Introduction
Gastric cancer (GC) is an aggressive malignancy and
remains a major health issue (1).
Chemotherapy improves survival and quality of life compared with
the best supportive care, but the median overall survival remains
poor (2). There is an urgent need
for more effective target agents for treating this disease.
Cetuximab is a recombinant human/mouse chimeric
monoclonal antibody against the epidermal growth factor receptor
(EGFR). Cetuximab was found to enhance the effect of oxaliplatin in
hypoxic GC cell lines (3). Several
phase II trials have evaluated cetuximab as a first-line treatment
in combination with various chemotherapy regimens (4–6).
However, Erbitux in combination with Xeloda and cisplatin in
advanced esophago-gastric cancer (EXPAND; NCT00678535) did not
significantly increase progression-free survival (PFS) in patients
with advanced GC (7).
In vitro and in vivo antitumor
activity of cetuximab in human GC cell lines is related to EGFR
expression and mutational phenotype (8,9). EGFR
mutations have been proven to be the most effective biomarker for
the prediction of superior efficacy for EGFR tyrosine kinase
inhibitors (TKIs) (10–12). The potential use of EGFR expression
as a marker has been widely investigated, with conflicting results
(13). Moreover, gene copy number
gain of EGFR is also a poor prognostic biomarker in GC (8). Unlike HER2 in GC, the predictive value
of increased EGFR copy number for tumor response is controversial
(14,15). Similarly, the relationship between
the level of EGFR amplification and the outcome of EGFR-positive GC
treated with first-line chemotherapy with cetuximab remains
unclear.
At present, there has been an increase in using
experimental models to predict the clinical activity of agents and
discover predictive biomarkers. A large collection of
patient-derived tumor xenografts (PDXs) reflects the diversity of
tumors in patient populations. We established GC PDXs by
transplanting surgically removed tumor tissues from patients into
immunocompromised BALB/c nude mice via subcutaneous inoculation, to
assess drug activity. Moreover, our previous data also suggested
that cases with a GC subtype with EGFR amplification and
overexpression benefit from cetuximab treatment (16). In the present study, our aim was to
determine whether the level of EGFR amplification significantly
predicts increased survival and response to therapy in GC treated
with cetuximab-based chemotherapy. We investigated the activity of
cetuximab in 20 GC-PDX models. After the therapeutic responders and
non-responders were identified, the correlation between EGFR
amplification, mRNA and protein expression level and tumor response
to cetuximab therapy in the GC xenografts were analyzed. Moreover,
we also investigated the survival of the GC PDX models in the
cetuximab-treated and control groups in regards to the different
levels of DNA amplification, mRNA and protein expression.
Materials and methods
Patient and tumor samples
All of the cases with freshly and surgically removed
tumor tissues included in the present study were diagnosed and
surgically treated at Peking University Cancer Hospital. The
tumor-node-metastasis (TNM) stage was classified according to the
7th edition of the classification recommended by the American Joint
Committee on Cancer (AJCC) (17).
All of the tumors were not previously treated with chemotherapy or
EGFR inhibitors. This investigation was performed after approval by
the Ethics Committee of Peking University Cancer Hospital.
Antitumor activity evaluation
The subcutaneous engraftment of patient tumor
fragments into immunocompromised mice was previously described
(18). When the tumor volume
reached 100–150 mm3, the mice were randomly grouped into
two groups of five mice with a similar average tumor volume.
Immediately after grouping, the control group was treated with
vehicle [phosphate-buffered saline (PBS), weekly intraperitoneal
injection or i.p. for 2 weeks], and the treatment groups were
injected with cetuximab (weekly i.p. injection for 2 weeks, 50
mg/kg; Merck KGaA, Darmstadt, Germany). The tumor growth was
monitored twice weekly, and ΔT/ΔC value was calculated for
assessing tumor response to the treatment (ΔT = tumor volume change
in the treatment group and ΔC = tumor volume change in the control
group). The total number of mice used for the xenografts was 200
(10 mice/model for 20 PDX models). Survival benefit was evaluated
by comparing the survival curves corresponding to the time for the
tumor to reach 600 mm3 (19,20).
All procedures were carried out under sterile conditions at Crown
Bioscience SPF facility and conducted in strict accordance with the
Guide for the Care and Use of Laboratory Animals of the National
Institutes of Health. The protocol was approved by the Committee on
the Ethics of Animal Experiments of Crown Bioscience (Crown
Bioscience IACUC Committee).
Immunohistochemistry (IHC)
staining
Four-micrometer sections from formalin-fixed
paraffin-embedded tissues were mounted on poly-L-lysine-coated
slides, and then deparaffinized in xylene and rehydrated through
alcohol to distilled water. Endogenous peroxidase activity was
blocked with 3% hydrogen peroxide for 15 min at room temperature.
After pressure cooking the slides in 10 mmol/l EDTA (pH 8.0) for 3
min, the sections were incubated overnight at 4°C with mouse rabbit
anti-human EGFR antibody (Cell Signaling Technology, Inc., Boston,
MA, USA) at final dilution of 1:200. Primary antibodies were
detected using a two-step EnVision System (Dako, Glostrup,
Denmark). Positive and negative immunohistochemistry controls were
routinely used. For negative controls, the primary antibody was
replaced by non-immune mouse serum to confirm its specificity.
Moreover, we used an internal positive control in
immunohistochemistry for quality assurance. The test specimens were
then independently scored by three investigators in a blinded
fashion: score 0 is when there was no specific membrane staining
within the tumor, and positive when there was any staining of the
tumor cell membrane above the background level. The positive cases
were further classified into 1+, 2+ and 3+ based on the staining
intensity of the membrane.
EGFR gene copy numbers and mRNA
expression detection
Quantitative RT-PCR was performed to determine the
relative EGFR gene expression level for all of the samples.
Extracted mRNA was subjected to amplification using human
EGFR-specific primers by TaqMan q-PCR (assay ID, Hs01076078_m1;
Applied Biosystems, Foster City, CA, USA). The human GAPDH gene was
used as a reference (assay ID, Hs99999905_m1; Applied Biosystems).
Each sample was run thrice. Expression of each gene was represented
as the ratio of expression of each target gene mRNA to that of
GAPDH mRNA. Moreover, Affymetrix HG-U219 array was also performed
following a standard protocol (http://media.affymetrix.com/support/downloads/manuals/3_ivt_express_kit_manual.pdf)
to detect EGFR mRNA expression.
In addition, EGFR gene copy numbers were determined
by quantitative PCR. Briefly, the same genomic DNAs were subjected
to amplification by TaqMan qPCR. The primers for EGFR (assay ID,
Hs04960197_cn) and RNase P as endogenous reference (part no.
4401631) were purchased from Applied Biosystems. The raw data were
transferred to CopyCaller software and analyzed.
FISH
Quantitative assessment of EGFR copy number was also
investigated by FISH. Dual-color, dual-target FISH assays were
carried out using the EGFR Spectrum Orange/CEP7 Spectrum Green
Probe (Vysis, Des Plaines, IL, USA). Three-micrometer thick tissue
sections were treated with the procedure provided by fluorescence
in-situ hybridization (FISH) detection kit (DakoCytomation,
Glostrup, Denmark). Briefly, samples were placed in pretreatment
solution for 30 min at 96°C, and digested with pepsin solution for
30 min at room temperature. Tissue sections, covered with 10-µl
probe solution, were incubated at 75°C for 5 min to co-denature the
EGFR and chromosome seven α-centromeric (CEP7) probes and allowed
to hybridize overnight at 37°C. Co-denaturation and hybridization
were carried out sequentially. Post-hybridization stringency wash
was carried out in a water bath at 65°C for 10 min. Then, tissue
sections were covered with DAPI II (Vysis) for chromatin
counterstaining. EGFR was visualized as a red signal with a
standard tetramethyl Rhodamine isothiocyanate (TRITC) filter, CEP7
as a green signal with a fluorescein isothiocyanate (FITC) filter,
and nuclei as a blue signal with a DAPI filter. Representative
images of the samples were acquired and then analyzed.
Two independent observers scored at least 100
non-overlapping interphase nuclei for the number of copies of EGFR
and CEP7 by use of predefined scoring guidelines. EGFR status was
scored as the number of EGFR signals/nucleus and as the ratio of
EGFR signals to CEP7 signals. Amplification was defined as the
presence of 2.5 or more signals/nucleus, i.e., EGFR copy number
≥2.5.
RNA in situ hybridization assays in
FFPE samples
In situ hybridization (ISH) was performed
using QuantiGene® ViewRNA ISH Tissue Assay (Affymetrix,
Santa Clara, CA, USA) following the manufacturer's protocol.
Briefly, FFPE slides were prepared according to the procedure
described. Then, sections were rehydrated and incubated with
Proteinase K. Standard probe design software was used to design
specific oligonucleotide probe sets for detecting target genes. A
no-probe sample was utilized as a negative control per the
Affymetrix manual's recommendations. The signal was amplified
before incubation with labeled probes and visualized. Hybridized
target mRNAs were visualized using confocal fluorescent microscopy,
or by bright field microscopy.
The test specimens were then independently scored by
three investigators in a blinded fashion: score 0 is when there was
no specific staining within the tumor, and positive when there was
any staining of the tumor cell membrane above the background level.
The positive cases were further classified into 1+, 2+ and 3+ based
on the staining intensity.
Statistical analysis
In order to investigate correlations between tumor
response (ΔT/ΔC value) and CN/mRNA/protein expression, the ΔT/ΔC
value, DNA and mRNA expression value was coded as one for
expression levels ranked as at or below the 25th percentile of the
total gene expression, two for levels above the 25th and at or
below the 50th percentiles, three for levels above the 50th and at
or below the 75th percentiles, and four for levels above the 75th
percentile. Spearman rank order correlations were performed to
analyze the correlation between tumor response and CN/mRNA/protein
expression. Hazard ratios from univariate Cox regression analysis
were used to determine whether the factor was associated with
death. Protective genes were defined as those associated with a
hazard ratio for death of <1; risk genes were defined as those
associated with a hazard ratio for death of >1. Kaplan-Meier
analysis was used to compare survival of the two groups of PDX
models (by 600 mm3) with the log-rank test. In all
analyses, P<0.05 was considered to indicate statistical
significance, and all tests were two-tailed. The statistical
analysis was carried out using SPSS V16.0 software (SPSS, Inc.,
Chicago, IL, USA).
Results
EGFR status in the GC xenografts
We established GC-PDX models by surgically
transplanting removed tumor tissues from GC patients into
immunocompromised BALB/c nude mice subcutaneously. qPCR, FISH (EGFR
and EGFR/CEP7), RNAish and immunohistochemical staining were
performed to investigate EGFR expression in the 20 GC xenografts.
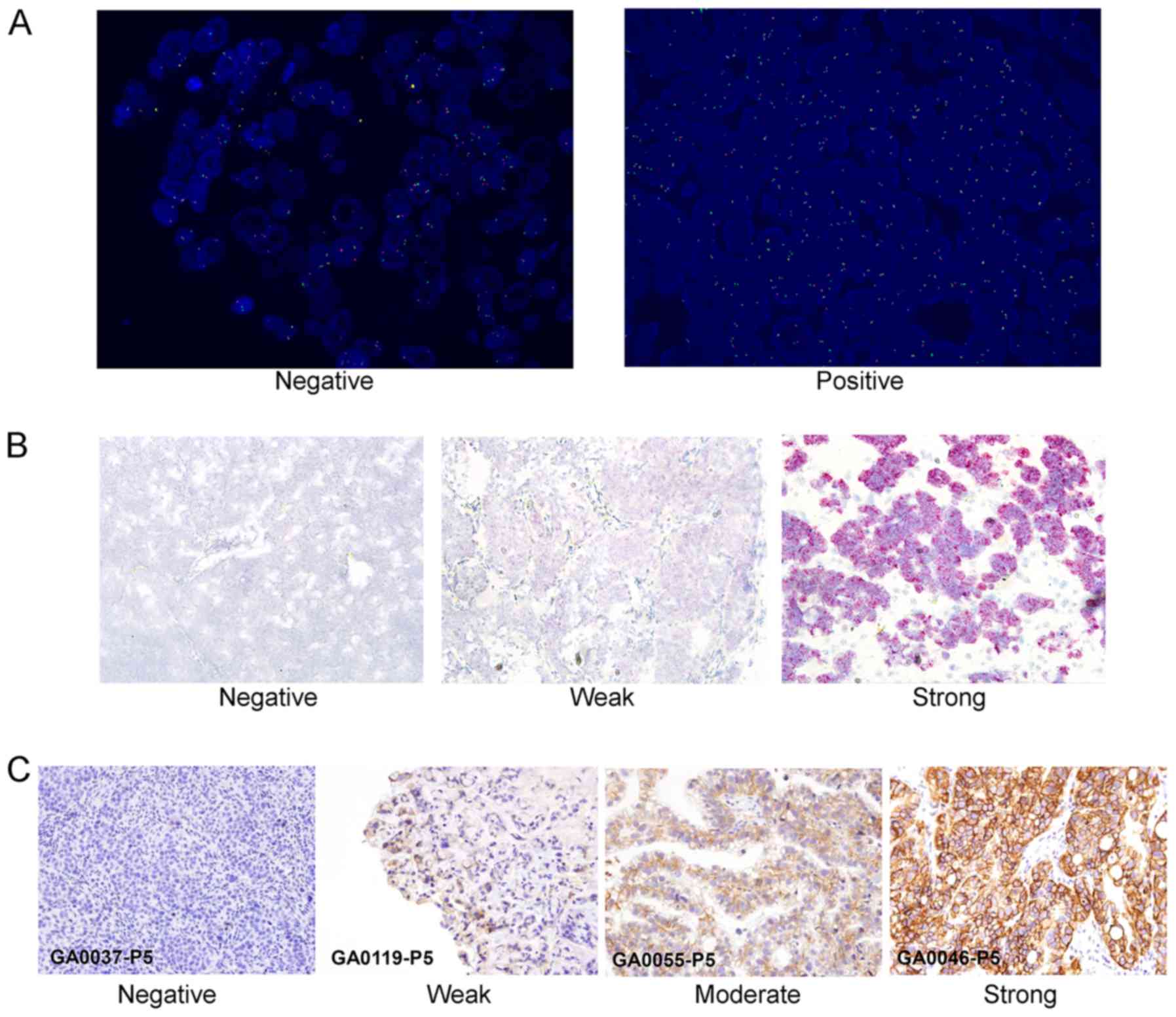
Amplification of EGFR by FISH was determined by EGFR or the
EGFR/CEP7 ratio with a range 1.9 to >15 or 0.83 to >2
(Fig. 1A). The ranges of EGFR CN by
qPCR were 1.4–1040.9. The ranges of mRNA by qPCR (EGFR/GAPDH) were
0.00–13.00. RNAish assay showed that 5 of 20 GC xenografts were
EGFR positive; one of them showed strong positive (Fig. 1B). For the 20 cases included in the
analysis, 7 (35%) were IHC 0, 7 (35%) were 1+, 2 (10%) were 2+, and
4 (20%) were 3+ (Fig. 1C).
Correlation between the tumor response
and EGFR status in the cetuximab-treated GC xenografts
The tested GC-PDXs fell into two distinct categories
according to the drug activity: 4 of 20 (20%) responded with a
nearly complete response (ΔT/ΔC value <0) to cetuximab
treatment; 16 of 20 (80%) did not, 1 of 20 had a partial response
and 15 had complete resistance (ΔT/ΔC value >30%).
The correlations between tumor response and the
level of DNA amplification, mRNA and protein expression in GC
xenografts treated with cetuximab were analyzed. Most of the IHC 3+
cases and mRNA overexpression by RNAish and qPCR had a significant
positive correlation with the tumor response to cetuximab
(Spearman's, RNAish, P=0.001; EGFR/GAPDH, P<0.001; EGFR IHC
score, P=0.003), while EGFR CN, either detected by qPCR or FISH
(EGFR, EGFR/CEP7), there was no significantly correlation between
the EGFR CN and tumor response (EGFR CN detected by qPCR, P=0.398;
FISH, P=0.119, 0.232, respectively; Table I).
 | Table I.Correlation between the DNA, RNA and
protein expression level of EGFR and cetuximab response. |
Table I.
Correlation between the DNA, RNA and
protein expression level of EGFR and cetuximab response.
| Detection method | Correlation | P-value |
|---|
| DNA CN |
| qPCR | 0.20 | 0.398 |
| FISH
(EGFR/CEP7) | 0.28 | 0.232 |
| FISH
(EGFR) | 0.36 | 0.119 |
| mRNA expression |
|
RNAish | 0.671 | 0.001 |
| U219
intensity | 0.707 | 0.001 |
| qPCR
(EGFR/GAPDH) | 0.720 | <0.001 |
| Protein |
| EGFR IHC
score | 0.630 | 0.003 |
Survival of the GC PDX models
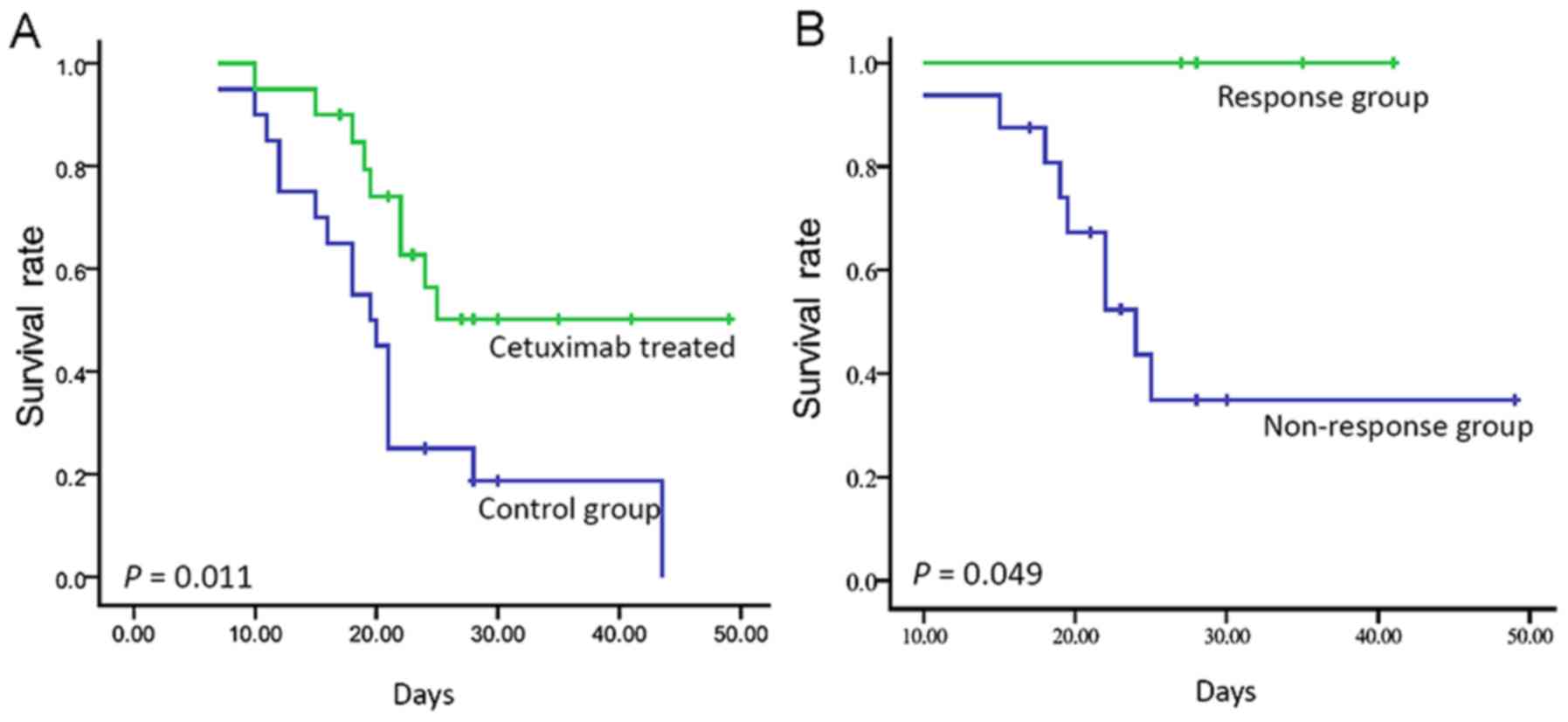
Survival of the GC PDX were compared between the
cetuximab-treated and control group. Our results suggested that the
PDX models treated with cetuximab had longer survival compared with
that noted in the control cases (median, 19.5 days vs. not
reached). This difference was statistically significant (log-rank,
P=0.011; Fig. 2A). In the
univariate Cox analyses, cetuximab was significantly associated
with survival [hazard ratio 0.371; 95% confidence interval (CI),
0.164 to 0.838; P=0.017]. Moreover, the PDX models responsive to
cetuximab had longer survival than those in the non-responsive
cases (log-rank, P=0.049; Fig.
2B).
Survival and different level of DNA
amplification, mRNA and protein expression in cetuximab-treated
cases
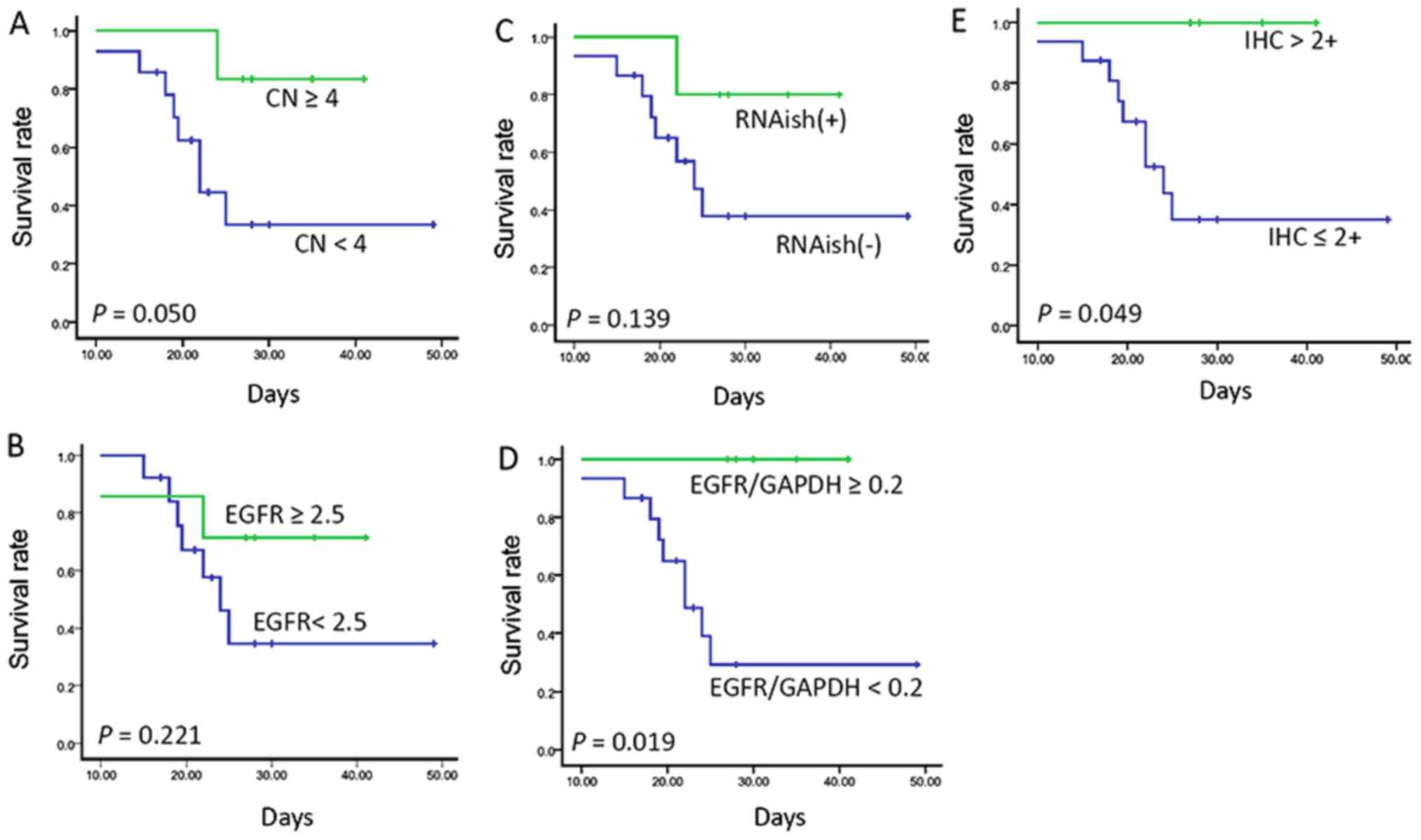
For the 20 cases, 6 were EGFR CN ≥4 as detected by
qPCR, 14 were EGFR/CEP7 ≥1 as detected by FISH, and 7 were EGFR
≥2.5 by FISH. PDX models treated with cetuximab considered CN <4
had shorter survival than those patients with CN ≥4 (median, 22.0
days vs. not reached; P=0.050; Fig.
3A). This difference was statistically significant by qPCR,
while there was no statistically significant difference detected by
FISH (EGFR/FISH, median, 24.0 days vs. not reached, log-rank;
P=0.221; Fig. 3B).
Then, we evaluated EGFR mRNA and protein expression
level and the survival following treatment with cetuximab. Of
these, 5 cases had EGFR mRNA overexpression as detected by RNAish
or qPCR (EGFR/GAPDH). Their survival was longer than that of those
with a lower mean (RNAish, median, 24.0 days vs. not reached,
log-rank; P=0.139; qPCR, log-rank; P=0.019, respectively; Fig. 3C and D). The EGFR protein expression
of 4 patients was IHC 3+; survival of these cases was better than
that of those with a lower expression (log-rank; P=0.049; Fig. 3E).
Survival and cetuximab therapy in
different EGFR copy number subgroup
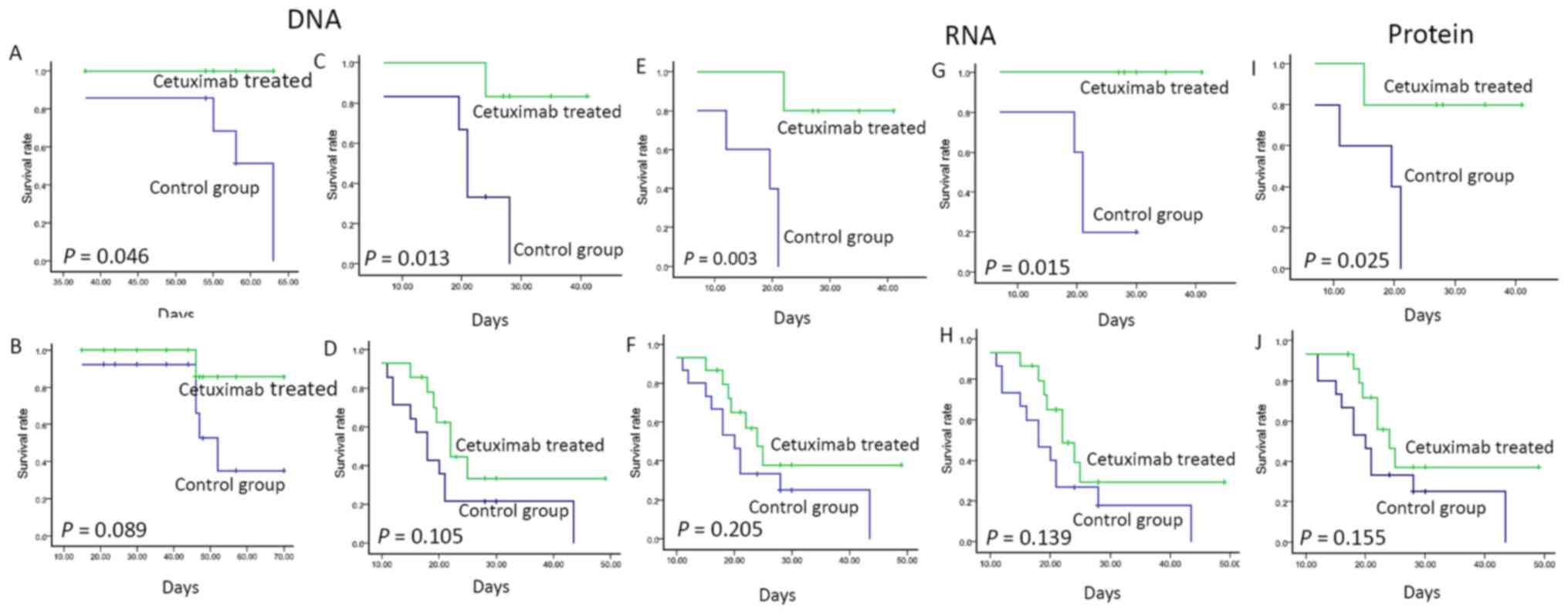
Amplification: for both EGFR/CEP7 ratio value, one
was used as the optimal cut-off value that discriminated between
the GC PDX model with longer survival and those who more probably
may respond to cetuximab-based therapy. As illustrated in Fig. 4A in the cases with EGFR ≥2.5, the
median survival of the cetuximab-treated group was significantly
longer than that of the non-treated group (log-rank; P=0.046).
Similarly, cases with EFGR CN >4 by qPCR showed an increased and
statistically significant benefit in survival (median, 21.0 vs. not
reached; log-rank, P=0.013; Fig.
4C). However, either in the group with EGFR <2.5 by FISH or
EFGR CN <4 by qPCR, the survival rate was not statistically
significant between the cetuximab-treated and control group
(EGFR/FISH, median, 52.0 days vs. not reached, log-rank; P=0.089;
EGFR CN/Q-PCR median, 18.0 vs. 22.0 days, log-rank; P=0.105,
respectively; Fig. 4B and D).
Survival and cetuximab therapy in the
different EGFR mRNA expression level subgroups
The mRNA expression of EGFR was detected by RNAish
and EGFR/GAPDH. In the EGFR expression-positive cases (detected by
RNAish), the median survival of the cetuximab-treated group was
significantly longer than that of the non-treated group (median,
19.5 days vs. not reached; log-rank, P=0.003; Fig. 4E), while in the RNAish-negative
cases, the survival rate did not differ significantly (median
survival, 20.0 vs. 24.0 days; P=0.205; Fig. 4F).
Similarly, in the EGFR/GAPDH ≥0.2
group, those in the cetuximab-treated group showed a longer
survival rate than those of the control cases (median survival;
P=0.015; Fig
4G), while in the EGFR/GAPDH <0.2 group, the
survival rate did not significantly differ in the cetuximab-treated
and control cases (median survival, 18.0 vs. 22 days; P=0.139;
Fig. 4H).
Survival and cetuximab therapy in the
different EGFR protein expression level subgroup
Moreover, the survival rate and cetuximab therapy in
the GC PDX models with different levels of EGFR protein expression
was also investigated. A score of 2+ was used as the optimal EGFR
IHC score cut-off value. In the EGFR protein overexpression cases,
the median survival rate of the cetuximab-treated group was
significantly longer than that of the non-treated group (median,
19.5 days vs. not reached; log-rank, P=0.025; Fig. 4I), while in the group with EGFR
expression IHC score <2 cases, the survival rate did not differ
significantly (median, 20.0 vs. 24.0 days; log-rank, P=0.155;
Fig. 4J).
Combined detection of DNA
amplification, mRNA and protein overexpression and the survival
rate and tumor response to cetuximab therapy
DNA amplification, mRNA and protein expression are
commonly detected by FISH, RNAish and IHC in clinical testing.
Combined detection is more meaningful for drug use. We found that 4
of the patients were positive for combined detection, which was
consistent with the nearly complete response cases; the others were
negative. In the combined positive cases, the median survival rate
of the cetuximab-treated group was significantly longer than the
negative cases (log-rank, P=0.049; data not shown).
Discussion
The potential use of EGFR expression as a marker has
been widely investigated, with conflicting results. In the present
study, our aim was to determine whether the level of EGFR gene
amplification, mRNA and protein level could significantly predict
some benefit in the survival and response to cetuximab in GC
xenografts. EGFR DNA amplification was detected by EGFR/CEP7 ratio
and qPCR, mRNA expression was detected by qPCR, RNAish, EGFR|U219
and protein overexpression was detected by IHC. EGFR protein and
mRNA expression levels allowed us to identify and discriminate
those patients with prolonged survival. We showed, for the first
time to the best of our knowledge, how the level of EGFR gene
amplification, mRNA and protein expression may be used as a
predictive factor for response to cetuximab-based treatment and
also for survival benefit in this subset of GC patients through PDX
models.
Benefit from the addition of an anti-EGFR agent to
chemotherapy could not be confirmed in the phase III trials
comparing chemotherapy with and without anti-EGFR agent, including
the randomized EXPAND and REAL3 trials. An important point is that
neither of these trials selected patients based upon biomarkers
(7,21). Various authors have suggested that
both IHC and FISH should be used to determine the HER2 target
chemotherapy status in GC (22).
However, EGFR status evaluation method mostly focused on the gene
mutations of downstream genes, such as KRAS and BRAF. EGFR protein
expression was found to be associated with the survival rate in GC
(23). In the present study, we
tested the RAS mutation status. There was no RAS mutation in these
tumor samples. Our results showed that cases with high EGFR mRNA
expression and immunohistochemistry score were more prone to
response to cetuximab. EGFR mRNA and protein overexpression were
associated with the survival rate in the cetuximab-treated PDX
models. Moreover, in the PDX models derived from mRNA or protein
overexpression cases, the survival rate of the cetuximab-treated
PDX models was significantly longer than that noted in the control
group, while the survival was not statistically different in the
other cases. The potential use of EGFR expression as a marker has
been widely investigated in other cancers, such as NSCLC, with
conflicting results. Previous report suggested that EGFR
overexpression is associated with improved response, longer time to
progression and improved survival in NSCLC patients treated with
gefitinib (24). Biomarker analysis
of the BR.21 study showed that survival among patients with high
EGFR expression was longer in the erlotinib arm vs. the placebo
arm, whereas a limited advantage of erlotinib treatment was noted
in patients with EGFR IHC-negative tumors (25). This result is similar with ours
detected in GC (25).
Gene copy number gain of EGFR is a poor prognostic
biomarker in GC (8). At present,
there are few studies that have focused on the association of EGFR
gene copy number and efficacy of cetuximab chemotherapy, including
evaluation of EGFR gene copy number as a predictive biomarker for
the efficacy of cetuximab in combination with chemotherapy in the
first-line treatment of recurrent and/or metastatic squamous cell
carcinoma of the head and neck: EXTREME study (15). The association of EGFR and tumor
response to cetuximab should be further studied. In the present
study, similar to Her2 amplification (2), we also found that EGFR gene copy
number was a predictive biomarker for the efficacy of cetuximab in
a GC PDX model, similar with the results in previous studies
(26). Compared to EGFR DNA
amplification, mRNA and protein overexpression may be more accurate
to determine the tumor response to cetuximab. This may be due to
other regulatory methods of EGFR expression, such as
epigenetics.
In summary, the level of EGFR gene amplification
significantly predicted the sensitivity to therapy and the survival
rate in GC cases treated with cetuximab-based chemotherapy. This
result supports the proposed combined use of IHC and in situ
hybridization in this setting. The study of the level of DNA
amplification, mRNA and protein expression as a continuous
biomarker is arguably a more rational approach for selecting
patients more likely to benefit from anti-EGFR-based therapies.
Acknowledgements
The present study was partially supported by the
National Natural Science Foundation of China (no. 81101879), the
National Key Technology R&D Program (2011ZX09307-001-05 and
2014AA020603), and the Beijing Municipal Science and Technology
Commission (Z121100007512010).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gomez-Martin C, Plaza JC, Pazo-Cid R,
Salud A, Pons F, Fonseca P, Leon A, Alsina M, Visa L, Rivera F, et
al: Level of HER2 gene amplification predicts response and
overall survival in HER2-positive advanced gastric cancer treated
with trastuzumab. J Clin Oncol. 31:4445–4452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luo HY, Wei W, Shi YX, Chen XQ, Li YH,
Wang F, Qiu MZ, Li FH, Yan SL, Zeng MS, et al: Cetuximab enhances
the effect of oxaliplatin on hypoxic gastric cancer cell lines.
Oncol Rep. 23:1735–1745. 2010.PubMed/NCBI
|
|
4
|
Pinto C, Di Fabio F, Siena S, Cascinu S,
Llimpe Rojas FL, Ceccarelli C, Mutri V, Giannetta L, Giaquinta S,
Funaioli C, et al: Phase II study of cetuximab in combination with
FOLFIRI in patients with untreated advanced gastric or
gastroesophageal junction adenocarcinoma (FOLCETUX study). Ann
Oncol. 18:510–517. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han SW, Oh DY, Im SA, Park SR, Lee KW,
Song HS, Lee NS, Lee KH, Choi IS, Lee MH, et al: Phase II study and
biomarker analysis of cetuximab combined with modified FOLFOX6 in
advanced gastric cancer. Br J Cancer. 100:298–304. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim C, Lee JL, Ryu MH, Chang HM, Kim TW,
Lim HY, Kang HJ, Park YS, Ryoo BY and Kang YK: A prospective phase
II study of cetuximab in combination with XELOX (capecitabine and
oxaliplatin) in patients with metastatic and/or recurrent advanced
gastric cancer. Invest New Drugs. 29:366–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lordick F, Kang YK, Chung HC, Salman P, Oh
SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, et
al: Arbeitsgemeinschaft Internistische Onkologie and EXPAND
Investigators: Capecitabine and cisplatin with or without cetuximab
for patients with previously untreated advanced gastric cancer
(EXPAND): A randomised, open-label phase 3 trial. Lancet Oncol.
14:490–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Higaki E, Kuwata T, Nagatsuma AK, Nishida
Y, Kinoshita T, Aizawa M5, Nitta H, Nagino M and Ochiai A: Gene
copy number gain of EGFR is a poor prognostic biomarker in
gastric cancer: Evaluation of 855 patients with bright-field dual
in situ hybridization (DISH) method. Gastric Cancer. 19:63–73.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hotz B, Keilholz U, Fusi A, Buhr HJ and
Hotz HG: In vitro and in vivo antitumor activity of cetuximab in
human gastric cancer cell lines in relation to epidermal growth
factor receptor (EGFR) expression and mutational phenotype. Gastric
Cancer. 15:252–264. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: North-East Japan Study Group: Gefitinib or chemotherapy for
non-small-cell lung cancer with mutated EGFR. N Engl J Med.
362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: West Japan Oncology Group: Gefitinib versus cisplatin plus
docetaxel in patients with non-small-cell lung cancer harbouring
mutations of the epidermal growth factor receptor (WJTOG3405): An
open label, randomised phase 3 trial. Lancet Oncol. 11:121–128.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mazières J, Brugger W, Cappuzzo F, Middel
P, Frosch A, Bara I, Klingelschmitt G and Klughammer B: Evaluation
of EGFR protein expression by immunohistochemistry using H-score
and the magnification rule: Re-analysis of the SATURN study. Lung
Cancer. 82:231–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dahabreh IJ, Linardou H, Kosmidis P,
Bafaloukos D and Murray S: EGFR gene copy number as a
predictive biomarker for patients receiving tyrosine kinase
inhibitor treatment: A systematic review and meta-analysis in
non-small-cell lung cancer. Ann Oncol. 22:545–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Licitra L, Mesia R, Rivera F, Remenár E,
Hitt R, Erfán J, Rottey S, Kawecki A, Zabolotnyy D, Benasso M, et
al: Evaluation of EGFR gene copy number as a predictive
biomarker for the efficacy of cetuximab in combination with
chemotherapy in the first-line treatment of recurrent and/or
metastatic squamous cell carcinoma of the head and neck: EXTREME
study. Ann Oncol. 22:1078–1087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang L, Yang J, Cai J, Song X, Deng J,
Huang X, Chen D, Yang M, Wery JP, Li S, et al: A subset of gastric
cancers with EGFR amplification and overexpression respond to
cetuximab therapy. Sci Rep. 3:29922013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang M, Shan B, Li Q, Song X, Cai J, Deng
J, Zhang L, Du Z, Lu J, Chen T, et al: Overcoming erlotinib
resistance with tailored treatment regimen in patient-derived
xenografts from naïve Asian NSCLC patients. Int J Cancer.
132:E74–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Julien S, Merino-Trigo A, Lacroix L,
Pocard M, Goéré D, Mariani P, Landron S, Bigot L, Nemati F,
Dartigues P, et al: Characterization of a large panel of
patient-derived tumor xenografts representing the clinical
heterogeneity of human colorectal cancer. Clin Cancer Res.
18:5314–5328. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schleich N, Po C, Jacobs D, Ucakar B,
Gallez B, Danhier F and Préat V: Comparison of active, passive and
magnetic targeting to tumors of multifunctional
paclitaxel/SPIO-loaded nanoparticles for tumor imaging and therapy.
J Control Release. 194:82–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waddell T, Chau I, Cunningham D, Gonzalez
D, Okines AF, Okines C, Wotherspoon A, Saffery C, Middleton G,
Wadsley J, et al: Epirubicin, oxaliplatin, and capecitabine with or
without panitumumab for patients with previously untreated advanced
oesophagogastric cancer (REAL3): A randomised, open-label phase 3
trial. Lancet Oncol. 14:481–489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fox SB, Kumarasinghe MP, Armes JE, Bilous
M, Cummings MC, Farshid G, Fitzpatrick N, Francis GD, McCloud PI,
Raymond W, et al: Gastric HER2 Testing Study (GaTHER): An
evaluation of gastric/gastroesophageal junction cancer testing
accuracy in Australia. Am J Surg Pathol. 36:577–582. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang D, Liu CY, Shen D, Fan S, Su X, Ye P,
Gavine PR and Yin X: Assessment and prognostic analysis of EGFR,
HER2, and HER3 protein expression in surgically resected gastric
adenocarcinomas. Onco Targets Ther. 8:7–14. 2014.PubMed/NCBI
|
|
24
|
Cappuzzo F, Hirsch FR, Rossi E, Bartolini
S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini
I, et al: Epidermal growth factor receptor gene and protein and
gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer
Inst. 97:643–655. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsao MS, Sakurada A, Cutz JC, Zhu CQ,
Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, et
al: Erlotinib in lung cancer - molecular and clinical predictors of
outcome. N Engl J Med. 353:133–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peled N, Yoshida K, Wynes MW and Hirsch
FR: Predictive and prognostic markers for epidermal growth factor
receptor inhibitor therapy in non-small cell lung cancer. Ther Adv
Med Oncol. 1:137–144. 2009. View Article : Google Scholar : PubMed/NCBI
|