Introduction
Hepatocellular carcinoma (HCC) is the second leading
cause of cancer-related death among males worldwide as well as in
less developed countries. It is estimated that ~782,500 new liver
cancer cases and 745,500 deaths occurred worldwide during 2012,
with China alone accounting for ~50% of the total number of cases
and deaths (1). Despite numerous
studies on multiple genes of HCC and advancements in the survival
rate of HCC patients over the past decades (2), the exact molecular mechanisms
underlying cancer initiation and progression are still unclear.
Therefore, identification of novel HCC-associated molecules may
provide insight into understanding the mechanism of
hepatocarcinogenesis.
The zinc-finger family of proteins is one of the
most common families of transcription factors in eukaryotic cells
and has more than 3,000 members in the human genome (3,4).
Approximately one-third of zinc-finger proteins contain the
Krüppel-associated box (KRAB-ZNFs) and 51% of KRAB-ZNFs are located
on chromosome 19q13 (5). The
functions of zinc-finger proteins are of great diversity, including
DNA recognition, RNA packaging, transcriptional activation and
regulation of apoptosis (6). Due to
their multiple functions, several zinc-finger proteins have been
identified as tumor-suppressors or oncogenes in the development of
tumors. For example, zinc-finger protein ZBTB20 has been
demonstrated to be a potential oncogene in non-small cell lung
cancer (NSCLC) and HCC (7,8). In addition, zinc-finger protein
X-linked (ZFX) has been identified as an oncogene in colorectal
cancer and is associated with poor prognosis (9). In contrast, ZNF382, ZNF569, ZNF331 and
ZNF545 have been identified as tumor-suppressors (10–13).
However, the role of most zinc-finger family members in tumor
development remains ambiguous including HCC (14). Therefore, identification of novel
zinc-finger proteins associated with tumor development may
contribute to the understanding of the mechanisms of HCC.
Zinc finger protein 307 (ZNF307), also known as
ZKSCAN4 (zinc-finger with KRAB and SCAN domains 4), ZSCAN36 and
ZNF427, cloned from human embryonic heart cDNA, is a zinc-finger
protein gene consisting of a Krüppel-associated box-A box, a SCAN
and a zinc-finger domains with seven Cys2His2. Initial study
demonstrated that in HEK-293 cells ZNF307 suppressed the
transcriptional activity of p53 and p21 by upregulating mRNA levels
of p53-binding protein MDM2 (MDM2) and E1A binding protein p300
(EP300) (15). However, Ecker et
al reported that ZNF307 was co-precipitated and interacted with
the glucocorticoid receptor in HEK293 cells (16). In addition, recent studies indicated
that ZNF307 was one of two susceptibility loci for schizophrenia at
6p21-p22.1 by a genome-wide association study (17). Later, ZNF307 was also identified as
one of nine risk SNPs distilled from genome-wide association
studies for schizophrenia (18).
However, the role of ZNF307 in tumor development and progression is
largely unknown.
In the present study, we detected the expression
level of ZNF307 in HCC and investigated its biological functions
and mechanisms in human HCC cell lines.
Materials and methods
Tissue samples
Thirty-three paired HCC and adjacent non-tumor
tissues (>2 cm distance from the margin of resection) were
randomly collected from patients who had not been pretreated with
radiotherapy or chemotherapy prior to hepatectomy at Nanfang
Hospital, Southern Medical University from April 2006 to Dec 2008
after obtaining informed consent. Specimens were frozen at −80°C
for RNA and protein isolation. The protocol of the present study
was approved by the Southern Medical University Ethics
Committee.
Cells and cell culture
L02, MHCC97L, HCCLM3, Huh7, QGY7701, QGY7703,
Bel7402 and Bel7404 cell lines were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA) or the Cell Bank of
the Chinese Academy of Sciences (Shanghai, China). All cells were
cultured in high glucose Dulbecco's modified Eagle's medium (DMEM)
(Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum
(Biological Industries, Beit HaEmek, Israel) at 37°C in a
humidified 5% CO2 atmosphere.
RNA extraction and reverse
transcription PCR
Total RNA from tissues or cells was extracted using
TRIzol reagent (Takara, Dalian, China), according to the
manufacturer's protocol. cDNA was synthesized from 1 µg of total
RNA using a reverse transcriptase cDNA synthesis kit (Takara).
Real-time PCR amplification was performed on the ABI 7500 Real-Time
PCR System (Applied Biosystems, Foster City, CA, USA), using a
SYBR-Green PCR kit (Takara). In brief, the reaction system (total
volume 20 µl) containing 500 ng cDNA, the forward primer,
5′-TCTCCCTTGGTGGTGAAATACA-3′ and the reverse primer,
5′-TTCCCTCAGGTGGCATATCTT-3′, was used to amplify a 98-bp PCR
product for human ZNF307 (NCBI; gene ID, 387032). The cycling
parameters were 94°C for 35 sec, 55°C for 40 sec and 72°C for 34
sec for 40 cycles. Real-time PCR was also carried out using the
primers for GAPDH to normalize each of the extracts for ZNF307. The
results are expressed as the ratio of copies of target genes to
GAPDH. Each experiment was repeated independently at least three
times. Relative expression levels of genes were calculated and
expressed as 2−ΔΔCt.
Western blotting
Cells and tissues were lysed on ice in RIPA buffer,
sonicated and protein concentrations were calculated using a
bicinchoninic acid (BCA) kit (both from KeyGen Biotech Co., Ltd.,
Nanjing, China). A total of 50 mg of protein lysates was separated
by sodium dodecyl sulphate-polyacrylamide gel electrophoresis
(SDS-PAGE), and then transferred onto a polyvinylidene difluoride
(PVDF) membrane (Millipore, Bedford, MA, USA) and blocked in 5%
non-fat dry milk in Tris-buffered saline, pH 7.5 (100 mM NaCl, 50
mM Tris and 0.1% Tween-20). Membranes were incubated with
anti-ZNF307 polyclonal antibody (LifeSpan BioSciences, Seattle, WA,
USA) and anti-GAPDH antibody (Proteintech, Wuhan, China) at 4°C
overnight, and then their respective secondary antibody. Signals
were detected by enhanced chemiluminescence (Fdbio Science,
Hangzhou, China).
Establishment of ZNF307-overexpressing
cells
A lentivirus containing the ZNF307 cDNA and a
lentivirus only-containing pCDF1-MCS2-EF1-copGFP mock vector were
purchased from GenePharma (Shanghai, China). Bel7402 and HCCLM3
cells were seeded into 24-well plates to 30–40% confluence and
transfected with the viral supernatant containing exogenous ZNF307
cDNA and negative control lentivirus, respectively. After 24 h of
incubation, the medium was replaced with fresh 10% fetal bovine
serum (FBS). The expression of report gene copGFP was examined
after 3–4 days of transfection. These two groups of cells were
selected using 5 µg/µl puromycin (MP Biomedicals, Santa Ana, CA,
USA) for two weeks. Then, overexpression of ZNF307 was confirmed by
real-time PCR and western blotting.
Establishment of ZNF307-knockdown
cells
ZNF307 short hairpin RNAs (shRNAs)
(5′-GGTTTCACTCGAACTTCAT-3′) and negative control shRNA
(5′-TTCTCCGAACGTGTCACGTTTC-3′) were designed and synthesized by
GenePharma. MHCC97L and QGY7701 cells were seeded into 24-well
plates to 30–40% confluence, and transfected with the viral
supernatant containing exogenous ZNF307 shRNA and negative control
lentivirus, respectively. After 24 h of incubation, the medium was
replaced with fresh 10% FBS. The expression of report gene copGFP
was examined after 3–4 days transfection. These two groups of cells
were selected using 5 µg/µl puromycin (MP Biomedicals) for two
weeks. Then, knockdown efficiency was evaluated after selection by
real-time PCR and western blotting.
Colony-formation assay
Target HCC cells, as well as their control cells,
were added to different wells of a 6-well culture plate at a final
density of 1×102 cells. After culturing for 2 weeks at
37°C, the cells were washed twice with phosphate-buffered saline
(PBS), fixed with methanol and stained with Giemsa solution.
Colonies with >50 cells/colony were counted. The experiment was
repeated three times independently and results are shown as the
means ± SEM.
Cell proliferation assay
Target HCC cells, as well as their control cells,
were re-plated in 96-well plates at a final density of
1×103 cells. Proliferation was measured using the Cell
Counting Kit-8 (CCK-8; Dojindo, Shanghai, China) according to the
manufacturer's instructions from day 1 to 7. The experiment was
repeated three times independently and the results are shown as the
means ± SEM.
Wound healing and invasion assays
Cell migration ability was assessed using wound
healing assays. When the cells grew to 80–90% confluence in 6-well
culture plates, three scratch wounds across each well were made
using a 10-µl pipette tip. The phase contrast images of the wounds
were recorded at 0, 24 and 48 h, respectively. Cells transfected
with the empty vector served as the controls. Invasion assay was
performed using a Matrigel invasion chamber (Falcon, Corning Inc.,
Corning, NY, USA) according to the manufacturer's instructions. A
total of 1×105 cells suspended in a serum-free medium
were then added to the upper chamber, while 10% FBS was added in
the lower chamber as chemoattractant. After 24 h of incubation,
noninvasive cells remaining in the upper chamber were removed by
cotton swabs and the lower surface of membranes was stained with
Giemsa solution. The permeating cells were counted under a inverted
microscope and photographed in at least six random microscopic
fields. The experiment was repeated three times independently and
results are shown as the means ± SEM.
In vivo tumorigenicity
Four- to six-week-old female BALB/c nude mice were
purchased from the Central Laboratory of Animal Science at Southern
Medical University (Guangzhou, China) and maintained in
laminar-flow cabinets under specific pathogen-free conditions. A
total of 1×107 targeted cells in 200 µl PBS were
subcutaneously injected into both flanks of nude mice. Tumor size
was measured by calipers every 5 days, up to 25 days after
injection. Tumor volume was calculated using the equation: V =
[length × (width)2]/2. The experiments were performed in
seven mice in each treatment group.
Apoptosis analysis
Apoptosis assay was performed according to the
manual of the Annexin V-FITC/propidium iodide (PI) apoptosis
detection kit (KeyGen Biotech Co., Ltd.). Briefly, the harvested
cells were washed twice with PBS, and then resuspended in binding
buffer. Then, these cells were stained with Annexin V-FITC and PI
at room temperature in the dark for 15 min. In addition, these
stained cells were analyzed using flow cytometry as soon as
possible (within 1 h). The experiment was repeated three times
independently and results are shown as the means ± SEM.
Statistical analysis
All measurements or variables are shown as means ±
SEM. Real-time PCR, western blotting, clone formation, wound
healing, invasion and apoptosis assays were examined using the
Student's t-test. To test statistical differences between in
vitro cell proliferation and in vivo tumorigenicity,
multiple-factor repetitive measurement and analysis of variance
were performed. A difference was considered statistically
significant at P-value <0.05. All statistical analyses were
conducted using SPSS software (version 20; SPSS, Inc., Chicago, IL,
USA).
Results
Downregulation of expression levels of
ZNF307 mRNA and protein in HCC
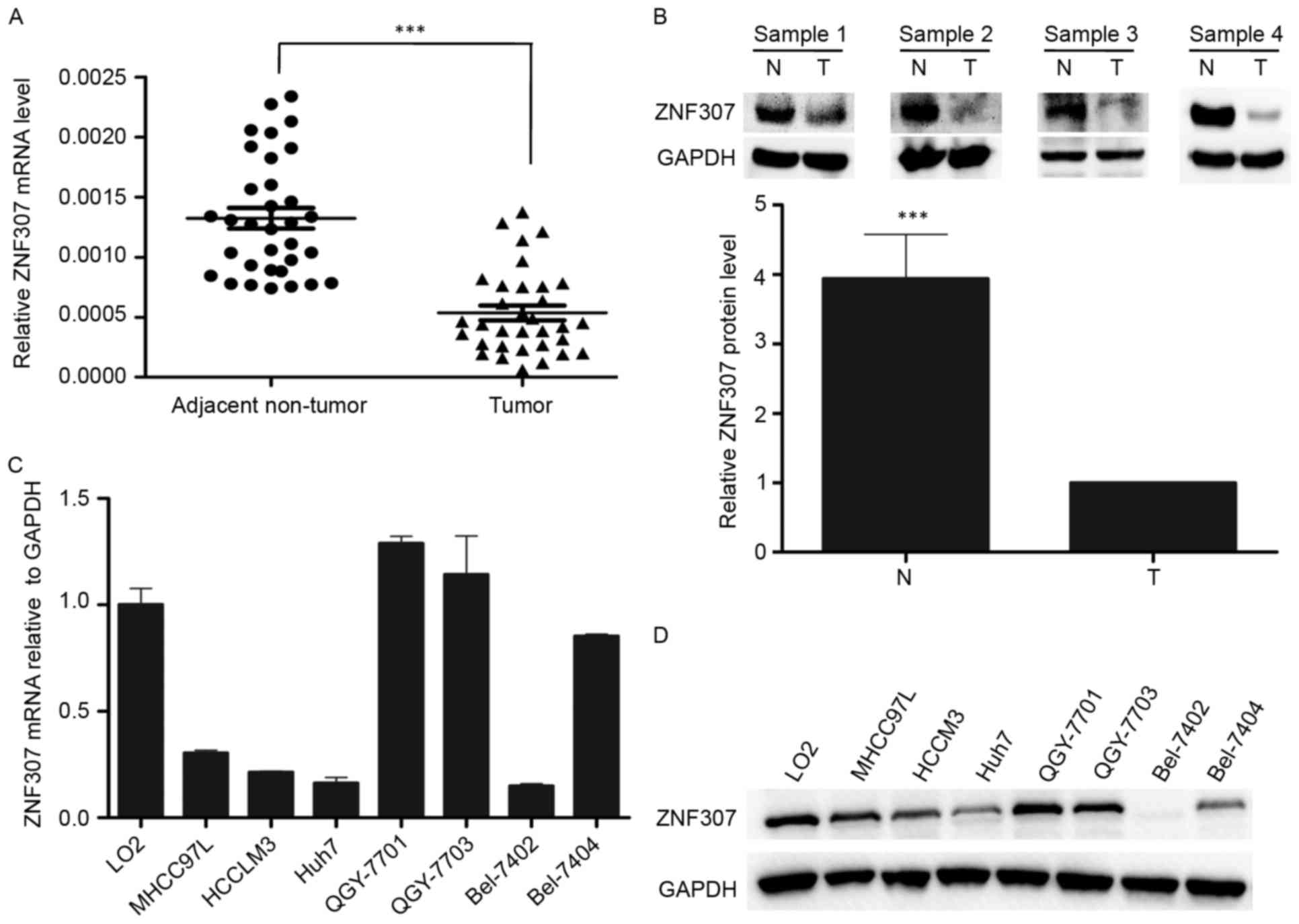
Firstly, ZNF307 mRNA levels were measured in
33-paired HCC and adjacent non-tumor tissues by real-time PCR. The
result revealed that ZNF307 mRNA levels were significantly
downregulated in the HCC tissues (P<0.001; Fig. 1A). In addition, western blot
analysis of 20-paired HCC tissues also confirmed the decreased
protein abundance of ZNF307 in HCC tissues (P<0.001; Fig. 1B). Moreover, we detected the ZNF307
mRNA expression in one human normal liver cell line (L02) and seven
HCC cell lines (MHCC97L, HCCLM3, Huh7, QGY7701, QGY7703, Bel7402
and Bel7404). The results showed that ZNF307 was significantly
downregulated in most of the HCC cell lines (Fig. 1C). The western blot analysis had a
result similar to that of real-time PCR (Fig. 1D).
Alteration of ZNF307 expression
regulates proliferation, migration, invasion and apoptosis of HCC
cell lines in vitro
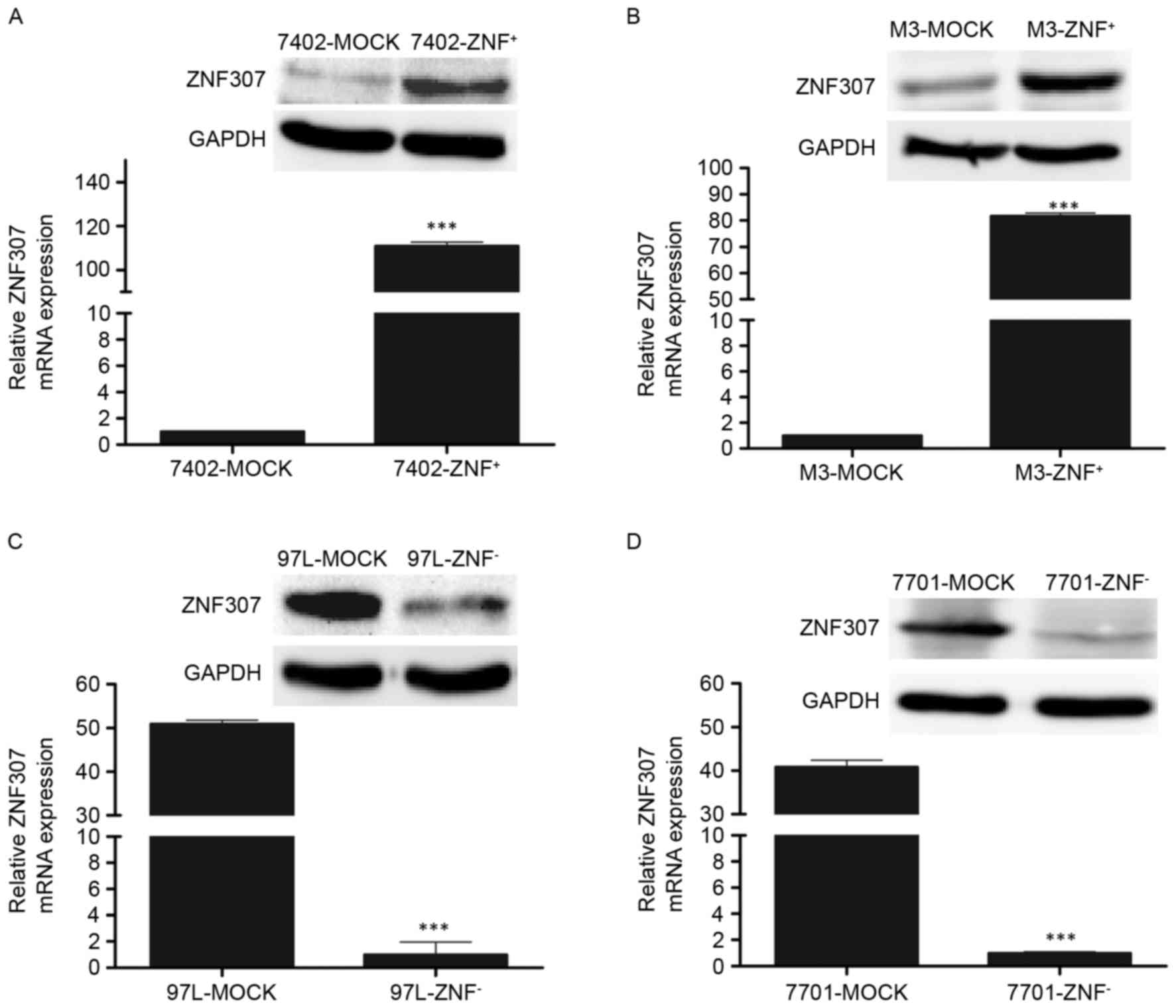
We successfully constructed two stable clones
overexpressing ZNF307 from parental Bel7402 and HCCLM3 cells.
Expression of ZNF307 after stable transfection was measured by
RT-PCR and western blotting (P<0.001; Fig. 2A and B). First, we found that
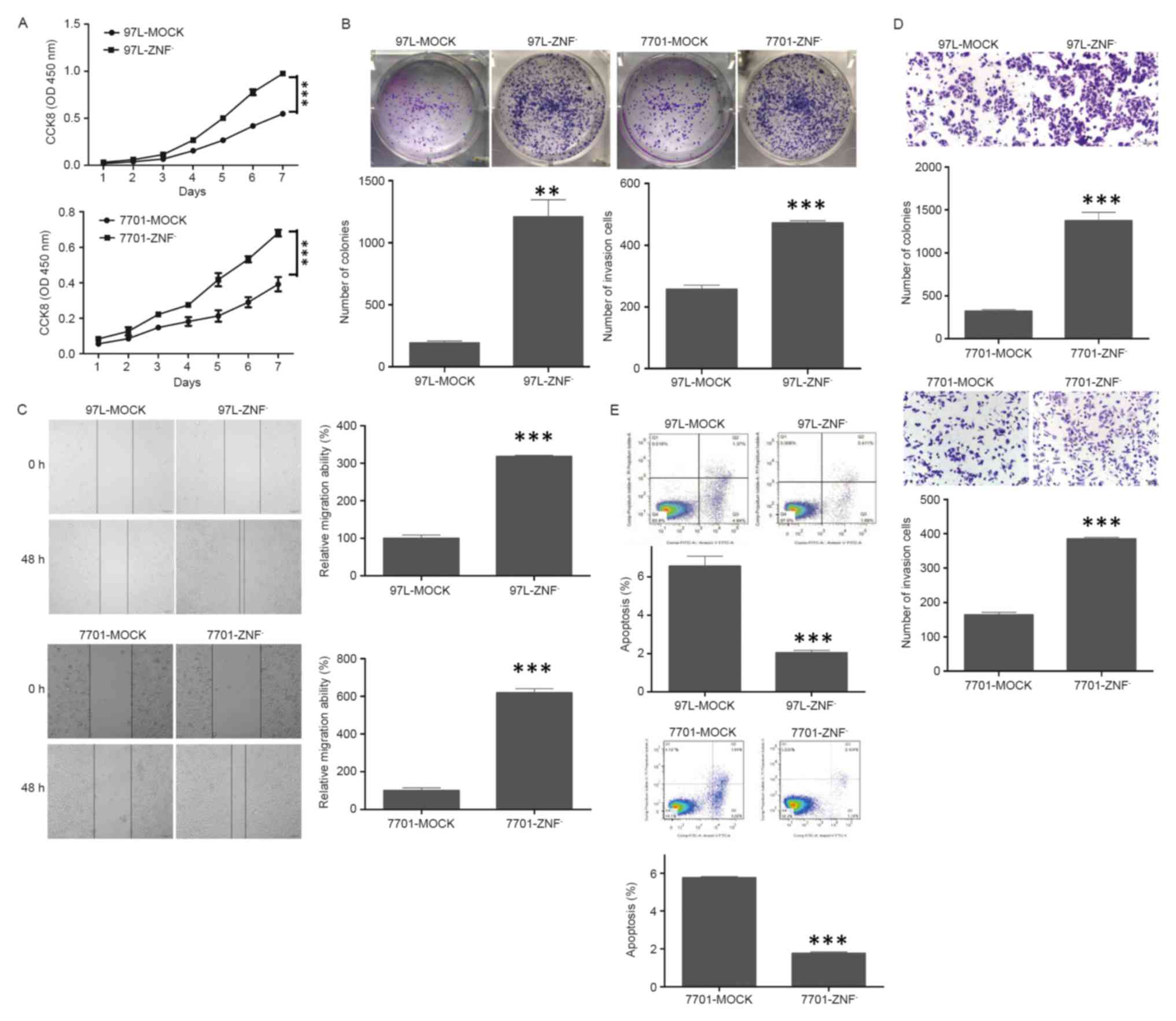
expression of ZNF307 suppressed the growth of both Bel7402 and
HCCLM3 cell lines by CCK-8 assay (P<0.001; Fig. 3A). This was confirmed by colony
formation assay. The colonies formed by Bel7402-ZNF307+
(P<0.001) and HCCLM3-ZNF307+ cells (P<0.01) were
significantly less and smaller than the control cells (Fig. 3B). In addition, ectopic expression
of ZNF307 markedly slowed cell migration at the edges of the
scratch wound in the Bel7402 and HCCLM3 cells. Quantitative
analyses at 48 h confirmed a significant reduction in wound closure
in the Bel7402-ZNF307+ (P<0.001) and
HCCLM3-ZNF307+ cells (P<0.01), compared with the
controls cells (Fig. 3C).
Furthermore, in the Matrigel invasion assay, the number of cells
that passed through the Matrigel in the Bel7402-ZNF307+
(P<0.01) and HCCLM3-ZNF307+ cells (P<0.001) was
significantly less than in the mock cells (Fig. 3D). Moreover,
Bel7402-ZNF307+ and HCCLM3-ZNF307+ cells had
an increased early and late apoptotic population compared to the
control group (P<0.001; Fig.
3E).
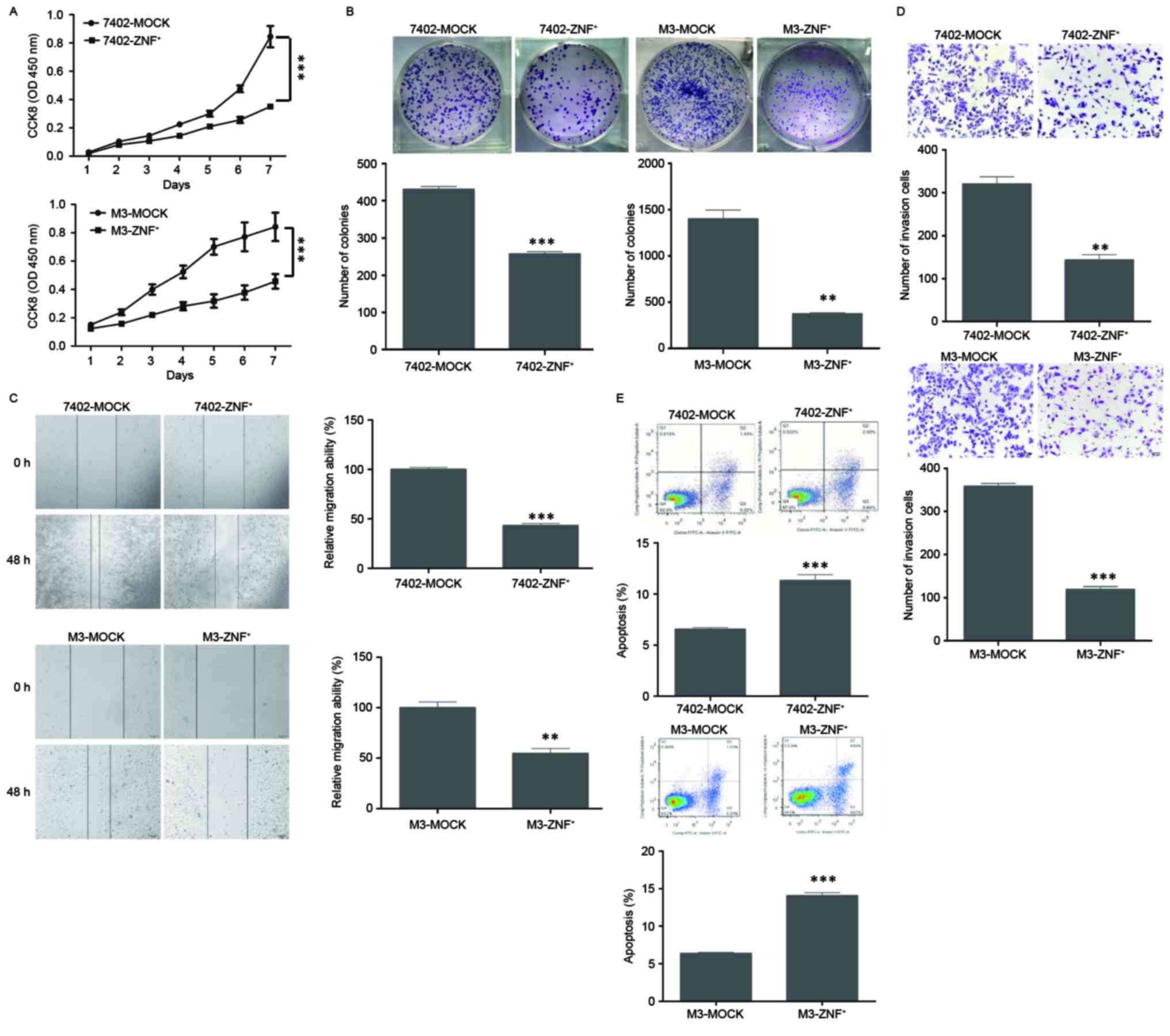 | Figure 3.Effect of ZNF307 overexpression on
cell growth, colony formation, migration, invasion and apoptosis in
HCC cell lines. (A) ZNF307 overexpression inhibited the cell
proliferation of Bel7402 (7402-ZNF+) and HCCM3
(M3-ZNF+) cell lines, as assessed by CCK-8 assay. (B)
ZNF307 overexpression suppressed the ability of colony formation in
the 7402-ZNF+ and M3-ZNF+ cells, as analyzed
by colony formation assay. (C) ZNF307 overexpression reduced the
ability of migration in the 7402-ZNF+ and
M3-ZNF+ cells, as assessed by wound-healing assay. (D)
ZNF307 overexpression decreased the number of invaded cells in the
7402-ZNF+ and M3-ZNF+ cells, as assessed by
Matrigel invasion assay. (E) ZNF307 overexpression increased the
incidence of apoptosis in the 7402-ZNF+ and
M3-ZNF+ cells, as assessed by flow cytometric apoptosis
assay. Data represent the mean ± SEM of three independent
experiments: **P<0.01, ***P<0.001, in comparison with the
control (MOCK). |
In contrast, the endogenous ZNF307 level in MHCC97L
and QGY7701 cells was stably knocked down by transfection of shRNA
(P<0.001; Fig. 2C and D).
Contrary to the effect of ZNF307 overexpression, downregulation of
endogenous ZNF307 in the MHCC97L and QGY7701 cells promoted the
growth and colony formation compared with their controls
(P<0.01; Fig. 4A and B).
Furthermore, an evident acceleration in the wound closure rate and
enhanced invasiveness were observed in the
MHCC97L-ZNF307− and QGY7701-ZNF307− cells
compared with their controls (P<0.001; Fig. 4C and D).
However, the proportion of early and late apoptotic
cells was significantly decreased in the MHCC97L-ZNF307−
and QGY7701-ZNF307− cells compared to the control group
(P<0.001; Fig. 4E).
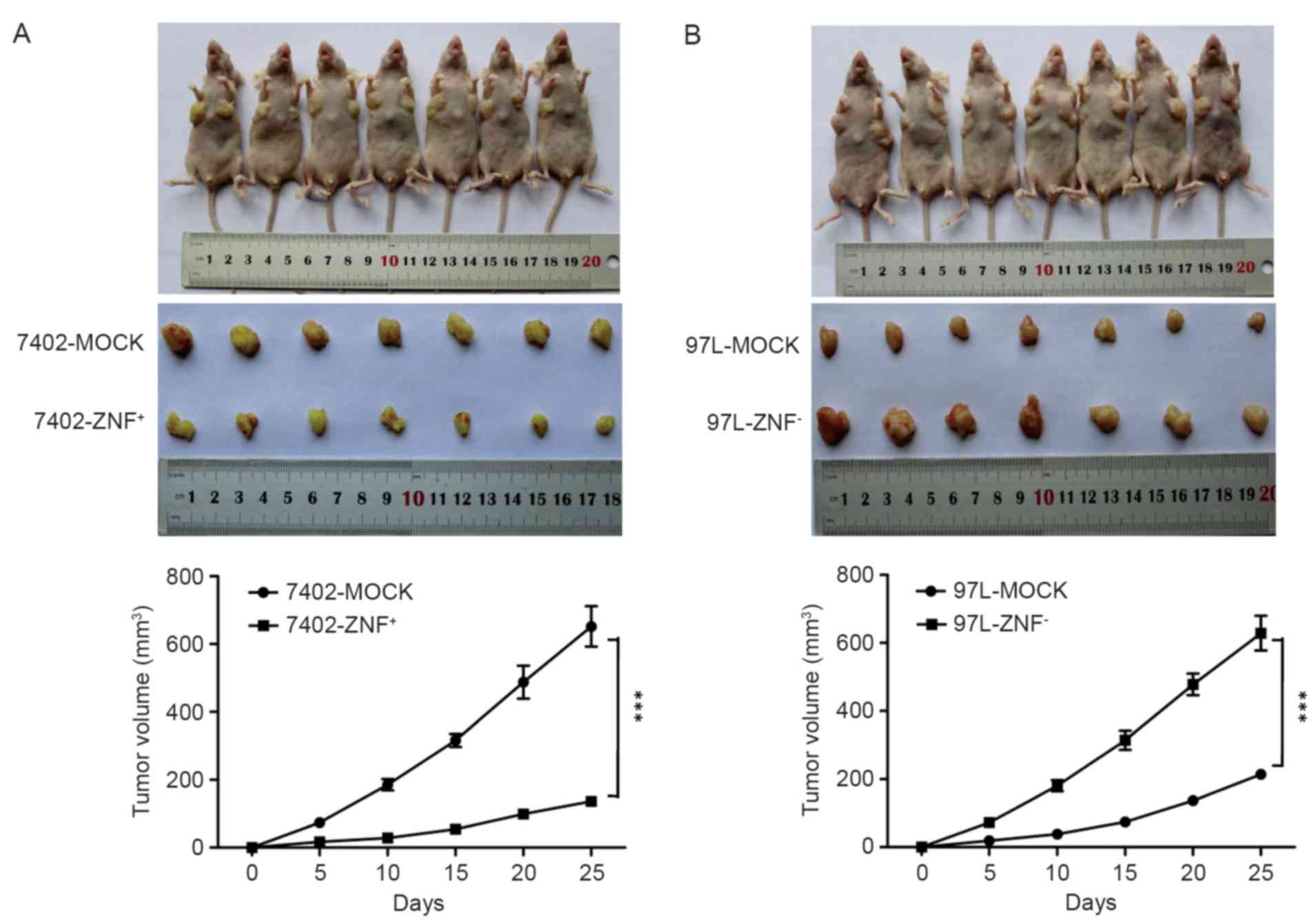
ZNF307 suppresses tumorigenicity in
vivo
The effect of ZNF307 on in vivo
tumorigenicity was investigated using a xenograft model in which
nude mice were subcutaneously injected with
Bel7402-ZNF+, MHCC97L-ZNF− and their
controls. We found that the volume of the tumors harvested from
Bel7402-ZNF+ cells was much smaller than that from the
control by day 25 after injection (P<0.001; Fig. 5A). In contrast, injection of
MHCC97L-ZNF− cells led to obvious larger tumor volumes
by day 25, compared with injection of MHCC97L-MOCK cells
(P<0.001; Fig. 5B).
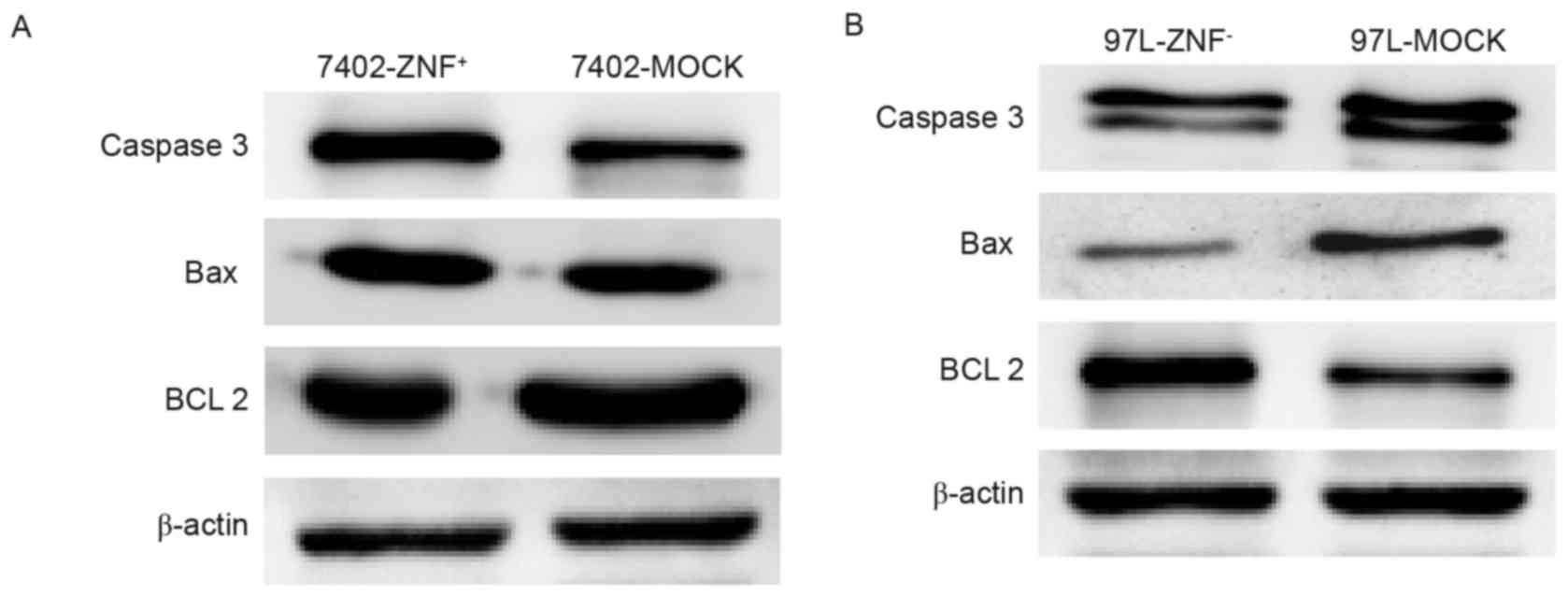
ZNF307 increases the expression of
caspase-3 and Bax, and decreases the expression of Bcl-2
We examined potential downstream target genes of
ZNF307 in Bel7402-ZNF+ and MHCC97L-ZNF− cells
using western blotting. Overexpression of ZNF307 led to increased
expression of caspase-3 and Bax while the expression of Bcl-2 was
significantly decreased (Fig. 6A).
In addition, the expression of Bcl-2 was upregulated and the
expression of caspase-3 and Bax were downregulated in the
MHCC97L-ZNF− cells, compared with the controls (Fig. 6B).
Discussion
Hepatocellular carcinoma (HCC) is the predominant
primary liver cancer worldwide and the incidence of HCC has
remained stable in the Asia-Pacific region except for Singapore and
Hong Kong over the past two decades (19). Thus, searching for new
HCC-associated molecules may provide clues to the study of the
mechanism of HCC.
Zinc-finger proteins, the largest transcription
factor family, are only present in tetrapod vertebrate genomes
(3,4). They are known to play a key role in
regulating expression of genes important for proliferation,
differentiation, apoptosis and tumorigenesis (5,6).
Several zinc-finger proteins, such as ZNF382, ZNF569, ZNF331 and
ZNF545, have been identified as tumor-suppressors, whereas others,
such as ZBTB20 and ZFX serve as oncogenic proteins (7–13).
ZNF307 is a novel zinc-finger protein and contains a
Krüppel-associated box-A box, a SCAN and zinc-finger domains with
seven Cys2His2. In HEK-293 cells, ZNF307 inhibits the
transcriptional activity of p53 and p21 and interacts with the
glucocorticoid receptor (15,16).
Recent two studies have demonstrated that ZNF307 is the
susceptibility loci for schizophrenia by genome-wide association
study (17,18). Therefore, the role of ZNF307 in the
development and progression of HCC is largely unknown.
In the present study, we examined the ZNF307 mRNA
levels in 33-paired HCC and adjacent non-tumor tissues. The results
revealed that ZNF307 mRNA levels were significantly downregulated
in the HCC tissues. In addition, we confirmed that ZNF307 protein
was expressed at a significantly lower level in the HCC tissues
compared with that in the adjacent non-tumor tissues in 20-paired
samples. Furthermore, we found that ZNF307 expression was
significantly higher in the L02 cell line than that in seven HCC
cell lines both at the mRNA and protein level. These results
suggest that the downregulation of ZNF307 expression may contribute
to HCC carcinogenesis.
In order to identify the function of ZNF307, we
constructed four target cell lines: Bel7402-ZNF307+,
HCCLM3-ZNF307+, MHCC97L-ZNF− and
QGY7701-ZNF− using a retroviral gene transfer method. We
observed an association between the upregulation of ZNF307 and
suppression of cell proliferation, migration and invasion in
vitro, as well as an inverse association following knockdown of
ZNF307. In addition, upregulation of ZNF307 expression in Bel7402
cells was related to suppression of tumorigenicity in mice.
Concordantly, downregulation of ZNF307 expression in MHCC97L cells
led to an enhancement of tumorigenicity in vivo.
Importantly, flow cytometric analysis showed that ZNF307
overexpression increased the incidence of apoptosis, while ZNF307
knockdown decreased the incidence of apoptosis.
Given the flow cytometric apoptosis results, we
examined the expression of apoptosis-related proteins in
Bel7402-ZNF+ and MHCC97L-ZNF− cells by
western blotting. We found that ZNF307 increased the transcription
of Bax and caspase-3, and decreased the transcription of Bcl-2.
Therefore, we confirmed that the suppression of HCC cell growth by
ZNF307 was mediated by induction of apoptosis and by regulation of
apoptosis-related proteins.
Unfortunately, we failed to examine the ZNF307
expression level of HCC tissues by immunohistochemistry. It is
likely that the ZNF307 protein expression level was too low to be
tested.
As a zinc-finger protein, ZNF307 may serve as a
transcriptional repressor in HCC. However, due to the absence of
specific chip-grade antibodies against ZNF307, the DNA sequences
binding ZNF307 under a biological state could not be analyzed by
ChIP or ChIP-seq. Systematic approaches (gene expression array or
ChIP-seq) may be useful to study the function of ZNF307 and may
reveal additional targets in the future.
According to the above mentioned results, ZNF307 may
be a tumor-suppressor in HCC. However, Li et al found that
in HEK-293 cells, ZNF307 suppressed the transcriptional activity of
p53 and p21, which indicates that ZNF307 may enhance tumorigenesis.
It is likely that ZNF307 may act as a tumor-suppressor or an
oncogene in different systems. Several genes have been reported to
function as a a tumor-suppressor or oncogene in different tumors.
For example, miR-506 has been demonstrated to play a role as an
oncogene in tumorigenesis and a tumor-suppressor in the progression
in pancreatic ductal adenocarcinoma (20). Lo et al found that FOXF2 has
a complex role in breast cancer and functions as either a
tumor-suppressor or an oncogene depending on the breast tumor
subtype (21). E2F1, a critical
tumor-suppressor molecule in cell cycle progression and induction
of apoptosis, has been identified to enhance invasion and
metastasis by activating growth receptor signaling pathways
(22). Therefore, all these
attempts addressing the function of ZNF307 in carcinomas require
further in-depth research in the future.
In the present study, we, for the first time,
demonstrated that ZNF307 functions as a tumor-suppressor in HCC by
inhibiting cell proliferation, migration and invasion, and it
induced apoptosis by regulating various genes (caspase-3, Bax and
Bcl-2). ZNF307 may serve as a potential tumor marker for HCC.
Acknowledgements
Grant sponsors of the present study were the Pearl
River S&T Nova Program of Guangzhou (grant no. 2014J2200015),
Guangdong Natural Science Funds for Distinguished Young Scholars
(grant no. 2015A030306015), the Excellent Young Teachers Program of
Higher Education of Guangdong Province (grant no. YQ2015036), the
Guangzhou Program for Support of Top-Notch Young Professionals
(grant no. 2015TQ01R279) and the National Natural Science
Foundation of China (grant no. 81672992).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanda M, Sugimoto H and Kodera Y: Genetic
and epigenetic aspects of initiation and progression of
hepatocellular carcinoma. World J Gastroenterol. 21:10584–10597.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Papavassiliou AG: Transcription factors:
Structure, function, and implication in malignant growth.
Anticancer Res. 15:891–894. 1995.PubMed/NCBI
|
|
4
|
Ladomery M and Dellaire G: Multifunctional
zinc finger proteins in development and disease. Ann Hum Genet.
66:331–342. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Urrutia R: KRAB-containing zinc-finger
repressor proteins. Genome Biol. 4:2312003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laity JH, Lee BM and Wright PE: Zinc
finger proteins: New insights into structural and functional
diversity. Curr Opin Struct Biol. 11:39–46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao JG, Ren KM and Tang J: Zinc finger
protein ZBTB20 promotes cell proliferation in non-small cell lung
cancer through repression of FoxO1. FEBS Lett. 588:4536–4542. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Q, Tan YX, Ren YB, Dong LW, Xie ZF,
Tang L, Cao D, Zhang WP, Hu HP and Wang HY: Zinc finger protein
ZBTB20 expression is increased in hepatocellular carcinoma and
associated with poor prognosis. BMC Cancer. 11:2712011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang J and Liu LY: Zinc finger protein
X-linked is overexpressed in colorectal cancer and is associated
with poor prognosis. Oncol Lett. 10:810–814. 2015.PubMed/NCBI
|
|
10
|
Cheng Y, Geng H, Cheng SH, Liang P, Bai Y,
Li J, Srivastava G, Ng MH, Fukagawa T, Wu X, et al: KRAB zinc
finger protein ZNF382 is a proapoptotic tumor suppressor that
represses multiple oncogenes and is commonly silenced in multiple
carcinomas. Cancer Res. 70:6516–6526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang X, Yuan W, Huang W, Bai Y, Deng Y,
Zhu C, Liang P, Li Y, Du X, Liu M, et al: ZNF569, a novel
KRAB-containing zinc finger protein, suppresses MAPK signaling
pathway. Biochem Biophys Res Commun. 346:621–628. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu J, Liang QY, Wang J, Cheng Y, Wang S,
Poon TC, Go MY, Tao Q, Chang Z and Sung JJ: Zinc-finger protein
331, a novel putative tumor suppressor, suppresses growth and
invasiveness of gastric cancer. Oncogene. 32:307–317. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao Y, Xiang T, Luo X, Li C, Li Q, Peng
W, Li L, Li S, Wang Z, Tang L, et al: Zinc-finger protein 545
inhibits cell proliferation as a tumor suppressor through inducing
apoptosis and is disrupted by promoter methylation in breast
cancer. PLoS One. 9:e1109902014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jen J and Wang YC: Zinc finger proteins in
cancer progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Wang Y, Fan X, Mo X, Wang Z, Li Y,
Yin Z, Deng Y, Luo N, Zhu C, et al: ZNF307, a novel zinc
finger gene suppresses p53 and p21 pathway. Biochem Biophys Res
Commun. 363:895–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ecker K, Lorenz A, Wolf F, Ploner C, Böck
G, Duncan T, Geley S and Helmberg A: A RAS recruitment screen
identifies ZKSCAN4 as a glucocorticoid receptor-interacting
protein. J Mol Endocrinol. 42:105–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH,
Zhang HX, Li WQ, Zhang YL, Zhang Y, Ma CC, et al: Genome-wide
association study identifies a susceptibility locus for
schizophrenia in Han Chinese at 11p11.2. Nat Genet. 43:1228–1231.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma L, Tang J, Wang D, Zhang W, Liu W, Wang
D, Liu XH, Gong W, Yao YG and Chen X: Evaluating risk loci for
schizophrenia distilled from genome-wide association studies in Han
Chinese from Central China. Mol Psychiatry. 18:638–639. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu RX, Seto WK, Lai CL and Yuen MF:
Epidemiology of hepatocellular carcinoma in the Asia-Pacific
region. Gut Liver. 10:332–339. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cheng RF, Wang J, Zhang JY, Sun L, Zhao
YR, Qiu ZQ, Sun BC and Sun Y: MicroRNA-506 is up-regulated in the
development of pancreatic ductal adenocarcinoma and is associated
with attenuated disease progression. Chin J Cancer. 35:642016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lo PK, Lee JS, Liang X and Sukumar S: The
dual role of FOXF2 in regulation of DNA replication and the
epithelial-mesenchymal transition in breast cancer progression.
Cell Signal. 28:1502–1519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engelmann D and Pützer BM: The dark side
of E2F1: In transit beyond apoptosis. Cancer Res. 72:571–575. 2012.
View Article : Google Scholar : PubMed/NCBI
|