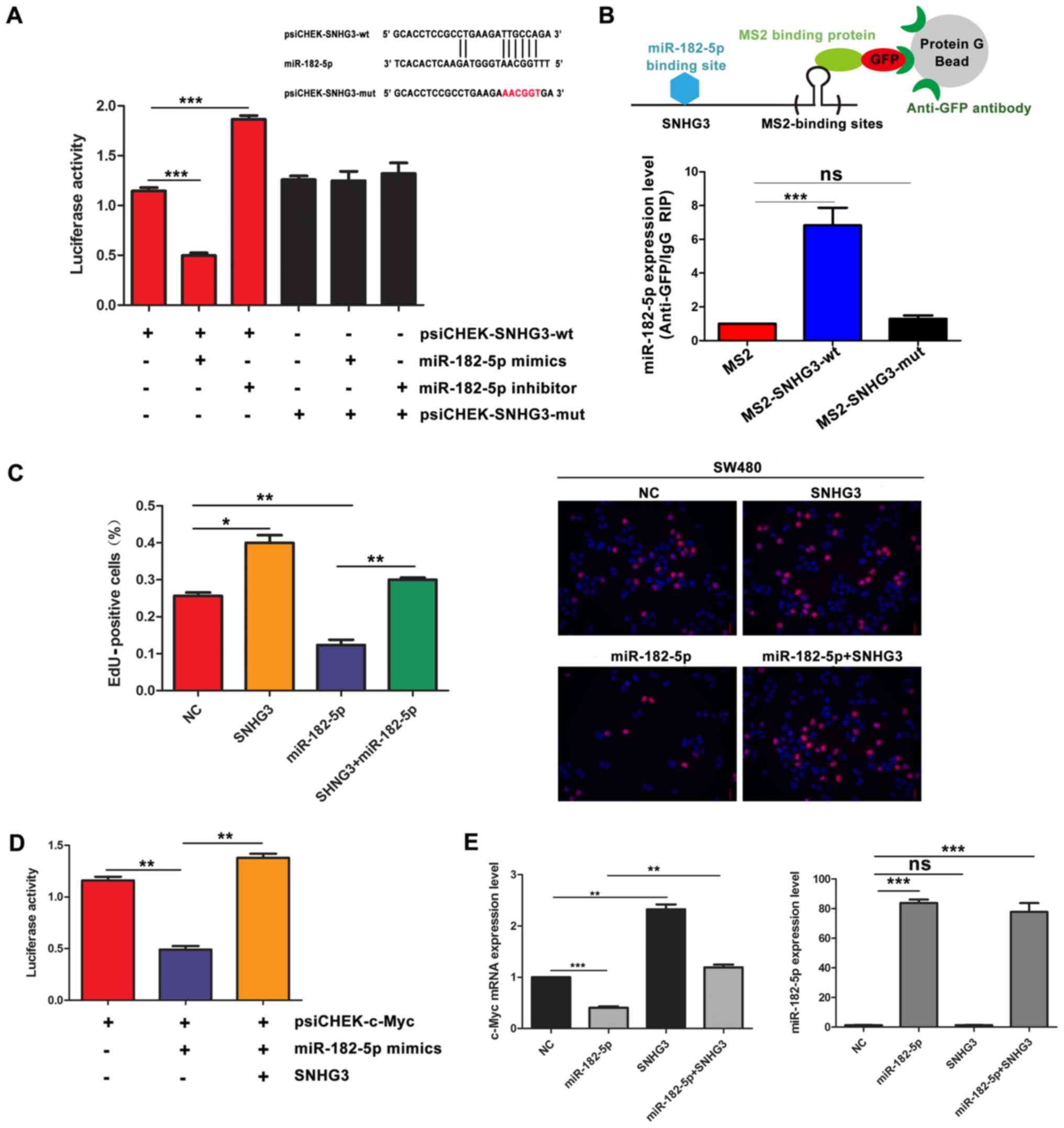
SNHG3 functions as a sponge for
miR-182-5p in CRC cells
Bioinformatics prediction suggested that both SNHG3
and c-Myc are targets of miR-182-5p. Furthermore, SNHG3 upregulated
c-Myc expression level in CRC cells. Based on these results, we
speculated that SNHG3 may function as a miR-182-5p sponge in CRC.
To determine whether miR-182-5p recognized the predicted target
site within SNHG3, luciferase vectors containing wild-type and
mutant SNHG3 (the binding motif for miR-182-5p was mutated) was
constructed. The dual-luciferase assays indicated that miR-182-5p
significantly decreased the wild-type SNHG3 (psiCHEK-SNHG3-wt), but
not the mutant SNHG3 (psiCHEK-SNHG3-mut) luciferase activities
(Fig. 5A). Consistently, inhibition
of miR-182-5p significantly increased the psiCHEK-SNHG3-wt
luciferase reporter activity. However, miR-182-5p inhibitor did not
have an effect on the psiCHEK-SNHG3-mut luciferase reporter
activity (Fig. 5A). Moreover,
MS2-RIP assay was performed to demonstrate the direct interaction
between SNHG3 and miR-182-5p. The results revealed that wild-type
SNHG3 (SNHG3-wt) rather than SNHG3 with the mutated miR-182-5p
binding site (SNHG3-mut) could be immunoprecipitated with
miR-182-5p, indicating that miR-182-5p can bind directly to SNHG3
through a miRNA recognition site (Fig.
5B). Notably, EdU assays were performed to determine whether
SNHG3 abolished the function of miR-182-5p on CRC cell
proliferation. As shown in Fig. 5C,
overexpression of SNHG3 prevented the inhibition of proliferation
of miR-182-5p overexpression in SW480 cells. Subsequently, the
psiCHEK-c-Myc plasmid was co-transfected with the SNHG3 plasmid and
miR-182-5p mimic to conduct another dual-luciferase assay.
Overexpression of SNHG3 totally abolished the decrease of the
luciferase activity induced by miR-182-5p overexpression (Fig. 5D). Furthermore, overexpression of
SNHG3 completely abrogated the decrease of c-Myc expression caused
by miR-182-5p overexpression without altering the expression of
miR-182-5p (Fig. 5E). These results
implied that SNHG3 sponged miR-182-5p and released c-Myc from
miR-182-5p.
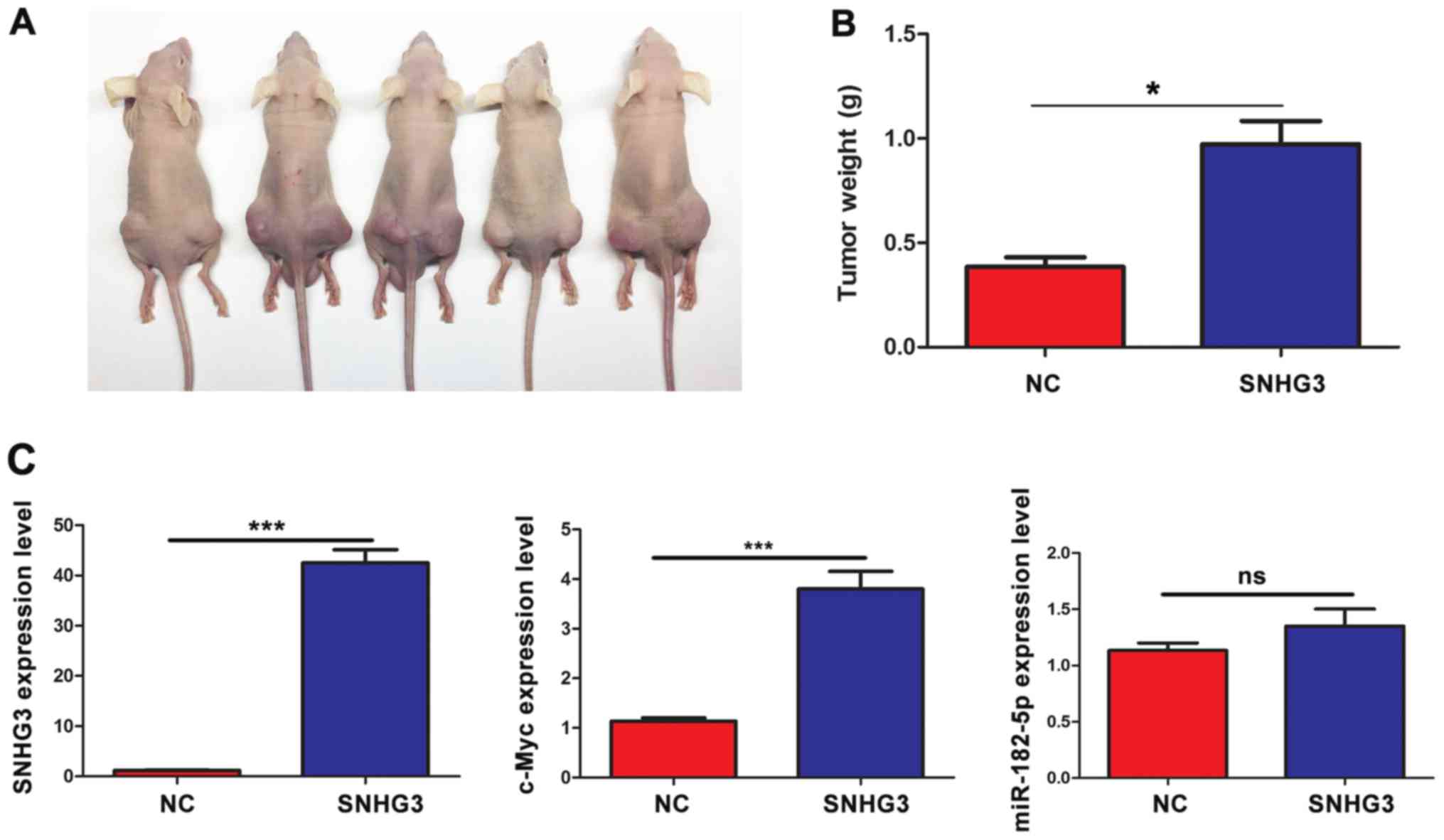
SNHG3 promotes tumor growth in
vivo
To further confirm whether SNHG3 promoted tumor
growth and the expression level of c-Myc in vivo, SW480
cells with SNHG3 overexpressing or control vector were injected
subcutaneously into the bilateral flanks of nude mice (n=5). As
shown in Fig. 6A and B, the tumor
burden and tumor weight in cells overexpressing SNHG3 were
significantly increased than those of the control cells.
Furthermore, SNHG3 markedly increased the expression of c-Myc
without altering the expression of miR-182-5p in vivo
(Fig. 6C).
Discussion
The results of the present study revealed that SNHG3
was significantly upregulated in CRC tissues and its high
expression was closely associated with the poor overall survival of
CRC patients. Functional investigation demonstrated that SNHG3
strongly promoted cellular proliferation ability. Mechanistically,
SNHG3 functioned as a ceRNA to sponge miR-182-5p and decrease the
binding of miR-182-5p to c-Myc, thereby contributing to the
increase of c-Myc.
Recently, Tay et al proposed a new regulatory
mechanism in which most RNA transcripts harbored miRNA-binding
sites capable of communicating and regulating each other's
expression level by competing for binding to shared miRNAs. These
RNA transcripts acted as ceRNAs (25). It's worthwhile to systematically
investigate whether lncRNAs function as ceRNA for the following
reasons: firstly, a large amount of human RNA transcripts are
lncRNAs; secondly, lncRNAs frequently express and function in more
disease-specific, stage-specific, and tissue-specific manners;
thirdly, more and more lncRNAs are found to be located in the
cytoplasm and involved in post-transcriptional gene regulation.
Additionally, growing evidence has proven that lncRNAs communicate
with miRNAs through a ceRNA crosstalk. For example, lncRNA
Unigene56159 directly bound to miR-140-5p and acted as a ceRNA to
regain the expression of Slug, a target gene of miR-140-5p
(26). lncRNA MALAT1 functioned as
a ceRNA to upregulate the expression of ANKXA2 and KRAS, and to
drive gallbladder cancer development (27). Increasing evidence suggests that a
new post-transcriptional regulation model has emerged in which
lncRNA transcripts control miRNA availability.
Based on our study, we suggested a ceRNA regulation
model including SNHG3, miR-182-5p, and c-Myc in CRC. In our study,
we demonstrated that SNHG3, sharing a miR-182-5p response element
with c-Myc, regulated the expression of c-Myc in both mRNA and
protein levels. c-Myc is a basic-helix-loop-helix-leucine zipper
protein which regulates abundant genes involved in the cell cycle,
protein synthesis, cell migration and adhesion, inflammation, and
DNA damage (28–30). GSEA analysis determined a
significant correlation between high expression of SNHG3 and c-Myc
gene signatures. Ectopic expression of SNHG3 increased the
expression of c-Myc and its targets including CCNB1, CCND2, CDK4,
and E2F1, whereas knockdown of SNHG3 had opposite effect on c-Myc.
Notably, by examining the expression data from TCGA database, we
found that SNHG3 was positively correlated with c-Myc, CCNB1,
CCND2, CDK4, and E2F1. Dual-luciferase reporter assays confirmed
that miR-182-5p bound to both SNHG3 and c-Myc, notably, SNHG3
overexpression decreased the interaction between miR-182-5p and
c-Myc. These results indicated that SNHG3 and c-Myc shared the same
miRNA-responsive element with miR-182-5p and promoted CRC
progression in a ceRNA manner.
In conclusion, high expression of SNHG3 predicted
poor prognosis for patients with CRC. SNHG3 acted as a ceRNA to
upregulate c-Myc by sponging miR-182-5p. Hence, SNHG3 may serve as
a novel prognostic approach or therapeutic target for CRC.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
GSEA
|
gene set enrichment analysis
|
|
ceRNA
|
competing endogenous RNA
|
|
lncRNA
|
long non-coding RNA
|
|
mRNA
|
message RNA
|
|
HCC
|
hepatocellular carcinoma
|
|
TICs
|
liver tumor-initiating cells
|
|
SNHG3
|
small nucleolar RNA host gene 3
|
|
ATCC
|
American Type Culture Collection
|
|
CCK-8
|
Cell Counting Kit-8
|
|
siRNA
|
small interfering RNAs
|
|
TCGA
|
The Cancer Genome Atlas project
|
|
GEO
|
Gene Expression Omnibus
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmoll HJ, Van Cutsem E, Stein A,
Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde
CJ, Balmana J, Regula J, et al: ESMO Consensus Guidelines for
management of patients with colon and rectal cancer. a personalized
approach to clinical decision making. Ann Oncol. 23:2479–2516.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cao D, Ding Q, Yu W, Gao M and Wang Y:
Long noncoding RNA SPRY4-IT1 promotes malignant development of
colorectal cancer by targeting epithelial-mesenchymal transition.
Onco Targets Ther. 9:5417–5425. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khorkova O, Hsiao J and Wahlestedt C:
Basic biology and therapeutic implications of lncRNA. Adv Drug
Deliv Rev. 87:15–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ballantyne MD, Pinel K, Dakin R, Vesey AT,
Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et
al: Smooth muscle enriched long noncoding RNA (SMILR) regulates
cell proliferation. Circulation. 133:2050–2065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun T and Wong N: Transforming growth
factor-β-induced long noncoding RNA promotes liver cancer
metastasis via RNA-RNA crosstalk. Hepatology. 61:722–724. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ranzani V, Arrigoni A, Rossetti G, Panzeri
I, Abrignani S, Bonnal RJ and Pagani M: Next-generation sequencing
analysis of long noncoding RNAs in CD4+ T cell
differentiation. Methods Mol Biol. 1514:173–185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Padua D, Mahurkar-Joshi S, Law IK,
Polytarchou C, Vu JP, Pisegna JR, Shih D, Iliopoulos D and
Pothoulakis C: A long noncoding RNA signature for ulcerative
colitis identifies IFNG-AS1 as an enhancer of inflammation. Am J
Physiol Gastrointest Liver Physiol. 311:G446–G457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia P, Cai H, Liu X, Chen J, Ma J, Wang P,
Liu Y, Zheng J and Xue Y: Long non-coding RNA H19 regulates glioma
angiogenesis and the biological behavior of glioma-associated
endothelial cells by inhibiting microRNA-29a. Cancer Lett.
381:359–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang F, Zhang H, Mei Y and Wu M:
Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the
Warburg effect. Mol Cell. 53:88–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Niknafs YS, Han S, Ma T, Speers C, Zhang
C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al:
The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1
in breast cancer progression. Nat Commun. 7:127912016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP
and Gao YF: LncSox4 promotes the self-renewal of liver
tumour-initiating cells through Stat3-mediated Sox4 expression. Nat
Commun. 7:125982016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang D, Cao C, Liu L and Wu D:
Up-regulation of lncRNA SNHG20 predicts poor prognosis in
hepatocellular carcinoma. J Cancer. 7:608–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng J, Huang X, Tan W, Yu D, Du Z, Chang
J, Wei L, Han Y, Wang C, Che X, et al: Pancreatic cancer risk
variant in LINC00673 creates a miR-1231 binding site and interferes
with PTPN11 degradation. Nat Genet. 48:747–757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang T, Cao C, Wu D and Liu L: SNHG3
correlates with malignant status and poor prognosis in
hepatocellular carcinoma. Tumour Biol. 37:2379–2385. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang W, Li J, Guo X, Zhao Y and Yuan X:
miR-663a inhibits hepatocellular carcinoma cell proliferation and
invasion by targeting HMGA2. Biomed Pharmacother. 81:431–438. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tichon A, Gil N, Lubelsky Y, Solomon T
Havkin, Lemze D, Itzkovitz S, Stern-Ginossar N and Ulitsky I: A
conserved abundant cytoplasmic long noncoding RNA modulates
repression by Pumilio proteins in human cells. Nat Commun.
7:122092016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang
Y, Liu M and Tang H: Long non-coding RNA Unigene56159 promotes
epithelial-mesenchymal transition by acting as a ceRNA of
miR-140-5p in hepatocellular carcinoma cells. Cancer Lett.
382:166–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng
MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA Malat1
promotes gallbladder cancer development by acting as a molecular
sponge to regulate miR-206. Oncotarget. 7:37857–37867. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sipos F, Firneisz G and Műzes G:
Therapeutic aspects of c-MYC signaling in inflammatory and
cancerous colonic diseases. World J Gastroenterol. 22:7938–7950.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pazos E, Garcia-Algar M, Penas C,
Nazarenus M, Torruella A, Pazos-Perez N, Guerrini L, Vázquez ME,
Garcia-Rico E, Mascareñas JL, et al: Surface-enhanced raman
scattering surface selection rules for the proteomic liquid biopsy
in real samples: Efficient detection of the oncoprotein c-MYC. J Am
Chem Soc. 138:14206–14209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng XH, Liang YY, Liang M, Zhai W and Lin
X: Direct interaction of c-Myc with Smad2 and Smad3 to inhibit
TGF-β-mediated induction of the CDK inhibitor p15(Ink4B). Mol Cell.
63:10892016. View Article : Google Scholar : PubMed/NCBI
|