Introduction
Esophageal cancer (ESC) is a common malignant tumor
in digestive tract system, and its incidence and mortality rank
eighth and sixth wordwide, respectively (1,2). ESC
mainly includes two subtypes: squamous cell carcinoma (ESCC) and
adenocarcinoma. The former accounts for 90% of patients with ESC in
China (3). In recent years,
although the clinical treatment for ESC has improved, the 5-year
survival rate of patients with ESC is still between 15 and 25%, and
this is strongly associated with the high recurrence and metastasis
(4,5). Most of the patients with ESC have
advanced stage disease when diagnosed, because the early stage of
ESC is difficult to discover (6).
Metastasis-associated colon cancer-1 (MACC1), a
tumor metastasis-related gene, was first identified in colon cancer
by Stein et al in 2009 (7).
Its encoding proteins include ZU5, SH3 binding domain, SH3
structural domain, and two death domains of the C-terminal tail
(8). MACC1 binds sp1 and further
locates in the promoter region of c-Met gene to promote the gene
transcription, thereby enhancing the proliferation and metastasis
of colon cancer cells (9). Recent
studies indicated that upregulation of MACC1 promoted cell
proliferation and metastasis in various cancers, such as gastric
cancer (10,11), hepatocellular carcinoma (12,13),
breast cancer (14), ovarian cancer
(15) and glioma (16). However, the role of MACC1 in ESC
development remains largely unclear.
Autophagy refers to a highly conserved biological
process in which cytoplasmic materials are delivered to lysosomes
for degradation and reuse, and this process plays an important role
in tumor recurrence, drug resistance, and distant metastasis
(17–19). Increasing evidence has shown that
deficiency of key autophagy-related genes induces tumorigenesis and
enhances survival ability of tumor cells (20,21).
On the contrary, accumulated autophagy causes irreversible damage
to organelles, resulting in tumor cell death (22). Autophagy is a double-edged sword in
tumor progress, but it still provides a new strategy for treatment
of tumor. In the present study, we found that MACC1 overexpression
induced autophagy to promote ESC cell proliferation, migration and
invasion, suggesting that MACC1 might be a theraputic target for
the treatment of ESC.
Materials and methods
Clinical specimens
Samples were collected from 51 patients with ESCC,
who underwent complete surgical resection at Department of
Cardiothoracic Surgery of the Affiliated Hospital of Southwest
Medical University from March 2011 to December 2014. This study was
approved by Ethics Review Board at Southwest Medical University.
After surgical resection, the tumor and matched normal tissues
(away from the tumor by >5 cm) were isolated and stored in
liquid nitrogen. Clinical characterization of the patients with
ESCC are shown in Table I.
 | Table I.Clinical characterization of patients
with ESCC. |
Table I.
Clinical characterization of patients
with ESCC.
| Index | n |
|---|
| Sex |
|
| Male | 45 |
|
Female | 6 |
| Age |
|
|
>60 | 19 |
| ≤60 | 32 |
| Tumor size |
|
| >5
cm | 10 |
| ≤5
cm | 41 |
| Location |
|
|
Middle | 44 |
|
Lower | 7 |
| Differentiation |
|
| Well | 25 |
|
Moderate | 21 |
| Poor | 5 |
| Invasion depth |
|
| Outer
layer | 39 |
| Inner
layer | 12 |
| Lymph node
metastasis |
|
| Yes | 29 |
| No | 22 |
Immunohistochemistry
Any five high-power fields (×200) were selected for
measurement, and density of the MACC1 staining was scored based on
two pathological experts. The scores were as follows: 0 for no
color, 1 for light yellow, 2 for yellow, and 3 for brown.
Percentage of positive cells was calculated under the view, and
scoring was performed according to the following standards: ≤5%, a
score of 0; 6–25%, a score of 1; 26–50%, a score of 2; and 51–100%,
a score of 3. The final score was obtained by multiplying the
average staining intensity of each slice by the average percentage
of positive cells, with 0–1 score for negative (−), 2–4 for weakly
positive (+), and 5–9 for strongly positive (++).
Cell culture
Normal human esophageal epithelial cells, and
esophageal squamous cell carcinoma cell lines including Eca109,
Eca9706, TE1, TE10, TE11 and Kyse150 were cultured in vitro.
Normal esophageal epithelium cells were cultured using EpiCGS-2
serum-free epithelial medium containing 1% P/S solution, and
esophageal squamous cell carcinoma cells were cultured in RPMI-1640
medium supplemented with 10% fetal bovine serum (FBS) at 37°C in 5%
CO2, respectively.
RNA isolation
Tissues were quickly frozen by liquid nitrogen and
completely pulverized. Tissue powder or ESC cells were lysated by
using TRIzol isolation reagent according to the manufacturer's
protocol. Total RNAs were analyzed using gel electrophoresis and
ultraviolet spectrophotometer, and then stored at −80°C.
Quantitative reverse transcription
polymerase chain reaction (qRT-PCR)
PrimeScript RT reagent kits (Takara) were used to
synthesize cDNA, and SYBR Green Real-time PCR Master Mix (Takara)
was used to perform the PCR assays accoding to the manufacturer's
protocols, respectively. Parameter of PCR reaction was as follows:
incubation at 94°C for 2 min, followed by 40 cycles of 94°C for 30
sec, 60°C for 1 min and 72°C for 30 sec, and after that incubation
at 72°C for 2 min. The sequences of primers of MACC1 and GAPDH are
as follows: MACC1 forward, 5′-GCAGACAAAGAATCAGAGAAAG-3′; reverse,
5′-GATGAGACGTGCGACTAACTC-3′.GAPDH forward,
5′-ATGCTGGCGCTGAGTACGTC-3′; reverse,
5′-GGTCATGAGTCCTTCCACGATA-3′.
Western blotting
Total proteins were extracted using RIPA lysis
buffer (Pierce Biotechnology) according to the manufacturer's
protocol. Forty micrograms of total proteins were separeted on 10%
SDS-PAGE gels and then transferred onto PVDF membranes. The
membranes were blocked in five percent skimmed milk for 1 h, and
then incubated with primary antibodies, MACC1 (1:1,000, Sigma) and
GAPDH (1:4000, Beyotime), at 4°C overnight. The membranes were
incubated with HRP-conjugated secondary antibodies (Zhongshan
Biotechnology) for 1 h, and finally analyzed by using enhanced
chemiluminescence.
Lentivirus vector construct
Eca9706 and Kyse150 cells were inoculated in
six-well plates with a density of 2×105 cells per well.
After the cells adhered to the plates, lentivirus
LV-GFP-PURO-shR-MACC1 and LV-GFP-PURO-shR-NC were added to infect
with the multiplicity of infection (MOI) value of 20. After 12 h,
fresh complete RPMI-1640 medium was replaced. At 72 h post medium
replacement, complete RPMI-1640 medium containing 1 µg of PURO was
added to the culture for 14 days. Efficiency of transfection was
measured by immunofluorescence microscopy and flow cytometry.
Silencing effect of MACC1 was measured by western blotting.
Cell proliferation
ESCC cells with a density of 1×104 per
well were seeded into 96-well plates. When the cells were cultured
for 24, 48 or 72 h, cell proliferation was analyzed using cell
counting kit 8 (CCK8, Beyotime) according to the manufacturer's
protocol.
Cell migration and invasion
For cell migration, 1×105 ESCC cells were
resuspended in 200 µl serum-free RPMI-1640 medium and added to the
upper compartment of Transwell chambers (8 µm), which were placed
into 24-well plates. RPMI-1640 medium (500 µl) supplemented with
10% FBS was added into each well of the 24-well plates, so that it
could be attracted to the lower compartment of Transwell chambers,
and then incubation at 37°C with 5% CO2 for 24 h. The
chambers were fixed with 4% formaldehyde at room temperature for 10
min, and then stained with Giemsa for 2 min. The cells detained in
the upper compartment were removed, and the cells passed through
the membrane were observed under a microscope. Five fields were
randomly selected and the cells were counted.
For cell invasion, the procedure was the same,
except that Matrigel (CA, USA) was added into the upper chamber of
Transwell chambers to simulate extracellular matrix, and the
incubation time continued for 72 h. Briefly, the original Matrigel
was diluted with serum-free RPMI-1640 medium at a ratio of 1:2.
Diluted Matrigel (50 µl) was added evenly to coat the upper
chamber, and incubated at 37°C for 60 min.
Cell apoptosis
The cells were collected and washed twice with
pre-cooled PBS. Cell apoptosis was analyzed using Annexin V
Fluorescein Isothiocyanate Apoptosis Detection kit (BD Bioscience)
according to the manufacturer's protocol. The single PE-Annexin
V-positive population and double-positive [PE and
7-amino-actinomycin D (AAD)] population detected by flow cytometry
were considered as early and late apoptotic cells, respectively.
The single PI-7-AAD-positive population was considered as necrotic
cells.
Confocal laser scanning microscopy and
transmission electron microscopy
For confocal laser scanning microscopy,
1×105 cells in each group were inoculated on a 20-mm
culture dish and cultured in RPMI medium supplemented with 10% FBS.
After the GFP-RFP-LC3B adenovirus (MOI, 20; purchased from Han Bio)
was added in the culture medium and co-cultured for 12 h, the
medium was replaced by a fresh medium and then cultured for 72 h.
Imagings were observed under a confocal fluorescence microscope
(SP8, Leica, Germany). The numbers of autophagosomes (green) and
autolysosomes (red) in random field were calculated.
For transmission electron microscopy, growing
adherent cells were directly scraped, and the cell precipitate was
collectet after using PBS to wash twice. The cells were fixed by
using 2.5% glutaraldehyde solution overnight at 4°C. Two days
later, imagings were observed under a transmission electron
microscope (Fucheng Biological Technology Co., Ltd., Shanghai,
China).
Statistical analyses
Continuous data are presented as mean ± standard
deviation. The comparison between two groups was performed by using
Student's t-test. A P<0.05 was considered statistically
significant. All experiments were repeated three times.
Results
MACC1 is frequently upregulated in
ESCC tissues and associated with lymph node metastasis
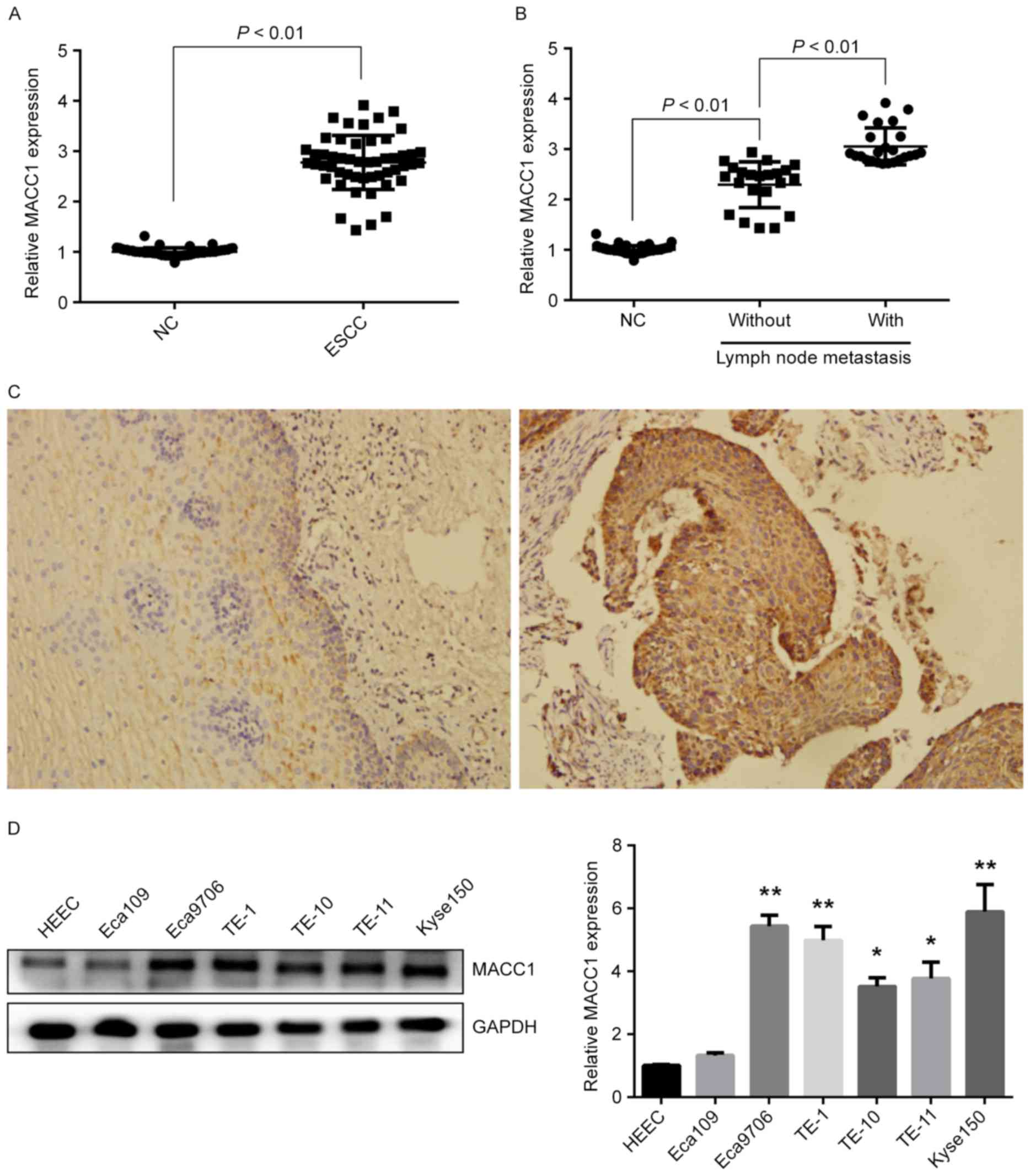
To determine MACC1 expression in ESCC, we detected
mRNA and protein expressions of MACC1 in 51 pairs of ESCC and
matched normal tissues. QRT-PCR experiments showed the mRNA
expression of MACC1 was significantly increased in ESCC tissues
compared with matched normal tissues (P<0.01) (Fig. 1A), and the expression was also
increased in ESCC tissues of patients with lymph node metastasis
compared to those without lymph node metastasis (P<0.01)
(Fig. 1B). Immunohistochemical
staining revealed that the protein expression of MACC1 was also
increased in ESCC tissues with 94.1% (48/51) positive rate compared
with matched normal tissues with 37.3% (19/51) positive rate
(Fig. 1C). Furthermore, mRNA and
protein expressions of MACC1 in ESCC cell lines were examined.
Western blotting and qRT-PCR assays showed high protein and mRNA
expression of MACC1 in Eca9706 and Kyse150 cells (P<0.01)
(Fig. 1D), suggesting that MACC1
expression is increased in ESCC and associated with lymph node
metastasis of ESCC patients.
MACC1 regulates proliferation and
apoptosis of ESCC cells
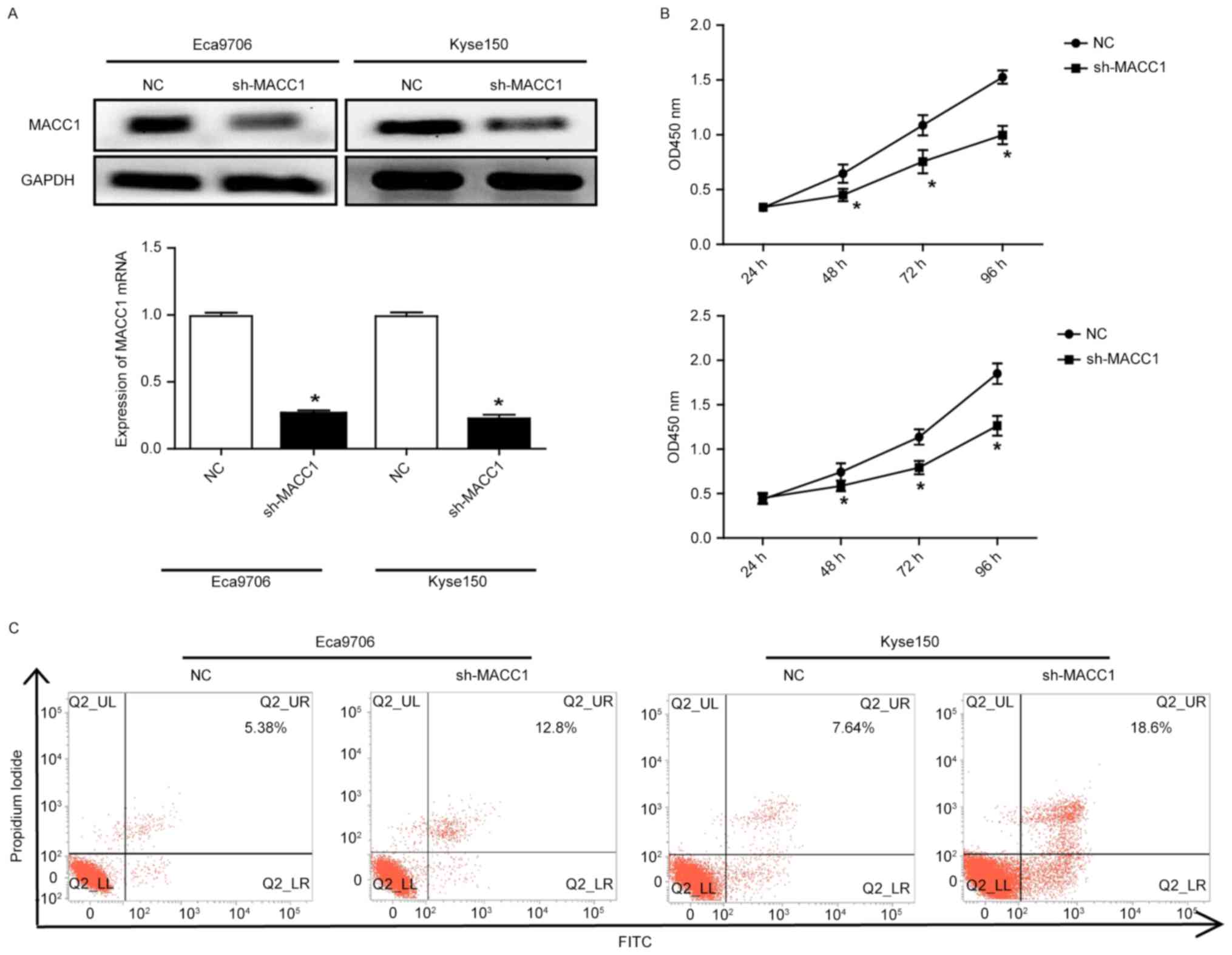
To determine the role of MACC1 in ESCC cells, we
selected Eca9706 and Kyse150 cells with high expression of MACC1 to
perform MACC1 knockdown using letivirus vectors containing the
specific shRNA for MACC1. Western blotting and qRT-PCR assays
showed that the protein and mRNA expression of MACC1 was
significantly decreased, respectively (P<0.05) (Fig. 2A). Cell count kit-8 experiments
indicated that MACC1 knockdown significantly decreased
proliferative capacity of the Eca9706 and Kyse150 cells (P<0.05)
(Fig. 2B). Flow cytometry revealed
that MACC1 knockdown significantly increased apoptosis of the
Eca9706 and Kyse150 cells (Fig.
2C).
Downregulation of MACC1 suppreses
migration and invasion of ESCC cells
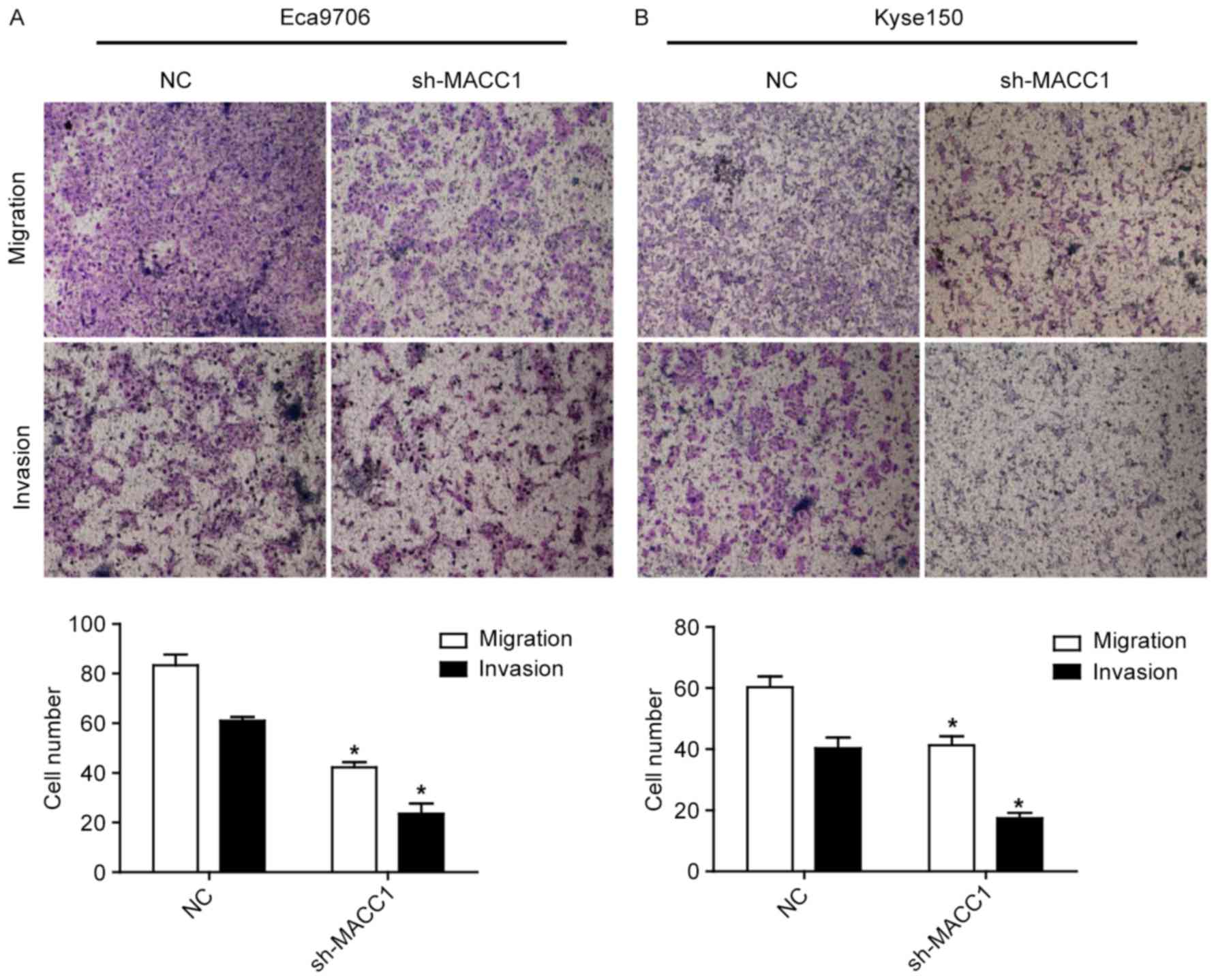
To determine whether MACC1 is involved in ESCC cell
metastasis, cell migration and invasion assays were performed. As
shown in Fig. 3, MACC1 knockdown
significantly repressed migration and invasion of the Eca9706 and
Kyse150 cells (P<0.05), suggesting that MACC1 is correlated with
tumor metastasis in ESCC.
Downregulation of MACC1 inhibits
autophagy in ESCC cells
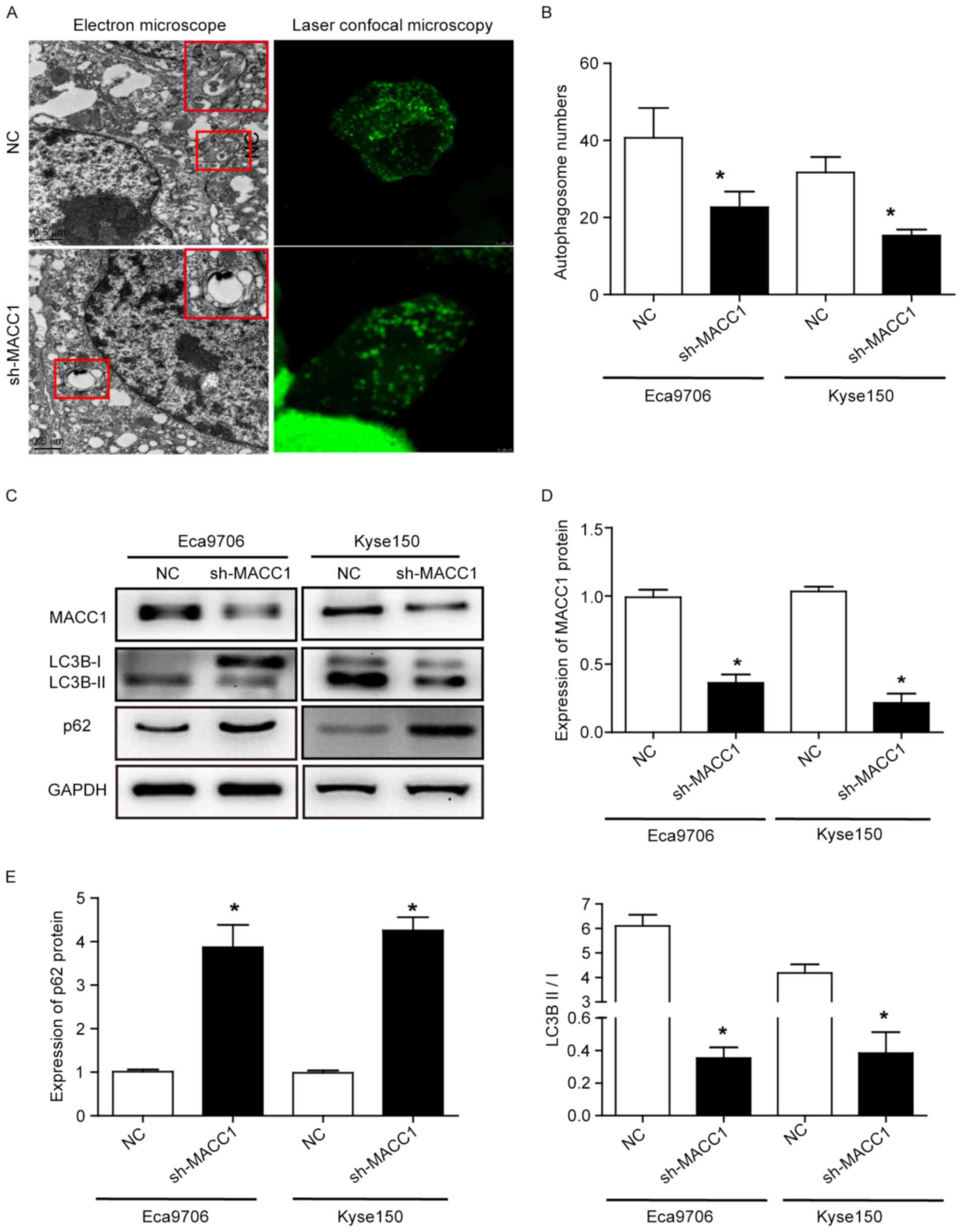
Autophagy is shown to be involved in the
proliferation, apoptosis and metastasis of tumor cells. Therefore,
we determined whether MACC1 is involved in ESCC cell autophagy.
Electron microscope and laser confocal microscopy assays showed
autophagy (Fig. 4A), and the number
of autophagosome was significantly decreased in the Eca9706 and
Kyse150 cells with downregulating MACC1 compared with those with
empty vector (P<0.05) (Fig. 4B).
Moreover, western blotting revealed that MACC1 knockdown
significantly decreased the ratio of LC3B II/I (P<0.05) and
increased the protein expression of p62 (P<0.05) (Fig. 4C-E). These data suggest that MACC1
knockdown inhibits autophagy in ESCC cells.
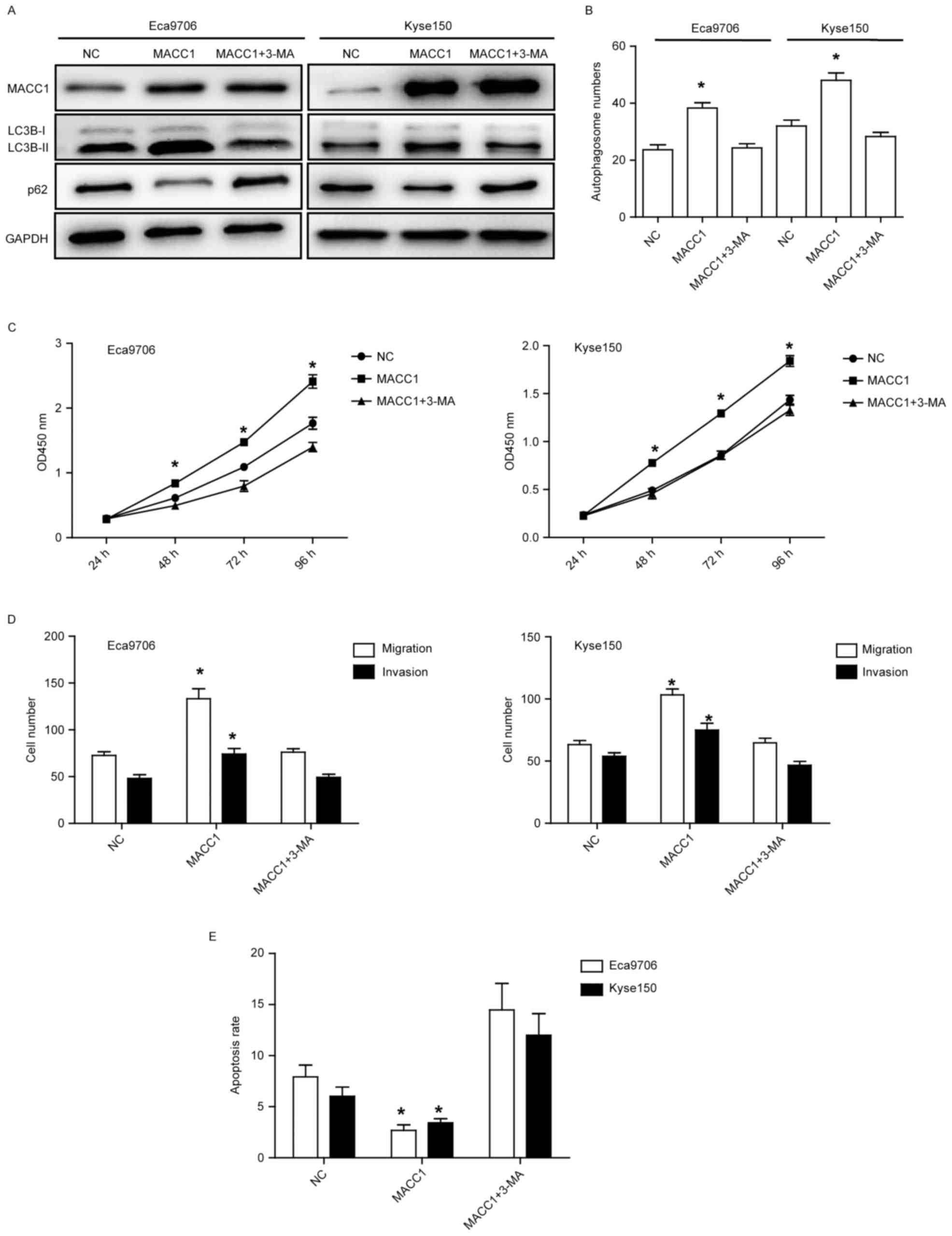
Three-methyladenine (3-MA) rescues
MACC1-induced ESCC cell proliferation, apoptosis, migration, and
invasion
To detemine whether autophagy plays a crucial role
in MACC1-induced phenotype in ESCC cells, we used 3-methyladenine
(3-MA), an inhibitor of autophagy, to inhibit autophagy in Eca9706
and Kyse150 cells. Western blotting and laser confocal microscopy
assays showed that 3-MA rescued the ratio of LC3B II/I and p62
expression, and the number of autopahosome, respectively
(P<0.05) (Fig. 5A and B),
suggesting that MACC1-induced autophagy was rescued by 3-MA
treatment. Furthermore, proliferation, apoptosis, migration, and
invasion of the Eca9706 and Kyse150 cells were also rescued with
3-MA treatment (P<0.05) (Fig.
5C-E), suggesting MACC1-induced phenotype in ESCC cells is
mainly through autophagy.
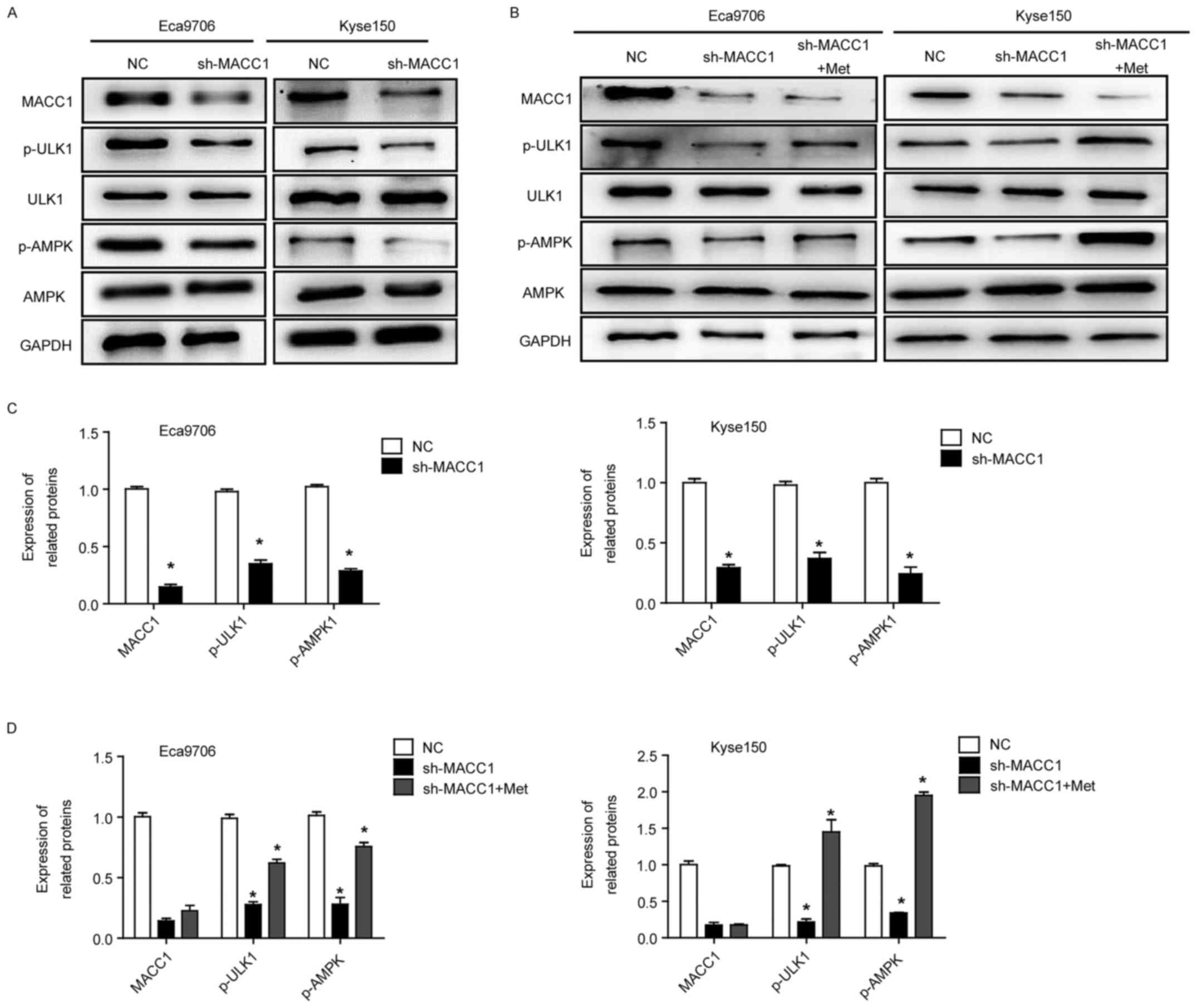
MACC1 regulates the AMPK signaling
pathway involved in autophagy
To understand the mechanism by which MACC1 regulates
ESCC cell autophagy, we detected AMPK-ULK1 signaling pathway that
is believed to be involved in cell autophagy. Western blot assays
showed that MACC1 knockdown significantly decreased phosphorylation
levels of p-ULK1 and p-AMPK in the Eca9706 and Kyse150 cells
(P<0.05) (Fig. 6A and C),
whereas protein expression of ULK1 and AMPK was not altered,
suggesting that MACC1-indued autophagy is associated with AMPK
signaling pathway. Furthermore, MACC1-induced autophagy in ESCC
cells was rescued when the cells were treated with metformin, an
AMPK activator (P<0.05) (Fig. 6B and
D), suggesting that AMPK signaling pathway is a key pathway in
MACC1-induced autophagy.
Discussion
This study found that MACC1 was overexpressed in
ESCC, and its upregulation was positively correlated with lymph
node metastasis of ESCC patients. Stable knockdown of MACC1
repressed ESCC cell proliferation, migration and invasion, while
enhanced cell apoptosis. Furthermore, MACC1 knockdown inhibited
ESCC cell autophagy. When 3-MA, an inhibitor for autophagy, was
used to treat ESCC cells, MACC1-induced autophagy was rescued,
resulting in ESCC cell proliferation, apoptosis, migration, and
invasion also being rescued to normal level, suggesting that MACC1
induced the aggressive progress of ESCC cells mainly through
affecting autophagy. AMPK signaling pathway was also confirmed to
be important in MACC1-induced autophagy.
MACC1 has been considered as an important molecule
for various cancer diagnosis, prognosis, and theraputic target
(10–15). Increasing evidence indicates that
ectopic expression of MACC1 is involved in tumorigenesis and cancer
development (16). However, the
role and mechnism of MACC1 in ESCC had not been investigated. Our
data demonstrated that MACC1 was upregulated in ESCC and correlated
with lymph node metastasis of ESCC patients, suggesting that the
expression signature of MACC1 might be a novel biomarker for ESCC
diagnosis and prognosis. The role of MACC1 was reported
inconsistently in variety of cancers, but in our data we first
showed MACC1 as an oncogene, promoting malignant phenotypes of ESCC
cells by gain- and loss-of-function.
Autophagy plays crucial role in tumorigenesis and
cancer development. For example, Beclin deficience could increase
the tumorigenesis risk of mice, such as lymphoma, lung cancer,
liver cancer and other tumors (23,24)
Weh et al found that Beclin-dependent autophagy is specific
and related with prognostic factors of EAC (25). Lee et al demonstrated that a
dominant-negative autophagy protein Atg5K130R could lead to
loss-of-heterozygosity of tp53 followed by promoting tumorigenesis
(26). In our study, we first found
that MACC1 induced ESCC cell autophagy by multiple experiments.
Importantly, 3-MA, an inhibitor for autophagy, was able to rescue
autophagy in ESCC cells, and further rescued MACC1-induced
malignant phenotype of ESCC cells, suggesting the important role of
autophagy in ESCC cells. However, 3-MA did not alter MACC1
expression, indicating that MACC1 is a upstream gene in autophagy
of ESCC cells. Consistant with a previous study (27), our data also revealed that autophagy
inhibition repressed ESCC cell proliferation, migration and
invasion, and enhanced cell apoptosis. Autophagy is involved in
several signaling pathways, such as AMPK and PI3K/AKT/mTOR
(28,29). Our results showed that MACC1
knockdown decreased phosphorylation levels of AMPK and ULK1,
suggesting that MACC1-induced autophagy is involved in AMPK
signaling pathway. AMPK directly phosphorylates VPS34 and ULK1,
resulting in autophagy formation. On the contrary, AMPK inhibits
autophagy by phosphorylating mTOR, so AMPK has different roles in
different cells. Moreover, when ESCC cells were treated using
metformin, an activator of AMPK, the autophagy was rescued,
suggesting MACC1-induced autophagy mainly through regulating AMPK
signaling pathway.
In conclusion, the present study showed that
upregulation of MACC1 was correlated with lymph node metastasis of
ESCC patients. MACC1 regulated ESCC cell proliferation, apoptosis,
migration and invasion mainly through affecting autophagy.
Acknowledgements
The authors wish to thank Mr. Biao Zhou, and KaiMing
He for assistance in obtaining patient tissue samples. This study
was supported by The Joint Fund of Technology Department, Sichuan
Province (2014TSX-0102), and by Youth Foundation of Affiliated
Hospital of South West Medical University (16025), and by Sichuan
Science and Technology Plan projects (no. 2016RZ0076).
References
|
1
|
Sawada G, Niida A, Uchi R, Hirata H,
Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y,
et al: Genomic landscape of esophageal squamous cell carcinoma in a
Japanese population. Gastroenterology. 150:1171–1182. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mao Y, Li L, Liu J, Wang L and Zhou Y:
miR-495 inhibits esophageal squamous cell carcinoma progression by
targeting Akt1. Oncotarget. 7:51223–51236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang HY, Yao ZH, Tang H, Zhao Y, Jin SL,
Zhou WP, Yao SN, Yang SJ, Liu YY and Luo SX: A retrospective
clinical study of comparing paclitaxel plus S-1 versus paclitaxel
plus cisplatin as the first-line treatment for patients with
advanced esophageal squamous cell carcinoma. Oncotarget.
8:7540–7547. 2017.PubMed/NCBI
|
|
4
|
Hu D, Lin X, Chen Y, Chang Q, Chen G, Li
C, Zhang H, Cui Z, Liang B, Jiang W, et al: Preoperative
blood-routine markers and prognosis of esophageal squamous cell
carcinoma: The Fujian prospective investigation of cancer (FIESTA)
study. Oncotarget. 8:23841–23850. 2017.PubMed/NCBI
|
|
5
|
Kim R, Keam B, Kwon D, Ock CY, Kim M, Kim
TM, Kim HJ, Jeon YK, Park IK, Kang CH, et al: Programmed death
ligand-1 expression and its prognostic role in esophageal squamous
cell carcinoma. World J Gastroenterol. 22:8389–8397. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Li M, Gao F and Ge X: Serum
microRNA-15a level acts as a potential diagnostic and prognostic
biomarker for human esophageal squamous cell carcinoma. Cancer
Biomark. 18:11–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stein U, Walther W, Arlt F, Schwabe H,
Smith J, Fichtner I, Birchmeier W and Schlag PM: MACC1, a newly
identified key regulator of HGF-MET signaling, predicts colon
cancer metastasis. Nat Med. 15:59–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stein U, Smith J, Walther W and Arlt F:
MACC1 controls Met: What a difference an Sp1 site makes. Cell
Cycle. 8:2467–2469. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shirahata A, Shinmura K, Kitamura Y,
Sakuraba K, Yokomizo K, Goto T, Mizukami H, Saito M, Ishibashi K,
Kigawa G, et al: MACC1 as a marker for advanced colorectal
carcinoma. Anticancer Res. 30:2689–2692. 2010.PubMed/NCBI
|
|
10
|
Ilm K, Fuchs S, Mudduluru G and Stein U:
MACC1 is post-transcriptionally regulated by miR-218 in colorectal
cancer. Oncotarget. 7:53443–53458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia J, Wang H, Huang H, Sun L, Dong S,
Huang N, Shi M, Bin J, Liao Y and Liao W: Elevated Orai1 and STIM1
expressions upregulate MACC1 expression to promote tumor cell
proliferation, metabolism, migration, and invasion in human gastric
cancer. Cancer Lett. 381:31–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie C, Wu J, Yun J, Lai J, Yuan Y, Gao Z,
Li M, Li J and Song L: MACC1 as a prognostic biomarker for
early-stage and AFP-normal hepatocellular carcinoma. PLoS One.
8:e642352013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qu JH, Chang XJ, Lu YY, Bai WL, Chen Y,
Zhou L, Zeng Z, Wang CP, An LJ, Hao LY, et al: Overexpression of
metastasis-associated in colon cancer 1 predicts a poor outcome of
hepatitis B virus-related hepatocellular carcinoma. World J
Gastroenterol. 18:2995–3003. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan W, Xie X, Li L, Tang H, Ye X, Chen L,
Tang W, Gao J, Pan L, Zhang X, et al: Diagnostic and prognostic
value of serum MACC1 in breast cancer patients. Oncotarget.
7:84408–84415. 2016.PubMed/NCBI
|
|
15
|
Li H, Zhang H, Zhao S, Shi Y, Yao J, Zhang
Y, Guo H and Liu X: Overexpression of MACC1 and the association
with hepatocyte growth factor/c-Met in epithelial ovarian cancer.
Oncol Lett. 9:1989–1996. 2015.PubMed/NCBI
|
|
16
|
Sun L, Li G, Dai B, Tan W, Zhao H, Li X
and Wang A: Silence of MACC1 expression by RNA interference
inhibits proliferation, invasion and metastasis, and promotes
apoptosis in U251 human malignant glioma cells. Mol Med Rep.
12:3423–3431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
DeVorkin L, Hattersley M, Kim P, Ries J,
Spowart J, Anglesio MS, Levi SM, Huntsman DG, Amaravadi RK, Winkler
JD, et al: Autophagy inhibition enhances sunitinib efficacy in
clear cell ovarian carcinoma. Mol Cancer Res. 15:250–258. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhan L, Zhang Y, Wang W, Song E, Fan Y, Li
J and Wei B: Autophagy as an emerging therapy target for ovarian
carcinoma. Oncotarget. 7:83476–83487. 2016.PubMed/NCBI
|
|
19
|
Ahn JS, Ann EJ, Kim MY, Yoon JH, Lee HJ,
Jo EH, Lee K, Lee JS and Park HS: Autophagy negatively regulates
tumor cell proliferation through phosphorylation dependent
degradation of the Notch1 intracellular domain. Oncotarget.
7:79047–79063. 2016.PubMed/NCBI
|
|
20
|
Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu
Y, Yi X, Ma J, Zhao T, Liu L, et al: Down-regulated miR-23a
contributes to the metastasis of cutaneous melanoma by promoting
autophagy. Theranostics. 7:2231–2249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carchman EH, Matkowskyj KA, Meske L and
Lambert PF: Dysregulation of autophagy contributes to anal
carcinogenesis. PLoS One. 11:e01642732016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jia YL, Xu M, Dou CW, Liu ZK, Xue YM, Yao
BW, Ding LL, Tu KS, Zheng X and Liu QG: P300/CBP-associated factor
(PCAF) inhibits the growth of hepatocellular carcinoma by promoting
cell autophagy. Cell Death Dis. 7:e24002016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al:
Promotion of tumorigenesis by heterozygous disruption of the beclin
1 autophagy gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weh KM, Howell AB and Kresty LA:
Expression, modulation, and clinical correlates of the autophagy
protein Beclin-1 in esophageal adenocarcinoma. Mol Carcinog.
55:1876–1885. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee E, Wei Y, Zou Z, Tucker K, Rakheja D,
Levine B and Amatruda JF: Genetic inhibition of autophagy promotes
p53 loss-of-heterozygosity and tumorigenesis. Oncotarget.
7:67919–67933. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu C and Xie C: Radiation-induced
autophagy promotes esophageal squamous cell carcinoma cell survival
via the LKB1 pathway. Oncol Rep. 35:3559–3565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamada E, Okada S, Bastie CC, Vatish M,
Nakajima Y, Shibusawa R, Ozawa A, Pessin JE and Yamada M: Fyn
phosphorylates AMPK to inhibit AMPK activity and AMP-dependent
activation of autophagy. Oncotarget. 7:74612–74629. 2016.PubMed/NCBI
|
|
29
|
He Y, Mo Q, Luo B, Qiao Y, Xu R, Zuo Z,
Deng J, Nong X, Peng G, He W, et al: Induction of apoptosis and
autophagy via mitochondria- and PI3K/Akt/mTOR-mediated pathways by
E. adenophorum in hepatocytes of saanen goat. Oncotarget.
7:54537–54548. 2016. View Article : Google Scholar : PubMed/NCBI
|