Introduction
Osteosarcoma is the most common type of primary bone
cancer (1,2), which typically occurs in children and
young adults. It is characterized by aggressive growth and a
tendency for early metastasis. The most common sites of
osteosarcoma are long bones of the limbs (3), particularly the distal ends of femur
and proximal tibia (4). Despite
advances in multimodal treatment strategies, including neoadjuvant
chemotherapy and surgery, the survival rate of these patients
remains abysmal (5). The five-year
survival rate for localized osteosarcoma is ~65–70%, while that for
metastatic disease is a lowly 20% (6–9).
Approximately one-third of all patients with osteosarcoma
experience recurrent or metastatic disease; the average survival
time after development of metastasis or recurrence is less than 1
year (10). The development of
targeted therapies over the last decade has brought a paradigm
change in cancer treatment. However, gaps in knowledge pertaining
to the complex molecular biology of osteosarcoma has been a barrier
to the development of targeted therapies for these patients.
Identification of genetic therapeutic targets to counter metastatic
and invasive growth characteristics of osteosarcoma is a potential
approach to new therapeutics for osteosarcoma.
TRIM29, also known as ataxia-telangiectasia
group D complementing gene (ATDC), is located at chromosome
11q23. It encodes a 588-amino acid protein which is a member of the
tripartite motif (TRIM) protein family (11). The TRIM family comprises multidomain
ubiquitin E3 ligases, which are characterized by a conserved
RING-finger domain, one or two B-box zinc-finger motifs and a
coiled-coil domain (12–14). Unlike most other TRIM family
proteins, there is no RING domain in TRIM29, which was earlier
considered indicative of its lack of E3 ubiquitin ligase activity.
However, a weak E3 ligase activity of TRIM29 mediated via B-box
domain has recently been reported (15). In addition, following DNA damage,
TRIM29 is phosphorylated and interacts with RNF8, an E3 ubiquitin
ligase, which promotes DNA repair and cell survival (16). The TRIM family proteins are involved
in a series of biological and physiological processes. However,
when altered they are implicated in several pathological processes,
including carcinogenesis (11,17).
Several studies have revealed a role of TRIM29 in carcinogenesis,
which suggests its potential as a therapeutic target for cancer
therapy (18). Recent studies have
revealed that TRIM29 overexpression exerts an oncogenic function in
several types of cancers, including lung cancer (19), hepatocellular carcinoma (20), pancreatic adenocarcinoma (21), gastric cancer (22), esophageal carcinoma (23), colorectal (24) and bladder cancer (25), nasopharyngeal carcinoma (26) and ovarian serous papillary tumors
(27). Upregulation of TRIM29 in
these cancers shows a significant correlation with pathological
grade, tumor invasion, metastasis, and poor prognosis. TRIM29 has
also been shown to act as a tumor suppressor in several types of
cancers, such as breast cancer (28,29).
However, the significance and prognostic value of TRIM29 expression
in osteosarcoma remain unclear. In the present study, we evaluated
the TRIM29 expression pattern and its relevance in
osteosarcoma.
Materials and methods
Patients and tissue samples
The present study was approved by the Research
Ethics Committee at The First Affiliated Hospital of Xi'an Jiaotong
University (Shaanxi, China). All patients or their guardians
provided informed consent for participation in the present study.
All specimens were handled and rendered anonymous according to
ethical and legal standards. All pathological specimens were
obtained from the Department of Pathology at The First Affiliated
Hospital of Xi'an Jiaotong University. Retrospective analysis of
data pertaining to 64 patients (40 men, 24 women; mean age ± SD:
23.6±10.9 years; range, 6–59 years) with osteosarcoma who underwent
surgery at our hospital from January 2005 to December 2014 was
conducted. Normal bone samples (femur, tibia, humerus, radius and
ulna) obtained from 30 patients who underwent surgery (internal
fixation of fracture, amputation, knee arthroplasty or hip
arthroplasty) at our hospital between January 2010 and December
2014 were used as controls. The diagnosis of osteosarcoma was based
on histopathological and radiographic findings. Data on clinical
variables such as age at diagnosis, sex, tumor size, tumor
location, tumor recurrence, visceral metastasis and survival time
after surgery were collected and analyzed. Overall survival time
was defined as the interval between initial surgery and death or
the end of follow-up (follow-up was terminated on September 1,
2016). The sites of osteosarcoma were femur (n=34), tibia (n=18),
humerus (n=4), radius and ulna (n=3), fibula (n=2), pelvis (n=2)
and sacrum (n=1). Tumor size was found to be >5 cm in 39 cases.
Visceral metastasis was present in 26 cases and absent in 38 cases.
Lung was the most common site of metastasis. Post-surgery tumor
recurrence occurred in 16 cases.
Immunohistochemical staining
Immunohistochemical staining for TRIM29 was
performed for all 64 samples of osteosarcoma and 30 samples of
normal bone. All biopsy specimens were embedded in paraffin and
4-µm thick sections were prepared. These sections were first
dehydrated by heating, dewaxed and rehydrated using a graded series
of xylene and ethanol, and blocked with 3%
H2O2 for 20 min in order to quench endogenous
peroxidase activity. Microwave antigen retrieval procedure was
performed in 10 mM citrate buffer. The sections were incubated
overnight at 4°C with TRIM29 primary antibody (GTX115749; 1:200;
GeneTex, San Antonio, TX, USA). Immunostaining was conducted using
the 3,3′-diaminobenzidine (DAB) kit. The sections were then stained
with hematoxylin, dehydrated, cleared and mounted.
Phosphate-buffered saline (PBS) was used as the negative control
(NC).
Evaluation of immunohistochemical
staining, immunohistochemical score (IHS)
The staining results were evaluated by 3 independent
pathologists. The intensity of staining was graded as score 0
(negative), 1 (pale yellow), 2 (dark yellow), and 3 (brown). The
staining extent was scored by the percentage of positive cells as 1
(0–25%), 2 (26–50%), 3 (51–75%) and 4 (76–100%). The final IHS was
obtained by multiplying the score of intensity and extent. Based on
the IHS, specimens were categorized into 1 of 4 groups: 0–1
(negative, -); 2–4 (weak positive, +); 5–8 (moderate positive, ++);
9–12 (strong positive, +++). Scores of 0–4 were designated as low
expression (−/+), while scores of 5–12 were designated as high
expression (++/+++).
Cell culture
Human osteoblast cell line hFOB1.19 and human
osteosarcoma cell lines MG-63, Saos-2, 143B and U-2OS were obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). hFOB1.19 cells were cultured in Dulbecco's modified Eagles
medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% fetal
bovine serum (FBS) (Gibco, Grand Island, NY, USA) at 34°C in 5%
CO2. MG-63, Saos-2, 143B and U-2OS cells were maintained
in DMEM supplemented with 10% FBS at 37°C in 5% CO2.
Cell transfection
The effects of TRIM29 knockdown was studied in the
143B cell line, while overexpression of TRIM29 was studied in the
Saos-2 cell line. In vitro transfections were achieved by
Lipofectamine 2000 reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) by following the manufacturer's protocols. Transfection
efficiency was estimated by western blotting and RT-qPCR. Small
interfering RNAs (siRNAs) to knock down TRIM29 were designed and
synthesized, and the scramble nonsense sequence was used as the
negative control (NC); a final concentration of 50 nM of siRNA and
NC-siRNA was used. The target siRNA sequence for TRIM29 was:
5′-CUGUGUUGUUUCUGCAGGAdTdT-3′; NC siRNA sequence was:
5′-UUCUCCGAACGUGUCACGUTT-3′. For overexpression of TRIM29, the
pcDNA3.1-HA expression vector containing a full-length human TRIM29
sequence (pcDNA3.1-HA-TRIM29) was synthesized and the empty
pcDNA3.1-HA plasmid was used as NC.
Reverse transcriptase quantitative-PCR
(RT-qPCR)
Total RNA from the cell lines was extracted using
TRIzol reagent (Invitrogen Life Technologies) according to the
manufacturer's instructions. Concentration and purity of total RNA
were evaluated by UV–Vis spectroscopy with the Bio-Rad SmartSpec
3000 system (Bio-Rad, Hercules, CA, USA) by optical density (OD)
readings at 260 nm and the ratio of 260/280, respectively. Then,
first-strand cDNA was synthesized using a PrimeScript RT reagent
kit (Takara, Tokyo, Japan). RT-qPCR was performed using SYBR-Green
Quantitative Real-Time Polymerase Chain Reaction Master Mix on the
ABI PRISM® 7500 Sequence Detection System (both from
Applied Biosystems, Foster City, CA, USA). GAPDH was used as an
internal control. Each sample was measured in triplicate. Relative
expression level of mRNA was quantified by the 2−ΔΔCt
method. The primer sequences are listed in Table I.
 | Table I.Primer sequences for RT-qPCR
analysis. |
Table I.
Primer sequences for RT-qPCR
analysis.
| Genes |
| Primer
sequences |
|---|
| TRIM29 | F |
5′-CATGTACCTGACACCCAAAG-3′ |
|
| R |
5′-CCGGGAGGTGTAGTTACCTT-3′ |
| E-cadherin | F |
5′-ATGAGTGTCCCCCGGTATCT-3′ |
|
| R |
5′-CAAACACGAGCAGAGAATCA-3′ |
| N-cadherin | F |
5′-ACAGTGGCCACCTACAAAGG-3′ |
|
| R |
5′-CCGAGATGGGGTTGATAATG-3′ |
| Vimentin | F |
5′-CGCCAGATGCGTGAAATGG-3′ |
|
| R |
5′-ACCAGAGGGAGTGAATCCAGA-3′ |
| ZEB1 | F |
5′-ACTTAAAGTGGCGGTAGATGGTA-3′ |
|
| R |
5′-CAACTGTTTGTAGCGACTGGATT-3′ |
| Snail | F |
5′-TTTACCTTCCAGCAGCCCTACGA-3′ |
|
| R |
5′-CAACTGTTTGTAGCGACTGGATT-3′ |
| GAPDH | F |
5′-GGGAAACTGTGGCGTGAT-3′ |
|
| R |
5′-GAGTGGGTGTCGCTGTTGA-3′ |
Western blot analysis
Total protein was collected from cells using RIPA
lysis buffer with protease inhibitors (Roche Diagnostics, Basel,
Switzerland) and phenylmethylsulfonyl fluoride (PMSF) on ice. The
insoluble debris was removed by centrifugation at 12,000 rpm for 30
min at 4°C. The concentration of each sample was quantified using
the Pierce BCA protein detection kit (Thermo Scientific, Hudson,
NH, USA). Protein samples were separated on SDS-PAGE gels, and then
electrophoretically transferred to polyvinylidene fluoride
membranes. The membranes were blocked with 5% fat-free milk in PBS
+ Tween-20 (PBST) buffer, and then incubated with primary
antibodies against TRIM29 (GTX115749; 1:1,000; GeneTex), HA tag
(ab9110; 1:4,000; Abcam, Cambridge, MA, USA), E-cadherin (3195T;
1:1,000), N-cadherin (13116T; 1:1,000), vimentin (5741T; 1:1,000)
[all from Cell Signaling Technology (CST), Danvers, MA, USA], Snail
(GTX100754; 1:1,000; GeneTex), ZEB1 (GTX105278; 1:1,000; GeneTex),
and GAPDH (5174S; 1:1,000; CST) overnight at 4°C. Then, the
membranes were washed and incubated with the HRP-conjugated
secondary antibody for 2 h [OB4050-05; 1:20,000; goat anti-rabbit
secondary antibody (Southern Biotech, Birmingham, AL, USA)] at room
temperature. The visual bands were obtained using enhanced
chemiluminescence (ECL) detection reagents (Pierce, Rockford, IL,
USA). GAPDH was used as an internal control.
Wound healing assays
Cells were seeded into 6-well plates. After
transfection, cells were cultured for 24 h until 95–100%
confluence. Then, pipette tips were used to make straight scratches
on the surfaces of confluent cell monolayers. After that, cells
were gently washed with PBS for 3 times to remove the detached
cells. Then, cells were cultured in serum-free medium with 10 µg/ml
mitomycin to inhibit cell proliferation. Images were obtained at 0,
24 and 48 h using an inverted microscope. Cell migration rates were
calculated from all the images using Image-Pro Plus 6.0 software
(version 6.0; Media Cybernetics, Rockville, MD, USA).
Transwell invasion assays
Cell invasion assays were performed on Transwell
chambers. The Transwell membrane was coated with 1:3 diluted
Matrigel (BD Biosciences, San Jose, CA, USA). Cells were
resuspended and trypsinized in serum-free medium, 100 µl of cell
suspension (1×105 cells) was added into the upper
chamber and 600 µl medium with 10% FBS was added into the lower
chamber. Then, cells were cultured at 5% CO2 at 37°C for
24 h. After that, the cells on the upper surface of the membrane
were wiped with a cotton swab, and the attached cells below the
membrane were fixed with paraformaldehyde for 15 min and stained
with crystal violet for 10 min. After washing with PBS, the number
of stained cells was calculated under a microscope.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 20.0; SPSS, Inc., Chicago, IL, USA).
Between-group differences with respect to categorical variables
(frequencies) were assessed using Chi-squared test. Those with
respect to continuous variables were assessed with t-test. Survival
analysis was performed using the Kaplan-Meier method, and
differences in survival were evaluated using the log-rank test. Cox
regression model was applied for multivariate survival analysis.
Differences were considered to be statistically significant at
P<0.05 or highly significant at P<0.01.
Results
Immunohistochemical expression of
TRIM29 in osteosarcoma and normal bone tissues
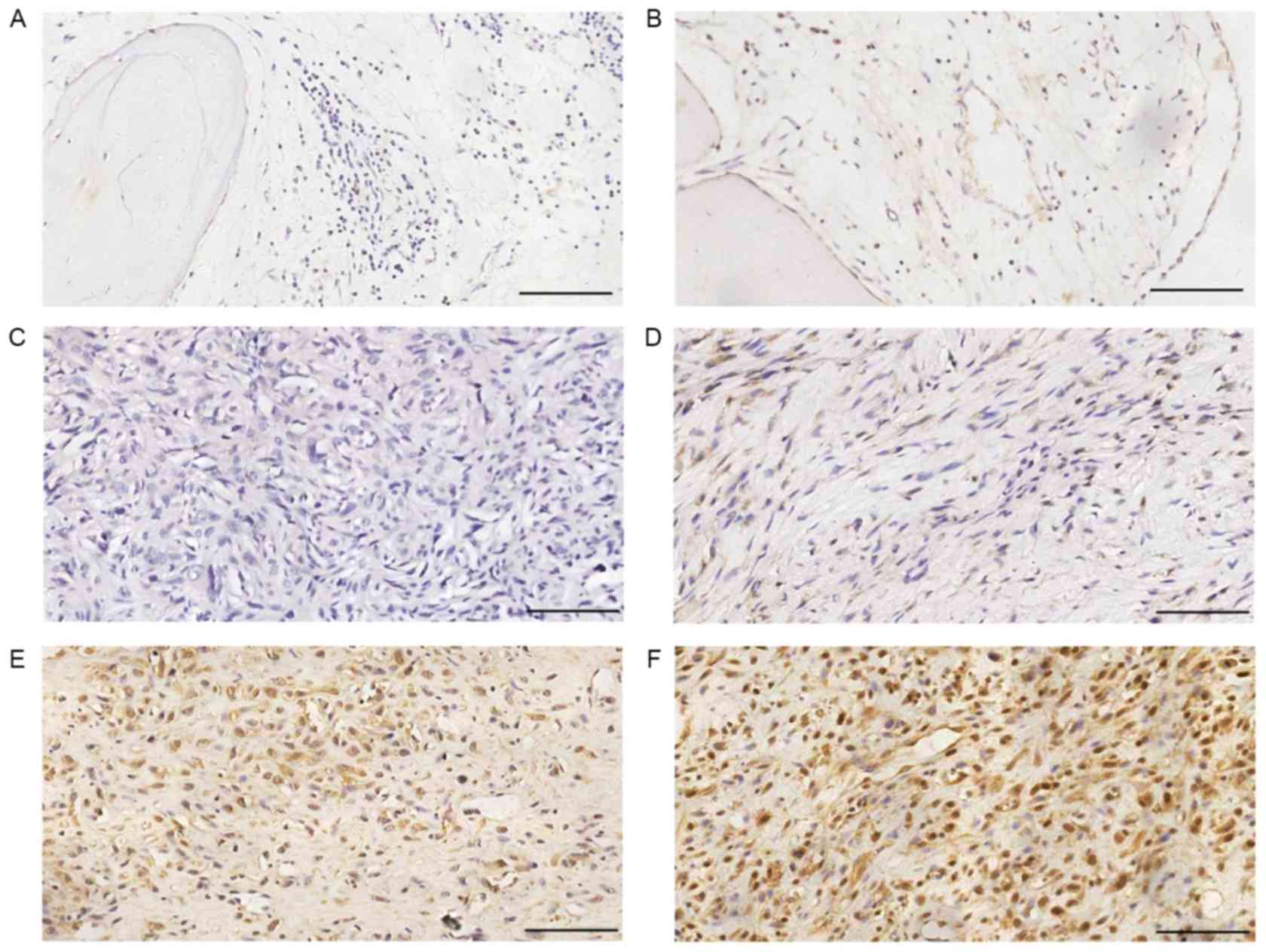
Fig. 1 shows
representative immunohistochemical staining images of TRIM29 in
human normal bone and osteosarcoma tissues. Forty-six out of 64
(71.88%) osteosarcoma specimens and 7 out of 30 (23.33%) normal
bone specimens stained positive for TRIM29. The between group
difference in this respect was statistically significant (Table II; P<0.01). None of the normal
bone tissue specimens showed high expression of TRIM29, while 33
out of 64 (51.56%) osteosarcoma specimens showed high expression
(Table II).
 | Table II.Immunohistochemical expression of
TRIM29 in osteosarcoma and normal bone tissues. |
Table II.
Immunohistochemical expression of
TRIM29 in osteosarcoma and normal bone tissues.
|
|
| Expression of
TRIM29 |
|
|
|---|
|
|
|
|
|
|
|---|
| Tissues | N | Negative (−) | Weak positive
(+) | Moderate positive
(++) | Strong positive
(+++) | Positive rate
(%) | P-value |
|---|
| Osteosarcoma | 64 | 18 | 13 | 19 | 14 | 71.88 | 0.000a |
| Normal bone | 30 | 23 | 7 | 0 | 0 | 23.33 |
|
Correlation between TRIM29 expression
and clinical parameters
Overall, there was no significant correlation
between TRIM29 expression and age at diagnosis (P=0.627), sex
(P=0.607) or tumor location (P=0.729) (Table III). However, a significant
correlation was found between TRIM29 expression and tumor size
(P<0.05), tumor recurrence (P<0.05), visceral metastasis
(P<0.01) and overall survival time after surgery
(P<0.05).
 | Table III.Correlation between TRIM29 expression
and clinical parameters. |
Table III.
Correlation between TRIM29 expression
and clinical parameters.
|
|
| Expression of
TRIM29 |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameters | N | Low (−/+) | High (++/+++) | χ2 | P-value |
|---|
| Age (years) |
|
>18 | 31 | 14 | 17 | 0.258 | 0.627 |
|
≤18 | 33 | 17 | 16 |
|
|
| Sex |
|
Male | 40 | 18 | 22 | 0.505 | 0.607 |
|
Female | 24 | 13 | 11 |
|
|
| Tumor location |
|
Femur | 34 | 15 | 19 | 0.631 | 0.729 |
|
Tibia | 18 | 10 | 8 |
|
|
|
Other | 12 | 6 | 6 |
|
|
| Tumor size
(cm) |
|
>5 | 39 | 14 | 25 | 6.286 | 0.020a |
| ≤5 | 25 | 17 | 8 |
|
|
| Tumor
recurrence |
|
Positive | 16 | 4 | 12 | 4.692 | 0.043a |
|
Negative | 48 | 27 | 21 |
|
|
| Visceral
metastasis |
|
Positive | 26 | 7 | 19 | 8.115 | 0.006b |
|
Negative | 38 | 24 | 14 |
|
|
| Overall survival
time (years) |
|
>5 | 32 | 21 | 11 | 7.570 | 0.012a |
| ≤5 | 32 | 10 | 22 |
|
|
Prognostic value of TRIM29
expression
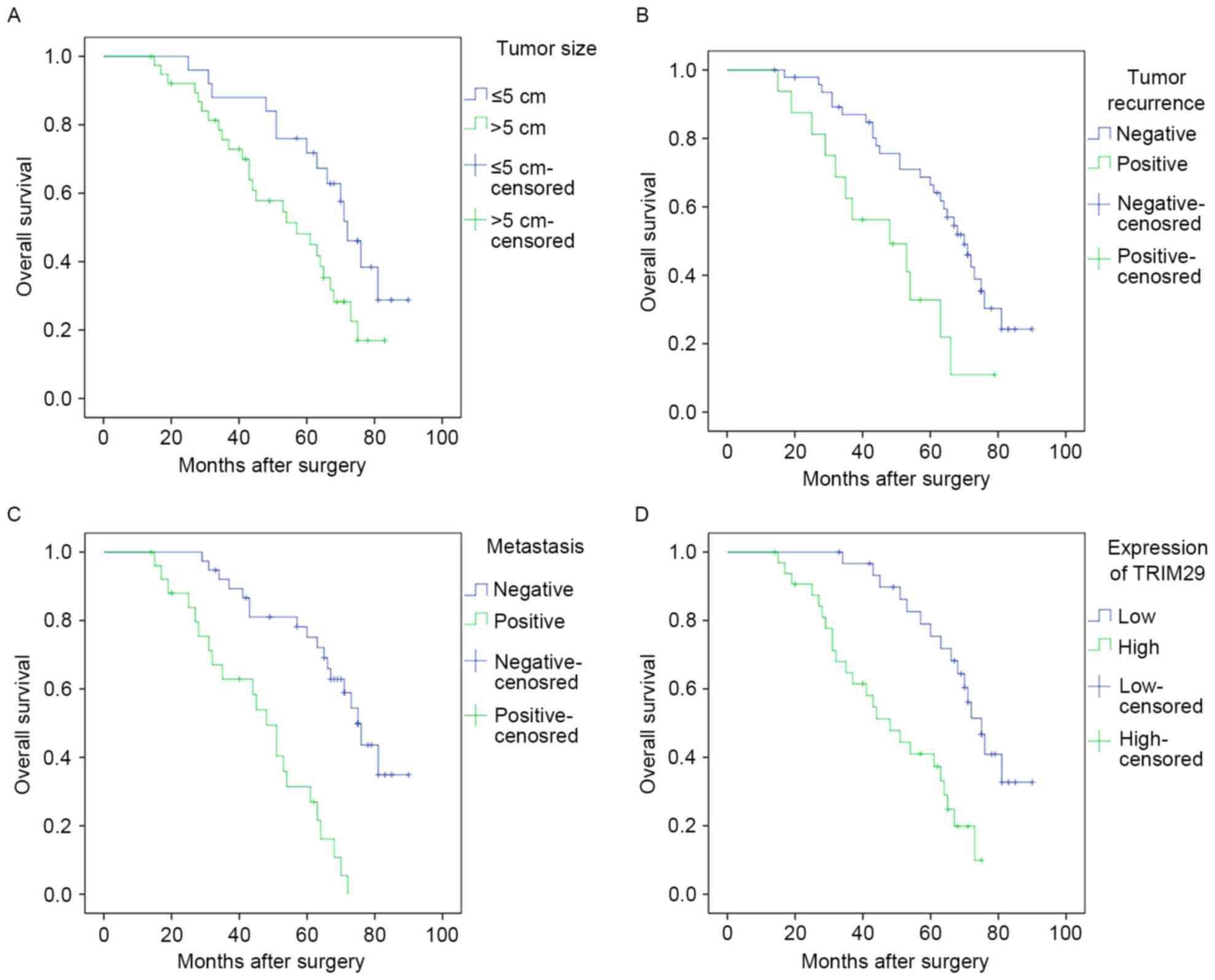
Upon Kaplan-Meier survival analysis, tumor size
(P<0.05), recurrence (P<0.01)and metastasis (P<0.01)
showed a significant direct correlation with overall survival
(Fig. 2A-C; Table IV). Additionally, high expression
of TRIM29 in osteosarcoma specimens showed a significant inverse
correlation with overall survival (P<0.01) (Fig. 2D; Table
IV). Nevertheless, no significant correlation was noted between
overall survival time and other clinical parameters, such as age at
diagnosis, sex and tumor location (Table IV). Variables that showed
significant association upon univariate analysis were included in
the multivariate analysis using Cox proportional hazard model. High
expression of TRIM29 and presence of metastasis were found to be
independent predictors of poor prognosis in patients with
osteosarcoma (Table V).
 | Table IV.Univariate analysis of overall
survival time in patients with osteosarcoma. |
Table IV.
Univariate analysis of overall
survival time in patients with osteosarcoma.
|
|
| Overall survival
(months) |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameters | N | Mean ± SD | Median | χ2 | P-value |
|---|
| Sex |
|
Male | 40 |
54.5±19 | 58 | 0.016 | 0.900 |
|
Female | 24 |
55.3±22.3 | 60.5 |
|
|
| Age (years) |
|
>18 | 31 |
55.6±19.7 | 60 | 0.256 | 0.613 |
|
≤18 | 33 |
54±20.9 | 61 |
|
|
| Tumor location |
|
Femur | 34 |
54.1±21.7 | 64 | 0.126 | 0.939 |
|
Tibia | 18 |
54.6±19.9 | 59.5 |
|
|
|
Others | 12 |
56.9±17.4 | 58.5 |
|
|
| Tumor size
(cm) |
|
>5 | 39 |
48.7±19.9 | 45 | 4.974 | 0.026a |
| ≤5 | 25 |
64.2±16.9 | 68 |
|
|
| Tumor
recurrence |
|
Positive | 16 |
43.8±17.9 | 44 | 7.555 | 0.006b |
|
Negative | 48 |
58.4±19.7 | 65 |
|
|
| Visceral
metastasis |
|
Positive | 26 |
42.7±18.6 | 44.5 | 25.592 | 0.000b |
|
Negative | 38 |
63±16.9 | 67.5 |
|
|
| Expression of
TRIM29 |
|
High | 33 |
44.6±19.1 | 43 | 15.022 | 0.000b |
|
Low | 31 |
65.6±15.2 | 70 |
|
|
 | Table V.Multivariate analysis of overall
survival time in patients with osteosarcoma. |
Table V.
Multivariate analysis of overall
survival time in patients with osteosarcoma.
| Parameters | HR | 95% CI of HR | P-value |
|---|
| Tumor size (>5
vs. ≤5 cm) | 1.661 | 0.816–3.379 | 0.162 |
| Tumor recurrence
(positive vs. negative) | 2.112 | 0.994–4.484 | 0.052 |
| Visceral metastasis
(positive vs. negative) | 4.911 | 2.339–10.310 | 0.000b |
| Expression of
TRIM29 (high vs. low) | 2.485 | 1.157–5.334 | 0.020a |
Expression of TRIM29 in human
osteoblast and osteosarcoma cell lines
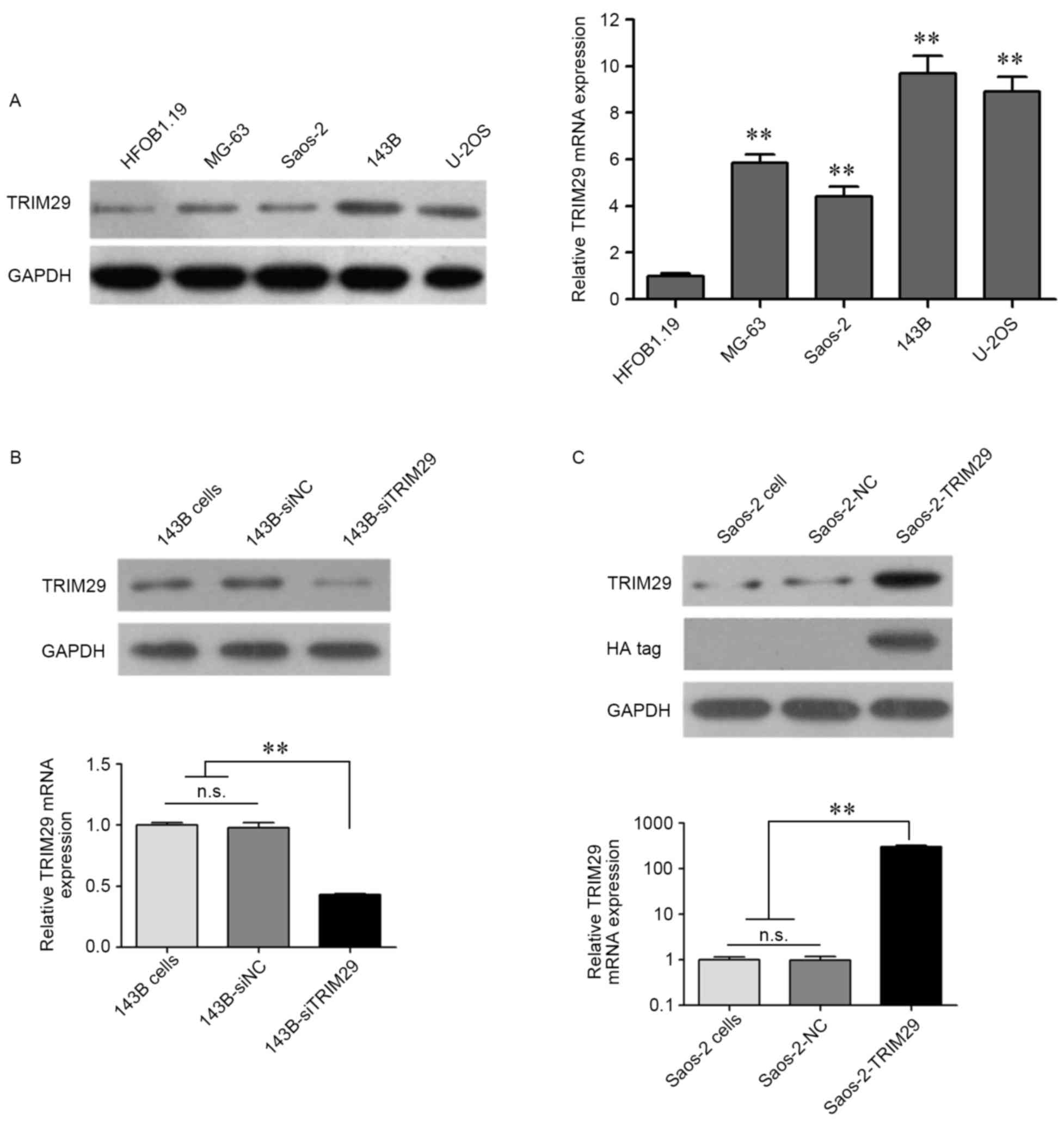
At the cellular level, both protein and mRNA
expression of TRIM29 were significantly higher in osteosarcoma cell
lines as compared to that in the osteoblast cell line hFOB1.19
(Fig. 3A). Moreover, in these 4
osteosarcoma cell lines, the protein and mRNA expression of TRIM29
were highest in 143B, and lowest in Saos-2. Coincidentally,
invasion and proliferation capability of 143B was much higher than
that of Saos-2. These findings indicate that TRIM29 may play
an important role in the progression of osteosarcoma. Knockdown of
TRIM29 in 143B cells and overexpression of TRIM29 in Saos-2 cells
were detected using western blotting and RT-qPCR (Fig. 3B and C). For confirmation of
exogenous TRIM29 expression, anti-HA tag antibody was used
(Fig. 3C). HA-TRIM29 protein was
detected only in Saos-2-TRIM29 cells (transfected with
pcDNA3.1-HA-TRIM29 plasmid).
TRIM29 enhances osteosarcoma cell
migration and invasion
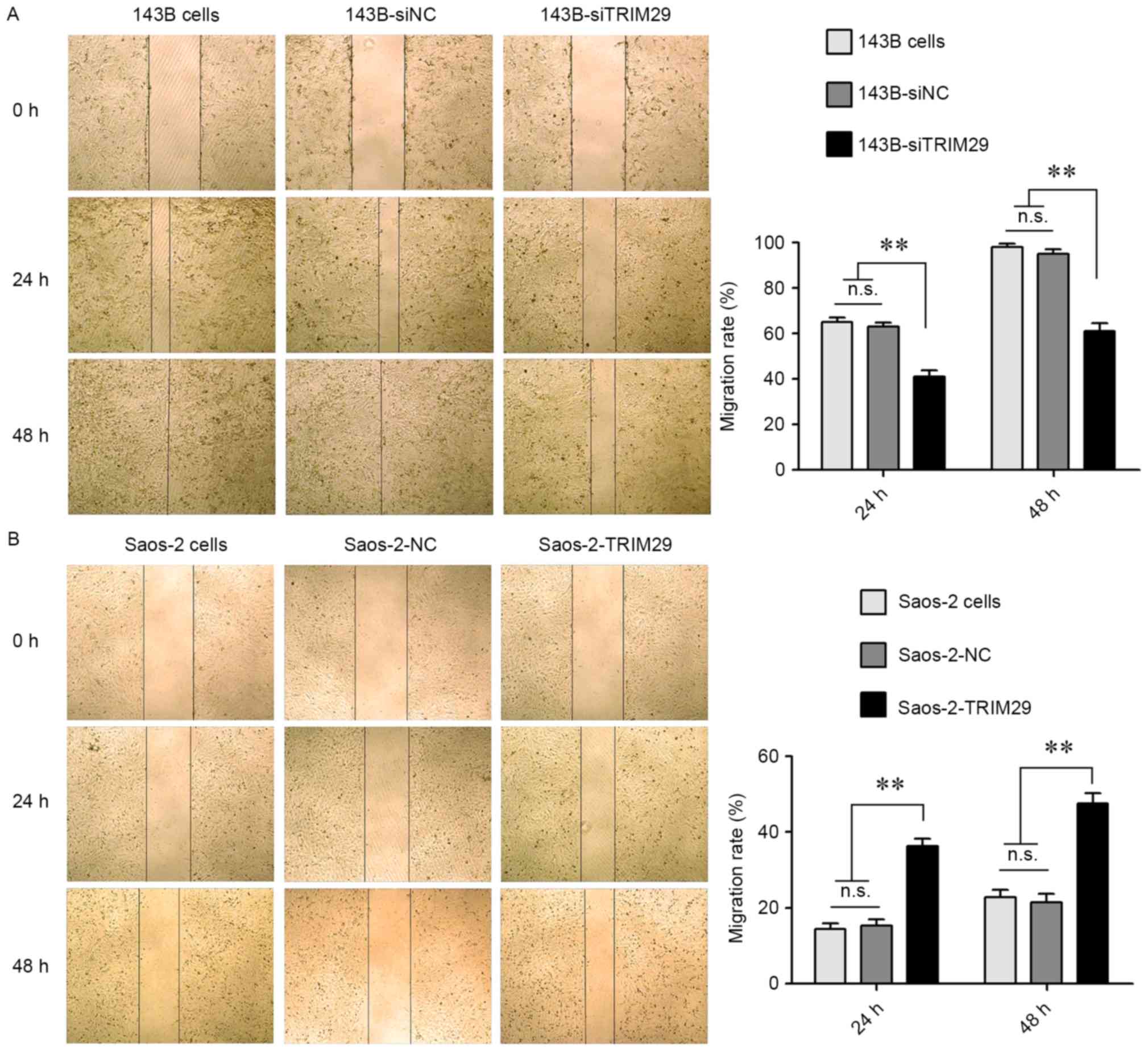
To study the functional relevance of TRIM29
in human osteosarcoma cells, wound healing assays were performed by
calculation of cell migration rates into the wounded area after
scratching, performed under serum-free conditions. Since these
assays were performed in the absence of serum or growth factors,
healing of the wounds occurred by cell migration and not as a
result of proliferation of cells. Fig.
4 depicts representative images obtained at 0, 24 and 48 h. As
shown in Fig. 4A, the migration
rates of 143B-siTRIM29 cells were significantly lower than that of
143B and 143B-siNC cells. The P-values for 143B-siTRIM29 group vs.
143B cells or 143B-siNC groups at both 24 and 48 h were all
<0.01. Moreover, overexpression of TRIM29 enhanced the migration
of the Saos-2 cells (Fig. 4B).
P-values for the Saos-2-TRIM29 group vs. Saos-2 cells or Saos-2-NC
groups at 24 and 48 h were also <0.01. Transwell invasion assays
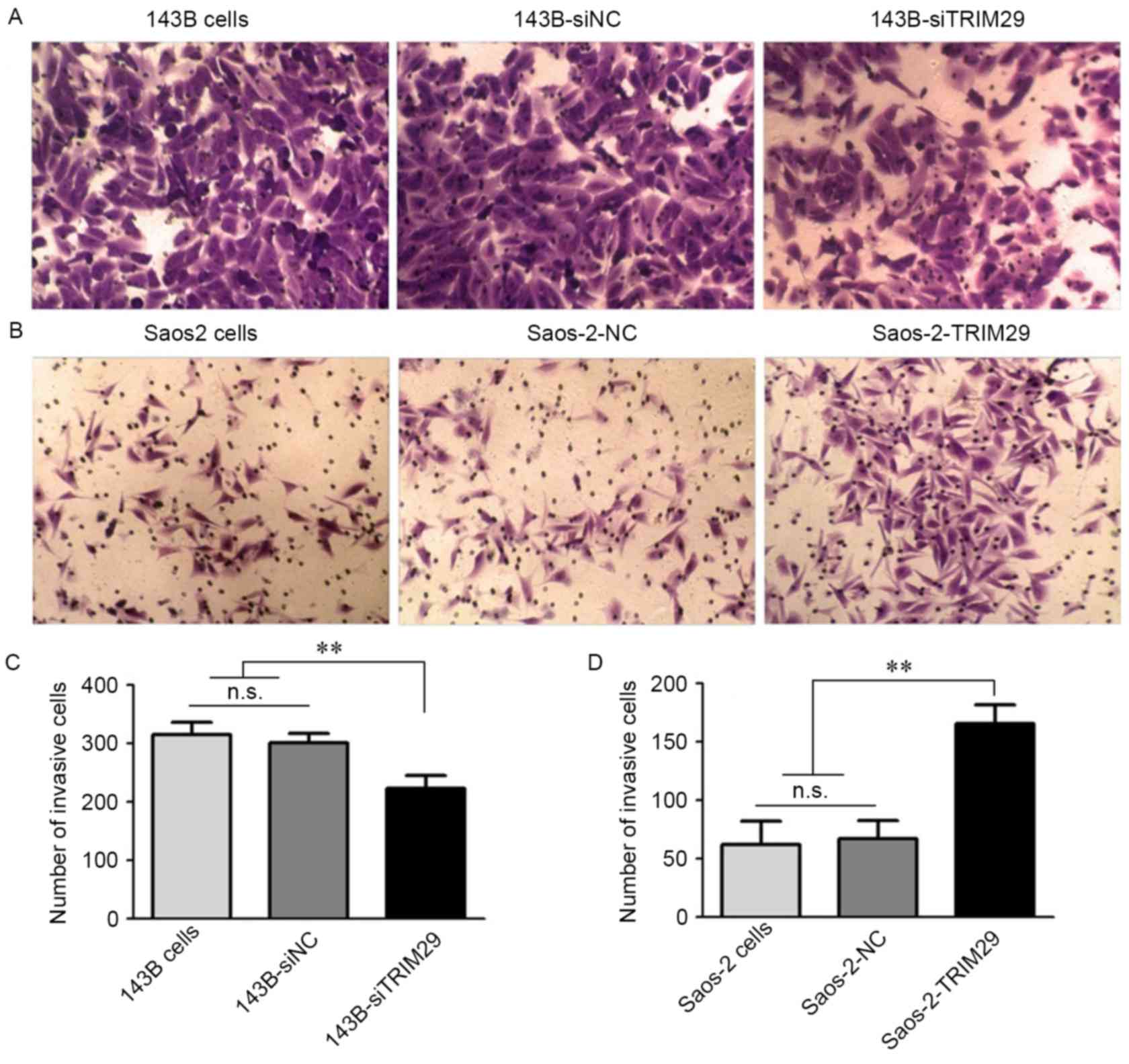
were also performed under serum-free conditions. As shown in
Fig. 5, TRIM29 knockdown in 143B
cells resulted in a significant reduction in invasive growth as
compared to that in the control groups (P=0.000 <0.01, P=0.004
<0.01, respectively). Furthermore, overexpression of TRIM29 in
Saos-2 cells significantly enhanced the invasion ability
(P=0.001<0.01, P=0.000<0.01, respectively). These results
suggest that TRIM29 markedly stimulates human osteosarcoma cell
motility.
TRIM29 promotes the migration and
invasive growth of osteosarcoma cells by inducing EMT
In cancer biology, cellular migration and invasion
properties are frequently correlated with EMT. To explore the
potential mechanisms of TRIM29-related cancer cell motility, we
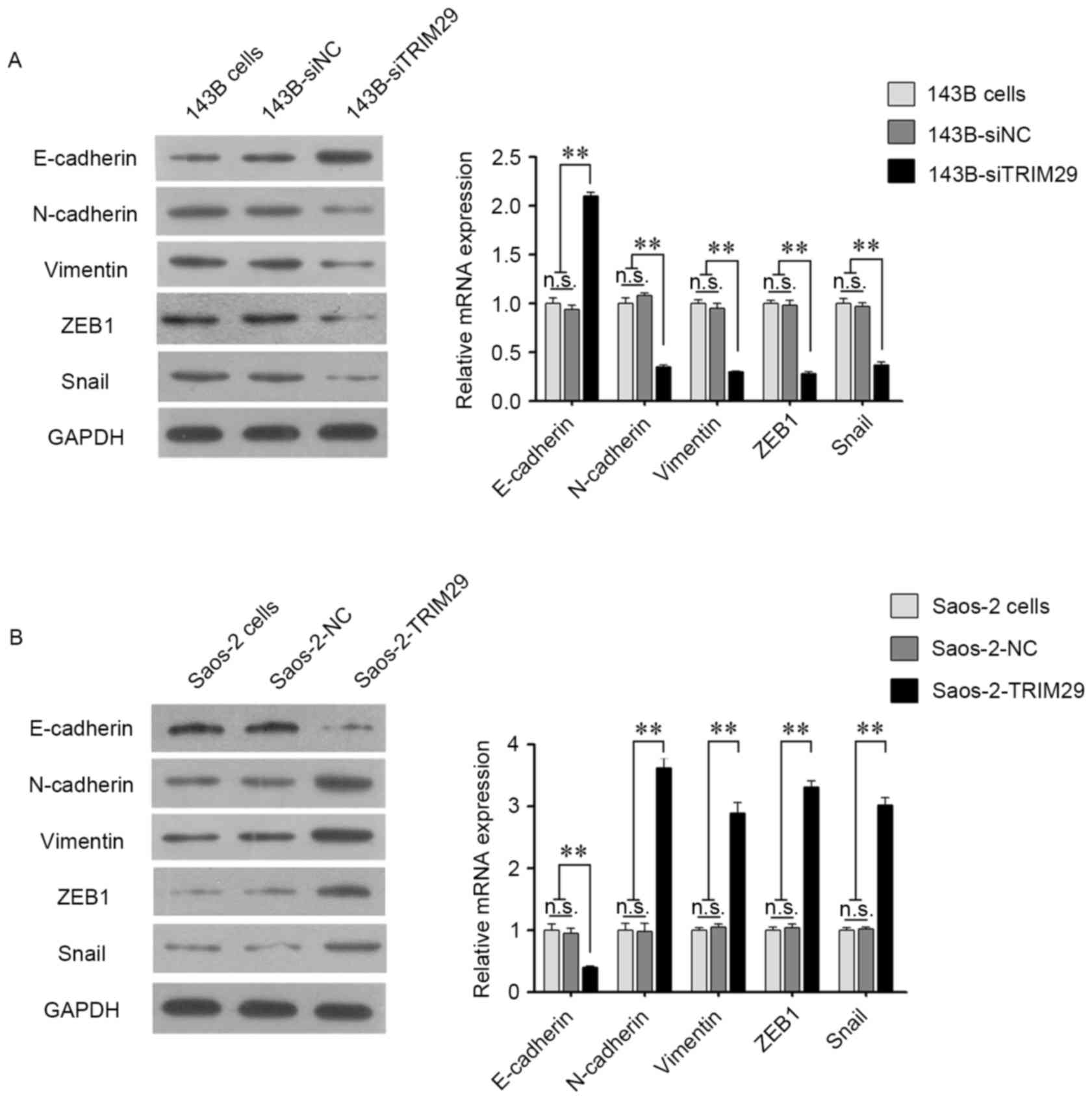
investigated the expression of key markers of EMT. As shown in
Fig. 6, knockdown of TRIM29
resulted in a significant upregulation of the epithelial marker
E-cadherin, and downregulation of vimentin, N-cadherin, ZEB1 and
Snail at both the mRNA and protein levels. Furthermore, opposite
results were observed in the Saos-2 cell line with overexpression
of TRIM29.
Discussion
Osteosarcoma is the most common type of primary bone
cancer with a high rate of mortality and distant metastasis
(30). Its complex biological and
molecular mechanisms make identification of an effective
therapeutic target difficult, hence it is a challenge to improve
clinical outcome of treatment (31). Therefore, it is critical to identify
the metastasis-associated and prognostic biomarkers of
osteosarcoma.
Invasive growth is a hallmark of malignant cancer
cells. It includes translocation of cancer cells from the primary
site into anatomically contiguous tissues as well as to distant
metastatic lesions via blood circulation or lymphatic system
(32). The association of EMT with
cancer metastasis is well-established (33–36).
The essential feature of EMT is the loss of polarized epithelial
traits and acquisition of mesenchymal characteristics, which
contributes to enhanced cell invasion and motility and release of
cells from the primary cancer site. EMT is characterized by the
downregulation of E-cadherin, a protein that provides physical
adhesion to adjacent epithelial cells that is critical for the
maintenance and establishment of the structural integrity and
polarity of epithelium. The transition is also marked by
accumulation of mesenchymal markers, such as N-cadherin and
vimentin (37,38). The process of EMT is governed by a
cohort of transcription factors, such as the ZEB, Snail and Twist
families (37,39,40).
These transcription factors regulate extensive gene expression and
cellular phenotypic switch. As core drivers of EMT, Snail and
zinc-finger E-box binding homeobox 1 (ZEB1) are transcriptional
repressors that directly inhibit the expression of E-cadherin and
several other intercellular adhesion components (39,41).
Given the role of TRIM29 in cancer
progression, it is a promising target gene for cancer therapy.
Recent studies have linked TRIM29 overexpression with
carcinogenesis and cancer progression. Increased TRIM29 expression
was associated with shorter overall survival and disease-free
survival; and was shown to be an independent prognostic factor in
patients with non-small cell lung cancer (19). Similarly, TRIM29 expression showed a
strong correlation with aggressive malignant behavior and was an
independent predictor of poor survival in patients with colorectal
cancer (24). Overexpression of
TRIM29 in gastric cancer cells was associated with histological
differentiation, lymph node metastasis, large tumor size, tumor
invasion and poor prognosis (42).
TRIM29 knockdown in gastric cancer cells downregulated the
Wnt/β-catenin pathway (22). TRIM29
was also found to stabilize β-catenin in pancreatic cancer via
DVL-2, a negative regulator of GSK3β (43). This finding indicates that TRIM29
promotes tumor progression via activation of the Wnt/β-catenin
signaling pathway in pancreatic cancer. Furthermore, TRIM29
promotes cancer cell proliferation by inhibiting the nuclear
activities of p53 (44,45). Furthermore, TRIM29 was shown to bind
with p53 and repress the expression of p53-mediated genes,
including NOXA and p21 (44,46).
Recent studies have shown that TRIM29 is involved in EMT
regulation. Overexpression of TRIM29 was shown to promote EMT,
metastasis and proliferation of nasopharyngeal carcinoma cells via
the PTEN/AKT/mTOR pathway (26). In
mouse and human PanIN lesions, TRIM29 was found to upregulate CD44
via activation of β-catenin signaling, which leads to induction of
the EMT phenotype characterized by expression of ZEB1 and Snail
(47). In cervical cancer cells,
knockdown of TRIM29 was shown to increase E-cadherin expression but
decrease N-cadherin and β-catenin expression, which also indicates
that TRIM29 promotes EMT (48).
The present study focused on TRIM29 and
investigated its potential role in osteosarcoma. To the best of our
knowledge, this is the first research to demonstrate a major role
of TRIM29 in osteosarcoma. The results of IHC showed that
TRIM29 expression in osteosarcoma tissues was significantly higher
than that in the normal bone tissues (P<0.01). Furthermore, we
observed that TRIM29 expression was significantly correlated with
tumor size, recurrence, metastasis, and overall survival time after
surgery, which suggests that TRIM29 may play an important
role in the malignant potential of osteosarcoma. Subsequently, we
determined the expression levels of TRIM29 in human osteoblast and
osteosarcoma cell lines. Results of RT-qPCR and western blot
analysis were consistent with those of immunohistochemical
analysis. Both mRNA and protein expression of TRIM29 was
significantly higher in osteosarcoma cell lines as compared to that
in the osteoblast cell line hFOB1.19. Moreover, in these 4
osteosarcoma cell lines, the expression levels of TRIM29 protein
and mRNA were highest in 143B, and lowest in Saos-2. Since the
invasion and proliferation capability of 143B was much higher than
that of Saos-2, these results showed that high expression of TRIM29
may have an adverse influence on the progression of osteosarcoma.
Then, we found that knockdown of TRIM29 in 143B cells significantly
reduced the migration rate in wound healing assays, and
overexpression of TRIM29 enhanced Saos-2 cell migration. In
accordance with the above data, TRIM29 also correlated well with
the invasive growth of osteosarcoma cells observed in Transwell
assays. Furthermore, we observed a significant upregulation of
E-cadherin and downregulation of vimentin, N-cadherin, ZEB1 and
Snail at both the mRNA and protein levels following knockdown of
TRIM29 in 143B cells; furthermore, overexpression of TRIM29 in
Saos-2 cells had an opposite effect. These results indicate that
TRIM29 promotes osteosarcoma cell migration and invasion via the
EMT process.
In conclusion, we demonstrated significantly higher
expression of TRIM29 in osteosarcoma tissues and cells as compared
to that in normal bone tissues and osteoblast cells. Expression of
TRIM29 was significantly associated with tumor size, recurrence,
metastasis, and overall survival in osteosarcoma patients. In
addition, the TRIM29 expression level was an independent prognostic
factor in patients with osteosarcoma. Moreover, TRIM29 promoted
migration and invasion of osteosarcoma cells by inducing EMT.
TRIM29 may serve as a useful prognostic biomarker in osteosarcoma
patients and a potential therapeutic target for osteosarcoma
treatment. However, the precise molecular mechanisms by which
TRIM29 contributes to tumorigenesis and progression of osteosarcoma
warrant further investigations.
References
|
1
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment - where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aung L, Tin AS, Quah TC and Pho RW:
Osteogenic sarcoma in children and young adults. Ann Acad Med
Singapore. 43:305–313. 2014.PubMed/NCBI
|
|
3
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dobrenkov K and Cheung NK: GD2-targeted
immunotherapy and radioimmunotherapy. Semin Oncol. 41:589–612.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lv YF, Dai H, Yan GN, Meng G, Zhang X and
Guo QN: Downregulation of tumor suppressing STF cDNA 3 promotes
epithelial-mesenchymal transition and tumor metastasis of
osteosarcoma by the Wnt/GSK-3β/β-catenin/Snail signaling pathway.
Cancer Lett. 373:164–173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bacci G, Longhi A, Versari M, Mercuri M,
Briccoli A and Picci P: Prognostic factors for osteosarcoma of the
extremity treated with neoadjuvant chemotherapy: 15-Year experience
in 789 patients treated at a single institution. Cancer.
106:1154–1161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Strauss SJ, Ng T, Mendoza-Naranjo A,
Whelan J and Sorensen PH: Understanding micrometastatic disease and
Anoikis resistance in Ewing family of tumors and
osteosarcoma. Oncologist. 15:627–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bielack S, Jürgens H, Jundt G, Kevric M,
Kühne T, Reichardt P, Zoubek A, Werner M, Winkelmann W and Kotz R:
Osteosarcoma: The COSS experience. Cancer Treat Res. 152:289–308.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moore DD and Luu HH: Osteosarcoma. Cancer
Treat Res. 162:65–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borden KL: RING fingers and B-boxes:
Zinc-binding protein-protein interaction domains. Biochem Cell
Biol. 76:351–358. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reddy BA, Etkin LD and Freemont PS: A
novel zinc finger coiled-coil domain in a family of nuclear
proteins. Trends Biochem Sci. 17:344–345. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reymond A, Meroni G, Fantozzi A, Merla G,
Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et
al: The tripartite motif family identifies cell compartments. EMBO
J. 20:2140–2151. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xing J, Weng L, Yuan B, Wang Z, Jia L, Jin
R, Lu H, Li XC, Liu YJ and Zhang Z: Identification of a role for
TRIM29 in the control of innate immunity in the respiratory tract.
Nat Immunol. 17:1373–1380. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang H, Palmbos PL, Wang L, Kim EH, Ney
GM, Liu C, Prasad J, Misek DE, Yu X, Ljungman M, et al: ATDC
(ataxia telangiectasia group D complementing) promotes
radioresistance through an interaction with the RNF8 ubiquitin
ligase. J Biol Chem. 290:27146–27157. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Napolitano LM and Meroni G: TRIM family:
Pleiotropy and diversification through homomultimer and
heteromultimer formation. IUBMB Life. 64:64–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hatakeyama S: Early evidence for the role
of TRIM29 in multiple cancer models. Expert Opin Ther Targets.
20:767–770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song X, Fu C, Yang X, Sun D, Zhang X and
Zhang J: Tripartite motif-containing 29 as a novel biomarker in
non-small cell lung cancer. Oncol Lett. 10:2283–2288.
2015.PubMed/NCBI
|
|
20
|
Jiang N, Chen WJ, Zhang JW, Xu C, Zeng XC,
Zhang T, Li Y and Wang GY: Downregulation of miR-432 activates
Wnt/β-catenin signaling and promotes human hepatocellular carcinoma
proliferation. Oncotarget. 6:7866–7879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun H, Dai X and Han B: TRIM29 as a novel
biomarker in pancreatic adenocarcinoma. Dis Markers.
2014:3178172014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qiu F, Xiong JP, Deng J and Xiang XJ:
TRIM29 functions as an oncogene in gastric cancer and is regulated
by miR-185. Int J Clin Exp Pathol. 8:5053–5061. 2015.PubMed/NCBI
|
|
23
|
Lai W, Zheng X, Huang Q, Wu X and Yang M:
Down-regulating ATDC inhibits the proliferation of esophageal
carcinoma cells. Eur Rev Med Pharmacol Sci. 18:3511–3516.
2014.PubMed/NCBI
|
|
24
|
Jiang T, Tang HM, Lu S, Yan DW, Yang YX
and Peng ZH: Up-regulation of tripartite motif-containing 29
promotes cancer cell proliferation and predicts poor survival in
colorectal cancer. Med Oncol. 30:7152013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan ST, Liu SY and Wu B: TRIM29
Overexpression promotes proliferation and survival of bladder
cancer cells through NF-κB signaling. Cancer Res Treat.
48:1302–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou XM, Sun R, Luo DH, Sun J, Zhang MY,
Wang MH, Yang Y, Wang HY and Mai SJ: Upregulated TRIM29 promotes
proliferation and metastasis of nasopharyngeal carcinoma via
PTEN/AKT/mTOR signal pathway. Oncotarget. 7:13634–13650. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Santin AD, Zhan F, Bellone S, Palmieri M,
Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, et al:
Gene expression profiles in primary ovarian serous papillary tumors
and normal ovarian epithelium: Identification of candidate
molecular markers for ovarian cancer diagnosis and therapy. Int J
Cancer. 112:14–25. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ai L, Kim WJ, Alpay M, Tang M, Pardo CE,
Hatakeyama S, May WS, Kladde MP, Heldermon CD, Siegel EM, et al:
TRIM29 suppresses TWIST1 and invasive breast cancer behavior.
Cancer Res. 74:4875–4887. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu J, Welm B, Boucher KM, Ebbert MT and
Bernard PS: TRIM29 functions as a tumor suppressor in
nontumorigenic breast cells and invasive ER+ breast
cancer. Am J Pathol. 180:839–847. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anderson ME: Update on Survival in
Osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou W, Hao M, Du X, Chen K, Wang G and
Yang J: Advances in targeted therapy for osteosarcoma. Discov Med.
17:301–307. 2014.PubMed/NCBI
|
|
32
|
Guarino M, Rubino B and Ballabio G: The
role of epithelial-mesenchymal transition in cancer pathology.
Pathology. 39:305–318. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hou CH, Lin FL, Hou SM and Liu JF: Cyr61
promotes epithelial-mesenchymal transition and tumor metastasis of
osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol
Cancer. 13:2362014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang L, Wu RL and Xu AM:
Epithelial-mesenchymal transition in gastric cancer. Am J Transl
Res. 7:2141–2158. 2015.PubMed/NCBI
|
|
36
|
Park SY, Korm S, Chung HJ, Choi SJ, Jang
JJ, Cho S, Lim YT, Kim H and Lee JY: RAP80 regulates
epithelial-mesenchymal transition related with metastasis and
malignancy of cancer. Cancer Sci. 107:267–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nantajit D, Lin D and Li JJ: The network
of epithelial-mesenchymal transition: Potential new targets for
tumor resistance. J Cancer Res Clin Oncol. 141:1697–1713. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Palma CS, Grassi ML, Thomé CH, Ferreira
GA, Albuquerque D, Pinto MT, Melo FU Ferreira, Kashima S, Covas DT,
Pitteri SJ, et al: Proteomic analysis of epithelial to mesenchymal
transition (EMT) reveals cross-talk between SNAIL and HDAC1
proteins in breast cancer cells. Mol Cell Proteomics. 15:906–917.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kosaka Y, Inoue H, Ohmachi T, Yokoe T,
Matsumoto T, Mimori K, Tanaka F, Watanabe M and Mori M: Tripartite
motif-containing 29 (TRIM29) is a novel marker for lymph node
metastasis in gastric cancer. Ann Surg Oncol. 14:2543–2549. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang L, Heidt DG, Lee CJ, Yang H, Logsdon
CD, Zhang L, Fearon ER, Ljungman M and Simeone DM: Oncogenic
function of ATDC in pancreatic cancer through Wnt pathway
activation and beta-catenin stabilization. Cancer Cell. 15:207–219.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuan Z, Villagra A, Peng L, Coppola D,
Glozak M, Sotomayor EM, Chen J, Lane WS and Seto E: The ATDC
(TRIM29) protein binds p53 and antagonizes p53-mediated functions.
Mol Cell Biol. 30:3004–3015. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sho T, Tsukiyama T, Sato T, Kondo T, Cheng
J, Saku T, Asaka M and Hatakeyama S: TRIM29 negatively regulates
p53 via inhibition of Tip60. Biochim Biophys Acta. 1813:1245–1253.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shibata MA, Yoshidome K, Shibata E, Jorcyk
CL and Green JE: Suppression of mammary carcinoma growth in vitro
and in vivo by inducible expression of the Cdk inhibitor p21.
Cancer Gene Ther. 8:23–35. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang L, Yang H, Abel EV, Ney GM, Palmbos
PL, Bednar F, Zhang Y, Leflein J, Waghray M, Owens S, et al: ATDC
induces an invasive switch in KRAS-induced pancreatic
tumorigenesis. Genes Dev. 29:171–183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu R, Hu J, Zhang T, Jiang C and Wang HY:
TRIM29 overexpression is associated with poor prognosis and
promotes tumor progression by activating Wnt/β-catenin pathway in
cervical cancer. Oncotarget. 7:28579–28591. 2016. View Article : Google Scholar : PubMed/NCBI
|