Introduction
ESR1, also known as ER-alpha or ERα, is an important
estrogen receptor (ER) and involves in the gene regulation in
various diseases and biological processes, including breast cancer
(1,2), osteosarcoma (3) and cell growth (4). 17β-estradiol (E2) is one of the most
representative estrogens responsible for the development and
reproductive capability of female characteristics (5). Besides, it participates in the
progression of many diseases, for example, E2 could regulate the
proliferation of breast cancer cells through focal adhesion and
chemokine signaling pathways (6);
through autophagy and apoptosis pathways, E2 plays important roles
in the regulation of renal cell carcinoma growth (7). Furthermore, ER, including ERα and ERβ,
are important media through which E2 involves the regulation of
many biological processes. So, exploration of interactions between
ER and E2 would promote our understanding of many types of complex
diseases and the identification of their potential therapeutic
targets.
Osteosarcoma is one of the most common malignant
neoplasms in children and adolescents with ~20% 5-year overall
survival rate after chemotherapy (8–10). In
primary osteosarcoma, adequate surgical margins could significantly
improve the prognosis, whereas, it is still a challenging surgical
technique (11). Moreover,
metastasis greatly affects prognosis for osteosarcoma due to the
propensity of early metastatic trend and ~80% of osteosarcoma
patients have metastasis at diagnosis (12,13).
Understanding its mechanisms and screening of potential biomarkers
would be helpful for its early diagnosis and improvement of
prognosis. The development of biotechnologies have promoted the
identification of diagnosis and therapeutic targets for
osteosarcoma. For example, microRNA-491-5p was considered to
suppress cell proliferation and promote apoptosis via FOXP4 in the
report of Yin et al (14)
through the quantification of miR-491-5p expression and cell
apoptosis; through immunohistochemistry and statistic analysis,
Zhao et al (15) identified
HIF1 as an independent prognostic biomarker in osteosarcoma;
furthermore, Zhou et al (16) even indicated that the level of
miR-199a-5p in serum/plasma might serve as biomarker for
osteosarcoma.
Estrone and estriol, also known as E1 and E3, are
another type of estrogens which have been proved to be involved in
the progression of several diseases, including osteosarcoma. For
example, Mochizuki et al proved that E3 is compatible with
E2 to induce the expression of IGF-1 mRNA and then affect the
proliferative potential of osteosarcoma cells (17). Besides, E1 sulfate represents the
most abundant estrogen in circulation of adults and plays a role in
human bone maturation and homeostasis (18), and it has been shown to influence
the progression of many types of cancers, such as colorectal cancer
(19) and breast cancer (20), so it might be also associated with
the development of osteosarcoma.
There are also several studies on E2 and ESR1 in
osteosarcoma (3,21,22).
While, no study has focused on the genome binding profiles of ESR1
with the presence of E2 and the E2-regulated genes through ESR1
simultaneously. In this study, a combined analysis of ESR1
chromatin immunoprecipitation coupled with high-throughput
sequencing (ChIP-seq) with the presence of E2 and gene expression
profiles of E2-regulated through ESR1 was conducted. This would
induce several potential target genes of ESR1 in osteosarcoma, and
here, the E2-regulated genes in balance. This would be helpful for
the identification of E2 targeted molecules in osteosarcoma and the
development of corresponding drugs.
Materials and methods
Genome binding and expression profile
datasets
All of the datasets in this study were obtained from
the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). For the analysis
of genome binding of ESR1, the data of Chen et al were used
with the accession number of GSE26110 which contains the SRC-1 and
ESR1 genome-wide binding profiles with or without the presence of
E2 in U2OS osteosarcoma cells. Another dataset (GSE1153), deposited
by Stossi et al (22) was
composed of U2OS cells expressing ERα (ESR1) or ERβ and treated
with vehicle or the same amount of E2 for 4, 8, 24 and 48 h, was
used for the determination of E2 targeted genes. The transcriptome
expression level of the samples were quantified via GPL91 [HG_U95A]
Affymetrix Human Genome U95A Array, a commercial affymetrix gene
microarray.
ChIP-seq data analysis
We first conducted quality control for the raw
sequencing data in fastq with FastQC, a quality control tool for
high-throughput sequence data. Bowtie 2 (23) was used for the fast and sensitive
read alignment with the default parameters setting after the
removal of reads with poor quality, i.e., length <20 and more
than 3 bases with the quality score <20 in this study. After the
removal of duplicate alignments, the binding sites, referred as
peak hereafter, of ESR1 were identified via the Model-based
Analysis for ChIP-seq (MACS) version 2 (24), a widely used peak caller for
transcription factor and histone modifier developed by Feng et
al. Finally, ChIPseeker (25)
and deepTools (26) were used for
the assignment of genomic features, such as gene ID and relative
location to transcription start site (TSS) to the peaks and
visualization of binding profiles of ESR1 respectively.
Microarray data analysis
Gene expression profile analysis mainly involved two
steps, i.e., preprocessing and identification of differential
expression genes (DEGs). Briefly, raw CEL data was imported to R
software and normalized through affy package (27) and the expression value was
logarithmically transformed based on 2 and summarized to gene
level; the identification of DEGs was performed through limma
package (28) with the thresholds
of fold change >1.41 and Bonferroni adjusted P-value
<0.05.
Functional enrichment analysis
To explore the functions involving DEGs, we
performed functional enrichment analysis through the Database for
Annotation, Visualization and Integrated Discovery (DAVID,
https://david.ncifcrf.gov/) (29). We considered Gene Ontology (GO)
terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
with P-value <0.05 and minimum gene hits >2 as significantly
enriched.
miRNA-Gene network analysis
In this study, the overlaps between target genes of
ESR1 in U2OS cells with the presence of E2 and DEGs in E2 treated
U2OS cells with expressed ESR1 were considered E2-regulated genes
through ESR1 in osteosarcoma. To further explore their roles in
osteosarcoma, we performed miRNA-Gene regulation analysis through
miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
(30) and the regulation
relationships were screened out only when they were found in four
out of the miRanda, miRDB, miRWalk, RNA22 and TargetScan databases.
Besides, Cytoscape (31), an open
source platform for complex network analysis and visualization, was
used for the presentation of miRNA-Gene network.
Results
ChIP-seq data analysis
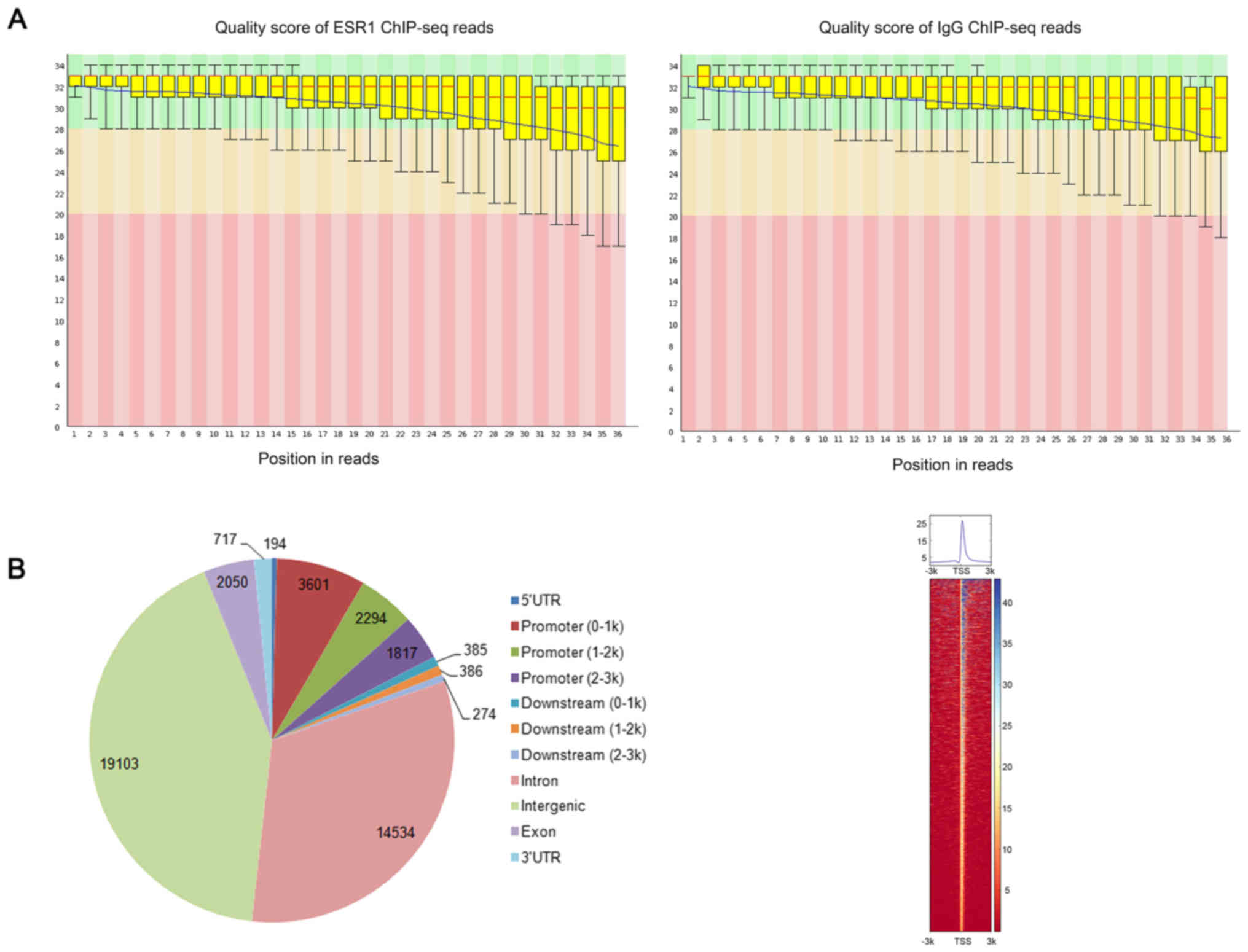
Quality control indicated high quality reads
suitable for the following analysis (Fig. 1A). Read alignments resulted in
88.76% (11,409,959 out of 12,854,413 reads) and 75.98% (9,744,663
out of 12,825,873 reads) overall alignment rate for the ChIP-seq
data of ESR1 and IgG control, respectively. Through MACS, we
obtained a total of 45,355 ESR1-specific binding peaks and their
genomic distribution are shown in Fig.
1B. Moreover, 7,712 peaks were assigned to the promoters of
4,541 unique genes, i.e., up- and down-stream 3,000 bp surrounding
the TSS, which were considered as the most confident target genes
of ESR1 and abbreviated as PGenes in this study. Besides,
significant enrichment of ESR1 binding peaks was obtained
surrounding TSS of its target genes and shown in Fig. 1C.
Microarray data analysis
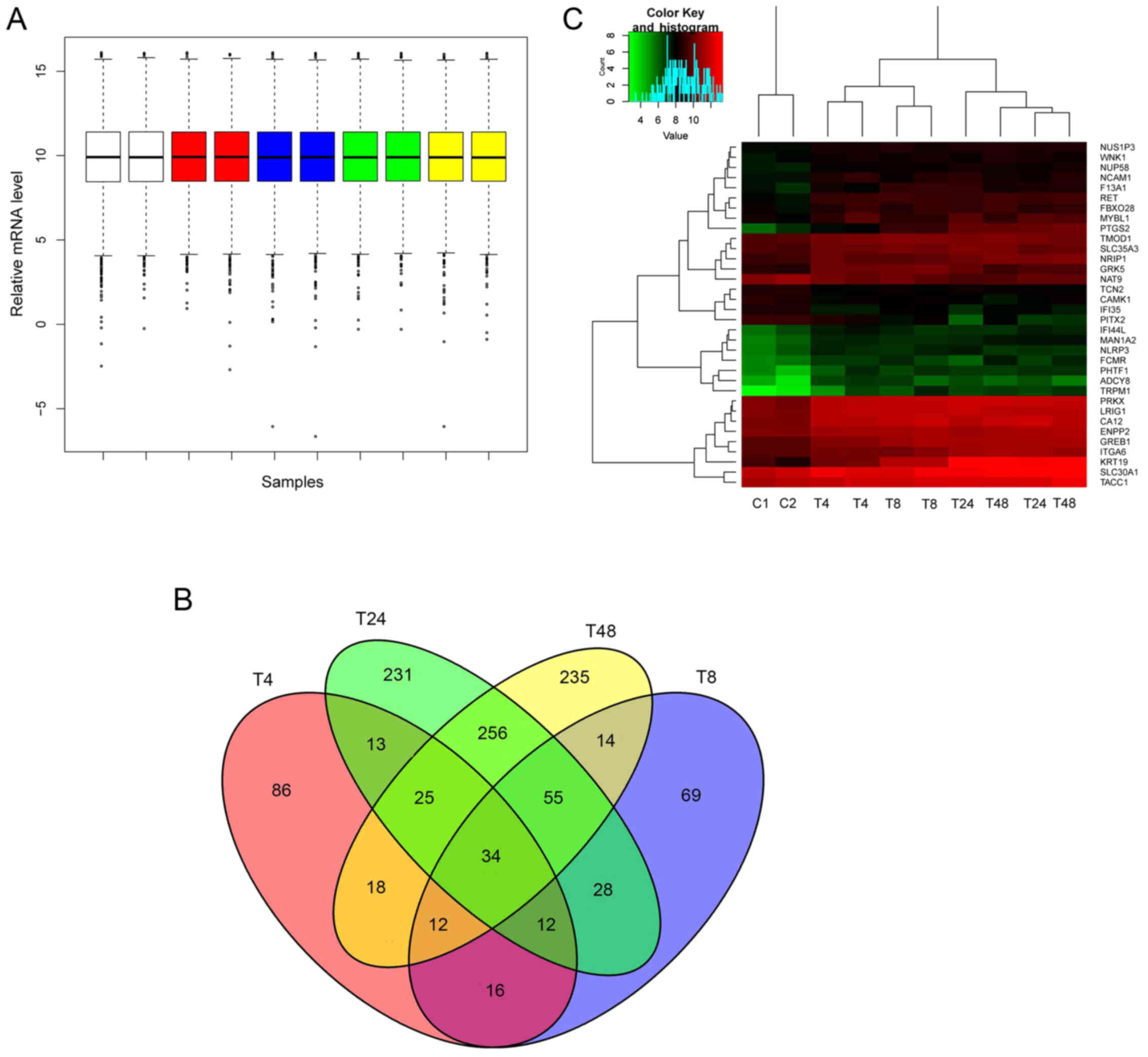
Preprocessing step resulted in comparable overall
expression values among all of the samples (Fig. 2A). DEGs in 4 h (DEG4), 8 h (DEG8),
24 h (DEG24) and 48 h (DEG48) E2 treated U2OS cells expressing ESR1
(U2OS-ERα) compared with U2OS-ERα treated with vehicle were
obtained and cross analyzed. As a result, we identified 216, 240,
654 and 649 hits for DEG4, DEG8, DEG24 and DEG48, respectively, and
34 overlaps were also obtained (Fig.
2B). The 34 overlaps are shown in Table I. Fig.
2C illustrated the supervised two-way hierarchical clustering
of the 34 overlaps and samples. As expected, 24 and 48 h E2 treated
U2OS-ERα samples were clustered together and the 4 h, and 8 h
treated, and vehicle treated U2OS-ERα were separately clustered in
their own groups.
 | Table I.The 34 overlapped genes among DEG4,
DEG8, DEG24 and DEG48. |
Table I.
The 34 overlapped genes among DEG4,
DEG8, DEG24 and DEG48.
| Gene name | log2FC_DEG4 | log2FC_DEG8 | log2FC_DEG24 | log2FC_DEG48 |
|---|
| ADCY8 | 2.68 | 2.31 | 2.36 | 1.62 |
| CA12 | 1.12 | 1.46 | 1.38 | 1.37 |
| CAMK1 | −0.72 | −0.64 | −0.75 | −0.87 |
| ENPP2 | 0.69 | 0.77 | 0.75 | 0.87 |
| F13A1 | 0.86 | 1.68 | 1.46 | 1.20 |
| FBXO28 | 0.95 | 0.98 | 0.95 | 1.18 |
| FCMR | 1.38 | 1.47 | 0.90 | 1.95 |
| GREB1 | 0.80 | 1.35 | 1.42 | 1.50 |
| GRK5 | 1.36 | 1.56 | 0.99 | 0.60 |
| IFI35 | −1.18 | −0.54 | −1.13 | −0.94 |
| IFI44L | 1.45 | 1.02 | 1.11 | 1.00 |
| ITGA6 | 0.56 | 0.72 | 1.44 | 1.36 |
| KRT19 | 1.22 | 2.46 | 4.10 | 4.08 |
| LRIG1 | 1.14 | 1.27 | 1.22 | 1.12 |
| MAN1A2 | 1.51 | 1.33 | 1.67 | 1.86 |
| MYBL1 | 1.14 | 0.71 | 1.52 | 1.06 |
| NAT9 | −0.66 | −0.76 | −1.08 | −1.05 |
| NCAM1 | 1.17 | 0.81 | 0.93 | 0.92 |
| NLRP3 | 1.32 | 1.31 | 1.38 | 0.99 |
| NRIP1 | 0.75 | 0.61 | 1.09 | 1.34 |
| NUP58 | 0.75 | 0.84 | 0.93 | 1.01 |
| NUS1P3 | 0.61 | 0.73 | 0.78 | 0.87 |
| PHTF1 | 1.90 | 2.03 | 2.17 | 2.11 |
| PITX2 | −0.57 | −1.21 | −2.69 | −1.65 |
| PRKX | 1.22 | 1.34 | 1.53 | 1.49 |
| PTGS2 | 1.39 | 2.58 | 3.62 | 3.72 |
| RET | 1.40 | 1.48 | 1.36 | 1.63 |
| SLC30A1 | 0.70 | 0.71 | 1.17 | 1.20 |
| SLC35A3 | 0.66 | 0.82 | 0.77 | 0.80 |
| TACC1 | 0.58 | 0.67 | 0.89 | 1.09 |
| TCN2 | −0.73 | −0.58 | −0.50 | −0.60 |
| TMOD1 | 0.71 | 0.89 | 0.78 | 0.73 |
| TRPM1 | 2.84 | 3.72 | 3.56 | 3.82 |
| WNK1 | 0.63 | 0.71 | 0.63 | 0.73 |
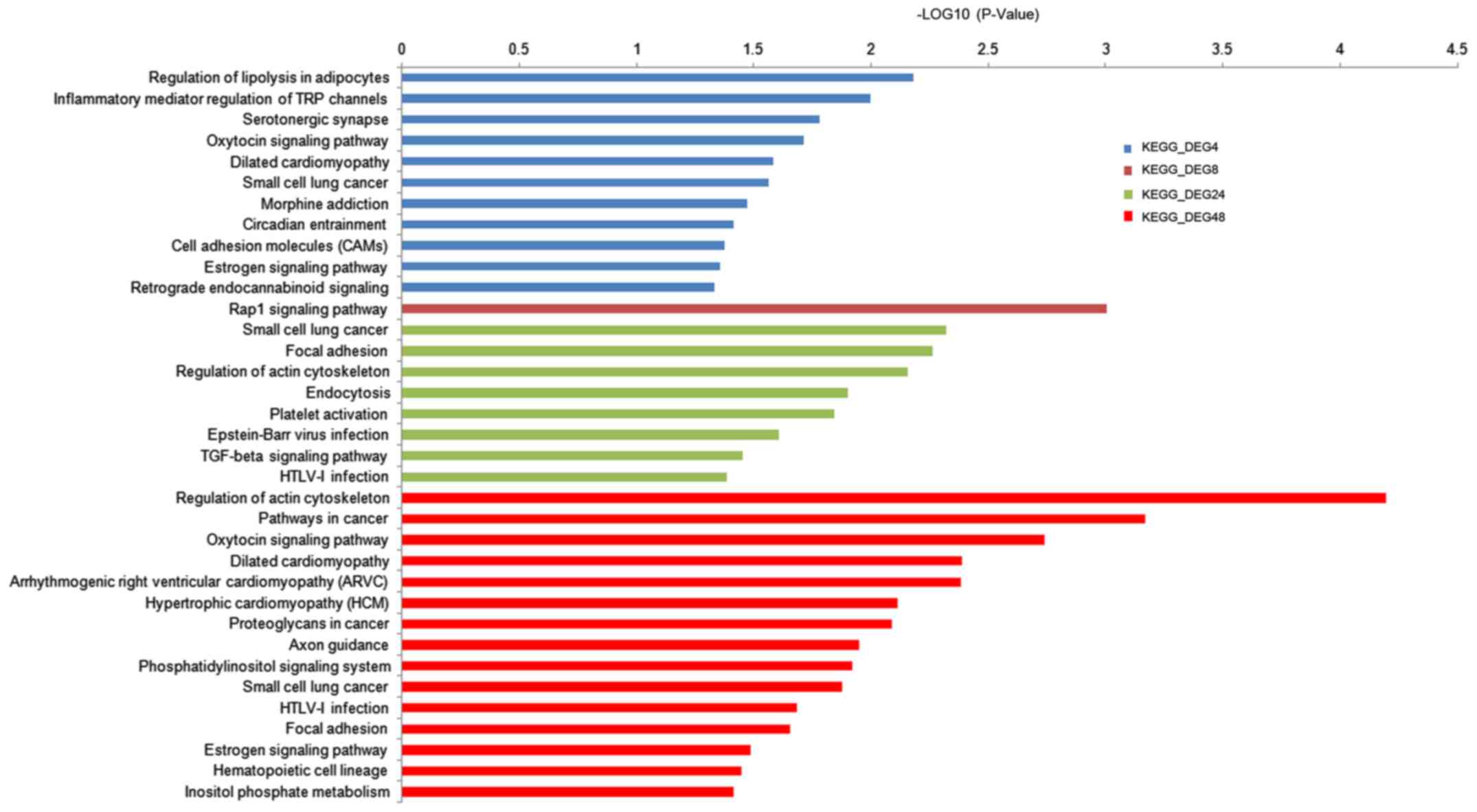
Enriched functions
We identified 50, 44, 175 and 171 significantly
enriched GO terms for DEG4, DEG8, DEG24 and DEG48, respectively.
Most of the GO terms were closely related with the processes of
cancer progression, cell adhesion, RNA binding and so on. Besides,
several estrogen, cancer and inflammatory associated KEGG pathways
were also obtained and shown in Fig.
3.
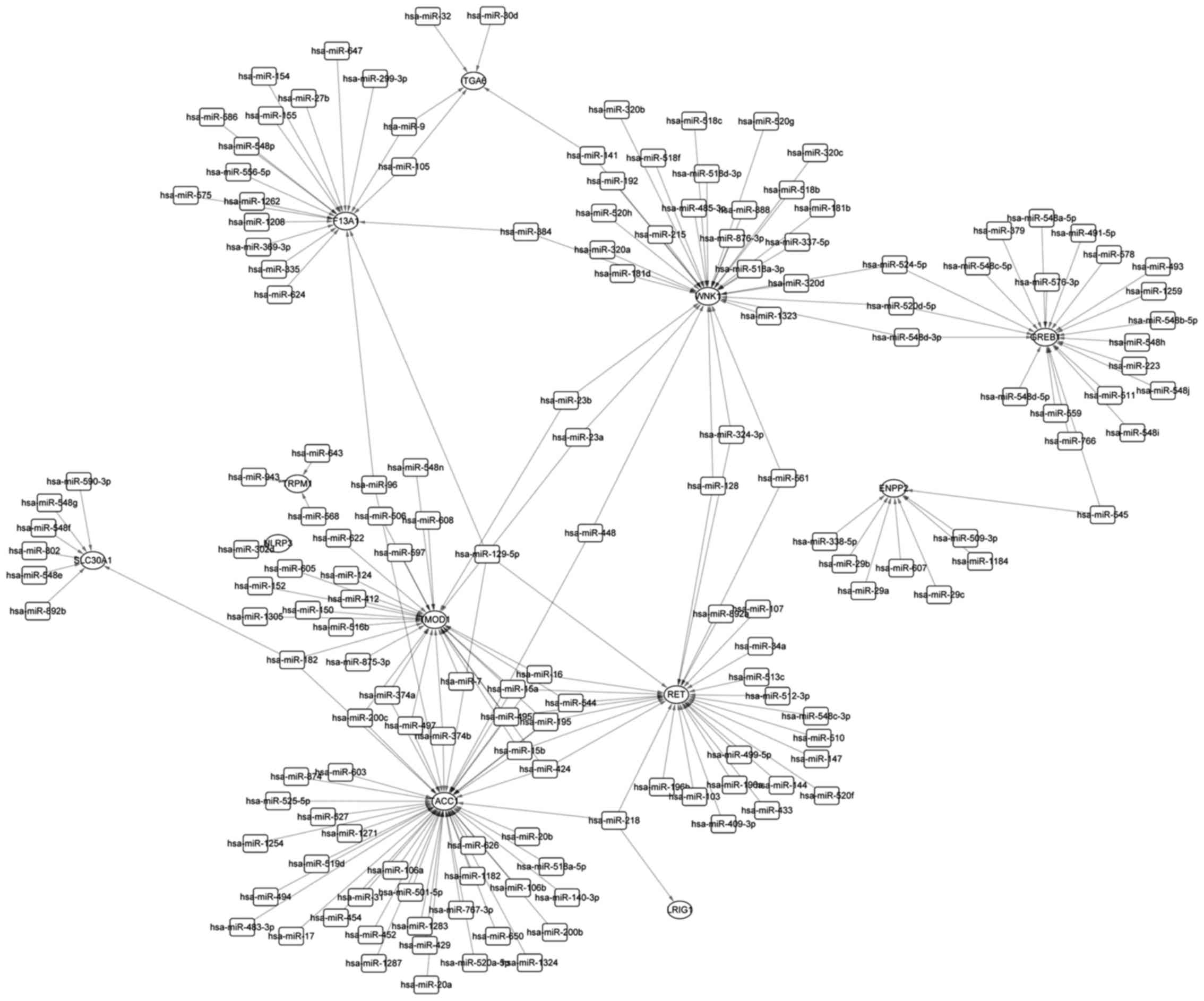
miRNA-Gene network
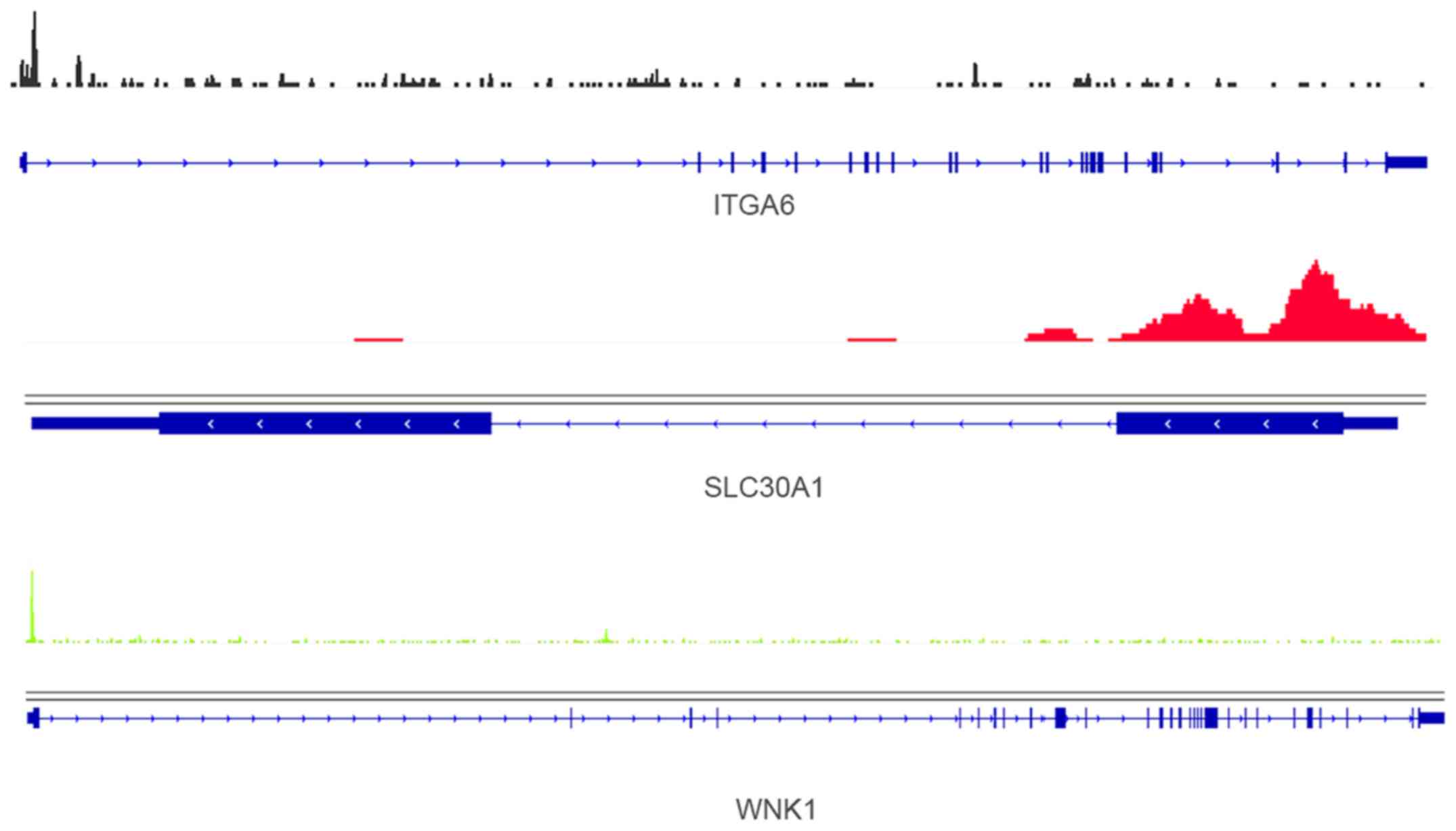
Cross analysis of PGenes and the 34 overlapped DEGs
identified 15 overlaps, i.e., RET, TRPM1, CA12, KRT19, GREB1,
F13A1, TACC1, LRIG1, ADCY8, TMOD1, SLC30A1, NLRP3, WNK1, ITGA6 and
ENPP2. Three examples of ESR1 binding peaks in the promoter of
ITGA6, SLC30A1 and WNK1 further confirmed their association with
ESR1 (Fig. 4).
miRNA is a kind of non-coding RNA with the length of
~22 nt. It can regulate gene expression through preventing
transcription or degradating protein. We further explored the
miRNA-Gene regulation relationships of the 15 overlaps for the
comprehensive understanding of gene regulation patterns in E2
treated U2OS-ERα cells. As a result, 12 out of the 15 overlaps
except CA12, KRT19 and ADCY8 were found to be regulated by one or
more miRNAs. Fig. 5 illustrates the
miRNA-Gene regulation network.
Discussion
Osteosarcoma is the most common bone tumor with high
morbidity and mortality. E2 was found to play important roles in
osteosarcoma through regulating physiological processes and gene
expression via ESR1 by many studies (3,32).
While, the combined analysis of E2-mediated gene expression and
regulation is still limited. In this study, we conducted gene
microarray and ChIP-seq analysis in E2 treated U2OS-ERα cell lines,
which would be useful to promote our understanding of the
E2-mediated gene regulation in osteosarcoma.
E2-activated ESR1 could directly or indirectly bind
to its target genes and then regulate the particular biological
processes or gene expression. In osteosarcoma, E2 is necessary for
the translocation of ESR1 to the nuclei. For example, Hu et
al showed that ESR1 could promote the binding of Sp1 to the
promoter of MALAT1, a long non-coding RNA, in U2OS cells to affect
physiological processes only with E2 stimulation (32); Tübel et al even proved that
the methylation status of ESR1 was affected by E2 (3). In this study, the ChIP-seq analysis of
E2 stimulated U2OS-ERα cell identified the potential target genes
of E2 mediated through ESR1. Besides, cross analysis with gene
microarray could further screen out more reliable target genes for
osteosarcoma.
Functional enrichment analysis resulted in the
identification of cancer, cell activity-related KEGG pathways,
which further confirmed our results. Estrogen signaling pathway was
found to be significantly enriched in DEG4 and DEG48, which
indicated roles of E2 in the activation of ESR1.
E2 could also regulate the expression level of
microRNA (miRNA) and affect cell proliferation, migration and
invasion in osteosarcoma. In turn, E2-mediated genes might also be
regulated by miRNA. In this study, we performed miRNA-Gene
regulation analysis for overlaps between PGenes and overlapped
genes among the four lists of DEGs. Finally, 12 out of the 15
overlaps were found to be regulated by miRNA and ACC1 with the
highest degree. ACC1 could act as anti- and pro-apoptotic
regulators which involves in several cellular activities, including
embryonic development, homeostasis, and tumorigenesis. Besides, it
has been proved as potential biomarker in many types of cancers,
such as gliomas, and melanoma (33,34).
Significant enrichment of ESR1-specific binding peaks were
identified in the promoter of WNK1 (Fig. 4, bottom), which was regulated by 31
miRNAs in this study. WNK1 and ESR1 were found to have differential
expression in osteoporosis simultaneously (35). GREB1 was found with differential
expression in U2OS-ERα cells at 4, 8, 24 and 48 h after E2 treated
and regulated by E2 through ESR1. Consistent with our study, Lin
and Lei reported that GREB1 was a estrogen-responsive gene and with
two canonical estrogen response elements (36).
In conclusion, this study identified the potential
E2-mediated genes through ESR1 in osteosarcoma via the combined
analysis of ChIP-seq and gene microarray analysis. It could be
helpful for the identification of therapeutic targets for
osteosarcoma.
Glossary
Abbreviations
Abbreviations:
|
GEO
|
Gene Expression Omnibus
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
U2OS-ERα
|
U2OS cells expressing ESR1
|
|
miRNA
|
microRNA
|
|
E2
|
17β-estradiol
|
References
|
1
|
Flodrova D, Toporova L, Macejova D,
Lastovickova M, Brtko J and Bobalova J: A comparative study of
protein patterns of human estrogen receptor positive (MCF-7) and
negative (MDA-MB-231) breast cancer cell lines. Gen Physiol
Biophys. 35:387–392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li T, Zhao J, Yang J, Ma X, Dai Q, Huang
H, Wang L and Liu P: A meta-analysis of the association between
ESR1 genetic variants and the risk of breast cancer. PLoS One.
11:e01533142016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tübel J, Kuntz L, Marthen C, Schmitt A,
Wiest I, Von Eisenhart-Rothe R, Jeschke U and Burgkart R:
Methylation of the ER-alpha promoter is influenced by its ligand
estrogen in osteosarcoma cells SAOS-2 in vitro. Anticancer Res.
36:3199–3204. 2016.PubMed/NCBI
|
|
4
|
Meng D, Li Z, Ma X, Fu L and Qin G:
MicroRNA-1280 modulates cell growth and invasion of thyroid
carcinoma through targeting estrogen receptor α. Cell Mol Biol
(Noisy-le-grand). 62:1–6. 2016.
|
|
5
|
Guo JJ, Yang DP, Tian X, Vemuri VK, Yin D,
Li C, Duclos RI Jr, Shen L, Ma X, Janero DR, et al: 17β-estradiol
(E2) in membranes: Orientation and dynamic properties. Biochim
Biophys Acta. 1858:344–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huan J, Wang L, Xing L, Qin X, Feng L, Pan
X and Zhu L: Insights into significant pathways and gene
interaction networks underlying breast cancer cell line MCF-7
treated with 17β-estradiol (E2). Gene. 533:346–355. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen KC, Lin CM, Huang CJ, Chen SK, Wu ST,
Chiang HS and Ku WC: Dual roles of 17-β estradiol in estrogen
receptor-dependent growth inhibition in renal cell carcinoma.
Cancer Genomics Proteomics. 13:219–230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He X, Gao Z, Xu H, Zhang Z and Fu P: A
meta-analysis of randomized control trials of surgical methods with
osteosarcoma outcomes. J Orthop Surg. 12:52017. View Article : Google Scholar
|
|
9
|
Zhang W, Han S and Sun K: Combined
analysis of gene expression, miRNA expression and DNA methylation
profiles of osteosarcoma. Oncol Rep. 37:1175–1181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ren W and Gu G: Prognostic implications of
RB1 tumour suppressor gene alterations in the clinical outcome of
human osteosarcoma: A meta-analysis. Eur J Cancer Care (Engl).
27–October;2015.(Epub ahead of print). PubMed/NCBI
|
|
11
|
Wang Y, Guo W, Shen D, Tang X, Yang Y, Ji
T and Xu X: Surgical treatment of primary osteosarcoma of the
sacrum: A case series of 26 patients. Spine. Dec 16–2016.(Epub
ahead of print).
|
|
12
|
Zhang Z, Wang F, Li Q, Zhang H, Cui Y, Ma
C, Zhu J, Gu X and Sun Z: CD151 knockdown inhibits osteosarcoma
metastasis through the GSK-3β/β-catenin/MMP9 pathway. Oncol Rep.
35:1764–1770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pu Y, Zhao F, Cai W, Meng X, Li Y and Cai
S: MiR-193a-3p and miR-193a-5p suppress the metastasis of human
osteosarcoma cells by down-regulating Rab27B and SRR, respectively.
Clin Exp Metastasis. 33:359–372. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin Z, Ding H, He E, Chen J and Li M:
Up-regulation of microRNA-491-5p suppresses cell proliferation and
promotes apoptosis by targeting FOXP4 in human osteosarcoma. Cell
Prolif. Oct 5–2016.(Epub ahead of print).
|
|
15
|
Zhao H, Wu Y, Chen Y and Liu H: Clinical
significance of hypoxia-inducible factor 1 and VEGF-A in
osteosarcoma. Int J Clin Oncol. 20:1233–1243. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou G, Lu M, Chen J, Li C, Zhang J, Chen
J, Shi X and Wu S: Identification of miR-199a-5p in serum as
noninvasive biomarkers for detecting and monitoring osteosarcoma.
Tumour Biol. 36:8845–8852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mochizuki S, Yoshida S, Yamanaka Y, Matsuo
H and Maruo T: Effects of estriol on proliferative activity and
expression of insulin-like growth factor-I (IGF-I) and IGF-I
receptor mRNA in cultured human osteoblast-like osteosarcoma cells.
Gynecol Endocrinol. 20:6–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Muir M, Romalo G, Wolf L, Elger W and
Schweikert HU: Estrone sulfate is a major source of local estrogen
formation in human bone. J Clin Endocrinol Metab. 89:4685–4692.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gilligan LC, Gondal A, Tang V, Hussain MT,
Arvaniti A, Hewitt AM and Foster PA: Estrone sulfate transport and
steroid sulfatase activity in colorectal cancer: Implications for
hormone replacement therapy. Front Pharmacol. 8:1032017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robarge JD, Desta Z, Nguyen AT, Li L,
Hertz D, Rae JM, Hayes DF, Storniolo AM, Stearns V, Flockhart DA,
et al: Effects of exemestane and letrozole therapy on plasma
concentrations of estrogens in a randomized trial of postmenopausal
women with breast cancer. Breast Cancer Res Treat. 71:453–461.
2017. View Article : Google Scholar
|
|
21
|
Fang D, Yang H, Lin J, Teng Y, Jiang Y,
Chen J and Li Y: 17β-estradiol regulates cell proliferation, colony
formation, migration, invasion and promotes apoptosis by
upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell
MG-63 in an estrogen receptor-independent manner. Biochem Biophys
Res Commun. 457:500–506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stossi F, Barnett DH, Frasor J, Komm B,
Lyttle CR and Katzenellenbogen BS: Transcriptional profiling of
estrogen-regulated gene expression via estrogen receptor (ER) alpha
or ERbeta in human osteosarcoma cells: Distinct and common target
genes for these receptors. Endocrinology. 145:3473–3486. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng J, Liu T, Qin B, Zhang Y and Liu XS:
Identifying ChIP-seq enrichment using MACS. Nat Protoc.
7:1728–1740. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu G, Wang LG and He QY: ChIPseeker: An
R/Bioconductor package for ChIP peak annotation, comparison and
visualization. Bioinformatics. 31:2382–2383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ramírez F, Dündar F, Diehl S, Grüning BA
and Manke T: deepTools: A flexible platform for exploring
deep-sequencing data. Nucleic Acids Res. 42W1:W187–W191. 2014.
View Article : Google Scholar
|
|
27
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy - analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for Annotation,
Visualization, and Integrated Discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
30
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - Database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu Q, Li S, Chen C, Zhu M, Chen Y and Zhao
Z: 17β-Estradiol treatment drives Sp1 to upregulate MALAT-1
expression and epigenetically affects physiological processes in
U2OS cells. Mol Med Rep. 15:1335–1342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
You G, Feng L, Yan W, Zhang W, Wang YZ, Li
SW, Li SW, Li GL, Song YJ, Kang CS, et al: BCL2A1 is a potential
biomarker for postoperative seizure control in patients with
low-grade gliomas. CNS Neurosci Ther. 19:882–888. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hind CK, Carter MJ, Harris CL, Chan HT,
James S and Cragg MS: Role of the pro-survival molecule Bfl-1 in
melanoma. Int J Biochem Cell Biol. 59:94–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiao P, Chen Y, Jiang H, Liu YZ, Pan F,
Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR, et al: In vivo
genome-wide expression study on human circulating B cells suggests
a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J
Bone Miner Res. 23:644–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin J and Lei Z: Chromatin
immunoprecipitation with estrogen receptor 1 and the promoter of
Greb1 in TM4 sertoli cells. Methods Mol Biol. 1366:67–77. 2016.
View Article : Google Scholar : PubMed/NCBI
|