Introduction
Hepatocellular carcinoma (HCC) is the fifth most
prevalent cancer and the third-leading cause of cancer-related
deaths worldwide (1). Despite great
progress in systemic management and treatment in recent years, the
5-year survival rate is still poor, ~15%, where locoregional and
distant recurrences remain primary issues due to widespread
intrahepatic and extrahepatic metastases (2). Therefore, it is of great urgency to
explore the molecular mechanisms responsible for HCC development
and progression, which may help to identify new antitumor
strategies against HCC.
Cancer stem cells (CSCs) are a minority cell
population within tumors and are characterized by unlimited
proliferation as well as the abilities of self-renewal and
differentiation into heterogeneous lineages of cancer cells that
constitute the major tumor population (3,4).
Emerging evidence has demonstrated that CSCs play a crucial role in
the progression of different types of cancer (5,6). The
existence of CSCs was first reported in acute myeloid leukemia
(7), and broad identification was
further demonstrated in several solid tumors, including colon
(8), pancreas (9) and breast cancer (10), indicating the ubiquitous existence
of CSCs in tumors. Furthermore, increasing evidence has suggested
that CSCs are the critical initiators of HCC. Lee et al
reported that CD24+ cells drive tumor initiation through
STAT3-mediated NANOG regulation in HCC (11). Moreover, a monoclonal antibody
against CSCs, 1B50-1, decreased self-renewal and tumor formation
capacities and induced apoptosis of CSCs in HCC (12). This evidence indicated that
therapeutic methods targeting CSCs may be efficient avenues in the
treatment of HCC.
MicroRNAs (miRNAs) are non-coding 17- to
25-nucleotide-long RNAs that function by binding to the
3′-untranslated (3UTR) region of downstream mRNAs and regulate gene
expression post-transcriptionally (13). Physiologically, miRNAs play
important roles in various cellular functions, including cell
apoptosis, proliferation and differentiation (14). Furthermore, miRNAs also play a
pivotal role in the stemness maintenance of CSCs, including HCC
(15,16). For example, Ma et al reported
that microRNA expression profiling of CD133+ and
CD133− cells from human HCC clinical specimens and cell
lines identified an overexpression of miR-130b in CD133+
CSCs. Ectopic expression of miR-130b in CD133− cells
enhanced self-renewal ability and tumorigenicity in vivo.
Conversely, antagonizing miR-130b in CD133+ CSCs
exhibited an opposing effect (17).
In addition, another study indicated that miR-181, which was found
to be overexpressed in EpCAM+ HCC cells isolated from
AFP+ tumors, promoted tumor-initiating ability. Notably,
inhibition of miR-181 led to a decrease in tumor-initiating ability
(18). Therefore, these studies
demonstrated that miRNAs play important roles in regulating
CSC-like phenotypes in HCC.
In the present study, we found that miR-217 was
upregulated in HCC tissue samples and cells. Moreover, upregulation
of miR-217 enhanced the stem cell properties of HCC cells via DKK1
targeting. Notably, the pro-CSC-like phenotype role of miR-217 was
attenuated by overexpression of dickkopf-1 (DKK1) in HCC cells.
Thus, our results revealed a novel mechanism by which miR-217
promoted a CSC-like phenotype in HCC, and anti-HCC therapy
targeting miR-217 may be a potential therapeutic strategy for the
treatment of HCC.
Materials and methods
Cell lines and cell culture
The human HCC cell lines 97H, HepG2, QGY-7703,
Hep3B, PLC, Huh7 and SMMC7721 were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA), and all human HCC
cell lines were cultured as described in the ATCC protocol. Human
liver immortal cell line L02 was purchased from Biomics
Biotechnologies (Nantong, China). The cell lines were maintained in
RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (FBS) (HyClone, Logan, UT, USA) and 100 U/ml
penicillin plus 100 µg/ml streptomycin. The cells were grown in a
humidified atmosphere of 5% CO2 at 37°C.
Plasmids, transfection and generation
of stable cell lines
The miR-217-expression plasmid was generated by
cloning the genomic pre-miR-217 gene, with a 300-bp sequence on
each flanking side, into retroviral-transfer plasmid pMSCV-puro
(Clontech Laboratories, Inc., Mounatin View, CA, USA) to generate
plasmid pMSCV-miR-217. pMSCV-miR-217 was cotransfected with the pIK
packaging plasmid in 293FT cells using the standard calcium
phosphate transfection method, as previously described (19). Thirty-six hours after the
co-transfection, supernatants were collected and incubated with
cells that were to be infected for 24 h in the presence of
Polybrene (2.5 µg/ml). After infection, puromycin (1.5 µ/ml) was
used to select stably transduced cells over a 10-day period. The
3′UTR of DKK1 was amplified and cloned downstream to the luciferase
gene in a modified pGL3 control vector. The reporter plasmids
containing wild-type (CCTTTGATC; TOPflash) or mutated (CCTTTGGCC;
FOPflash) TCF/LEF DNA binding sites were purchased from Upstate
Biotechnology. AntagomiR-217, small interfering RNA (siRNA) for
DKK1 knockdown, overexpressing DKK1 plasmid and respective control
RNA were synthesized and purified by RiboBio Co., Ltd. (Guangzhou,
China). Transfection of miRNAs, siRNAs and plasmids was performed
using Lipofectamine 3000 (Life Technologies, Grand Island, NY, USA)
according to the manufacturer's instructions.
RNA extraction, reverse transcription
and real-time RT-PCR
Total RNA was extracted from cultured cells using
the mirVana miRNA Isolation kit (Ambion Life Technologies,
Carlsbad, CA, USA). cDNA was synthesized from total RNA with
specific stem-loop primers and the TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems Life Technologies, Foster
City, CA, USA). The expression of miRNAs was analyzed by real-time
PCR using the TaqMan MicroRNA Assay kit (Applied Biosystems Life
Technologies). Detection of mRNA was performed as previously
described (20). The sequences of
the primers are listed in Table
I.
 | Table I.Probe sequences used for real-time
PCR. |
Table I.
Probe sequences used for real-time
PCR.
| Name |
| Sequence (5′ to
3′) |
|---|
| DKK1 | F |
TTTCCTCAATTTCTCCTCGG |
|
| R |
ATGCGTCACGCTATGTGCT |
| c-Myc | F |
CACCGAGTCGTAGTCGAGGT |
|
| R |
TTTCGGGTAGTGGAAAACCA |
| BMI1 | F |
TCGTTGTTCGATGCATTTCT |
|
| R |
CTTTCATTGTCTTTTCCGCC |
| OCT4 | F |
GGTTCTCGATACTGGTTCGC |
|
| R |
GTGGAGGAAGCTGACAACAA |
| Nanog | F |
ATGGAGGAGGGAAGAGGAGA |
|
| R |
GATTTGTGGGCCTGAAGAAA |
| Sox2 | F |
GCTTAGCCTCGTCGATGAAC |
|
| R |
AACCCCAAGATGCACAACTC |
| TCF1 | F |
GACTTGACCATCTTCGCCAC |
|
| R |
CCTCAAAGAGCTGGAGAACCT |
| LEF1 | F |
CACTGTAAGTGATGAGGGGG |
|
| R |
TGGATCTCTTTCTCCACCCA |
| GAPDH | F |
AGAGGCAGGGATGATGTTCTG |
|
| R |
AGAGGCAGGGATGATGTTCTG |
Patients and tumor tissues
A total of 58 paraffin-embedded, archived separate
HCC tissues and 6 paired HCC tissues with the matched adjacent
normal tissues were obtained during surgery at the First Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China) between 2006
and 2010. Patients were diagnosed based on clinical and
pathological evidence, and the specimens were immediately
snap-frozen and stored in liquid nitrogen tanks. For the use of
these clinical materials for research purposes, prior informed
patient consent and approval from the Institutional Research Ethics
Committee were obtained.
Western blotting
Western blotting was performed according to a
standard method, as previously described (21). The following primary antibodies were
used: anti-DKK1, anti-β-catenin, and α-tubulin (BD Pharmingen; BD
Biosciences, San Diego, CA, USA) and p84 (Abcam, Cambridge, MA,
USA). Nuclear extracts were prepared using the Nuclear Extraction
kit (Active Motif, Carlsbad, CA, USA), according to the
manufacturer's instructions.
Luciferase reporter assay
Cells were seeded in triplicate in 24-well plates,
and allowed to settle for 24 h. Indicated plasmids plus 1 ng pRL-TK
Renilla plasmid were transfected into the cells using
Lipofectamine 3000 reagent (Life Technologies). Forty-eight hours
after transfection, a Dual-Luciferase Reporter Assay (Promega,
Madison, WI, USA) was performed according to the manufacturer's
instructions, as previously described (22).
Tumor xenografts
All experimental procedures were approved by the
Institutional Animal Care and Use Committee of Sun Yat-sen
University. Six-week-old BALB/c-nu mice were randomly divided into
4 groups (n=6/group). The cells (1×105) were
subcutaneously inoculated along with Matrigel (final concentration
of 25%) into the inguinal folds of the nude mice. The tumor volume
was determined using an external caliper and calculated using the
equation (L × W2)/2. The mice were sacrificed 35 days
after inoculation and the tumors were excised and subjected to
pathological examination.
Side population analysis
The cell suspensions were labeled with Hoechst 33342
(molecular probes, #H-3570) dye for side population (SP) analysis
as per standard protocol (23).
Briefly, cells were resuspended at 1X pre-warmed Opti-MEM
(containing 2% FBS) (both from Gibco, Grand Island, NY, USA) at a
density of 106 cells/ml. Hoechst 33342 dye was added at
a final concentration of 5 µg/ml in the presence or absence of
verapamil (50 µmol/l; Sigma-Aldrich, St. Louis, MO, USA) and the
cells were incubated at 37°C for 90 min with intermittent shaking.
At the end of the incubation period, the cells were washed with
Opti-MEM containing 2% FBS and centrifuged at 4°C, and subsequently
resuspended in ice-cold Opti-MEM containing 2% FBS and 10 mmol/l
HEPES. Then, propidium iodide (Sigma-Aldrich) at a final
concentration of 2 µg/ml was added to the cells to gate viable
cells. The cells were filtered through a 40-µm cell strainer to
obtain a single cell suspension before sorting. Analysis and
sorting was carried out on a FACSAria I (Becton-Dickinson, Franklin
Lakes, NJ, USA). The Hoechst 33342 dye was excited at 355 nm and
its dual-wavelength emission at the blue and red regions was
plotted to get the SP scatter.
miRNP immunoprecipitation
Cells were co-transfected with HA-Ago1 along with
100 nM miR-217, followed by HA-Ago1 immunoprecipitation using an
HA-antibody. Real-time PCR analysis of the IP material was used to
assess the association of the mRNA of DKK1 with the RISC
complex.
Statistical analysis
All statistical analyses were carried out using the
SPSS 16.0 statistical software package. Comparisons between groups
for statistical significance were performed using Chi-square and
Fisher's exact tests. In all cases, P<0.05 was considered
significant. All the experiments were repeated 3 times.
Results
miR-217 is upregulated in HCC tissues
and cells
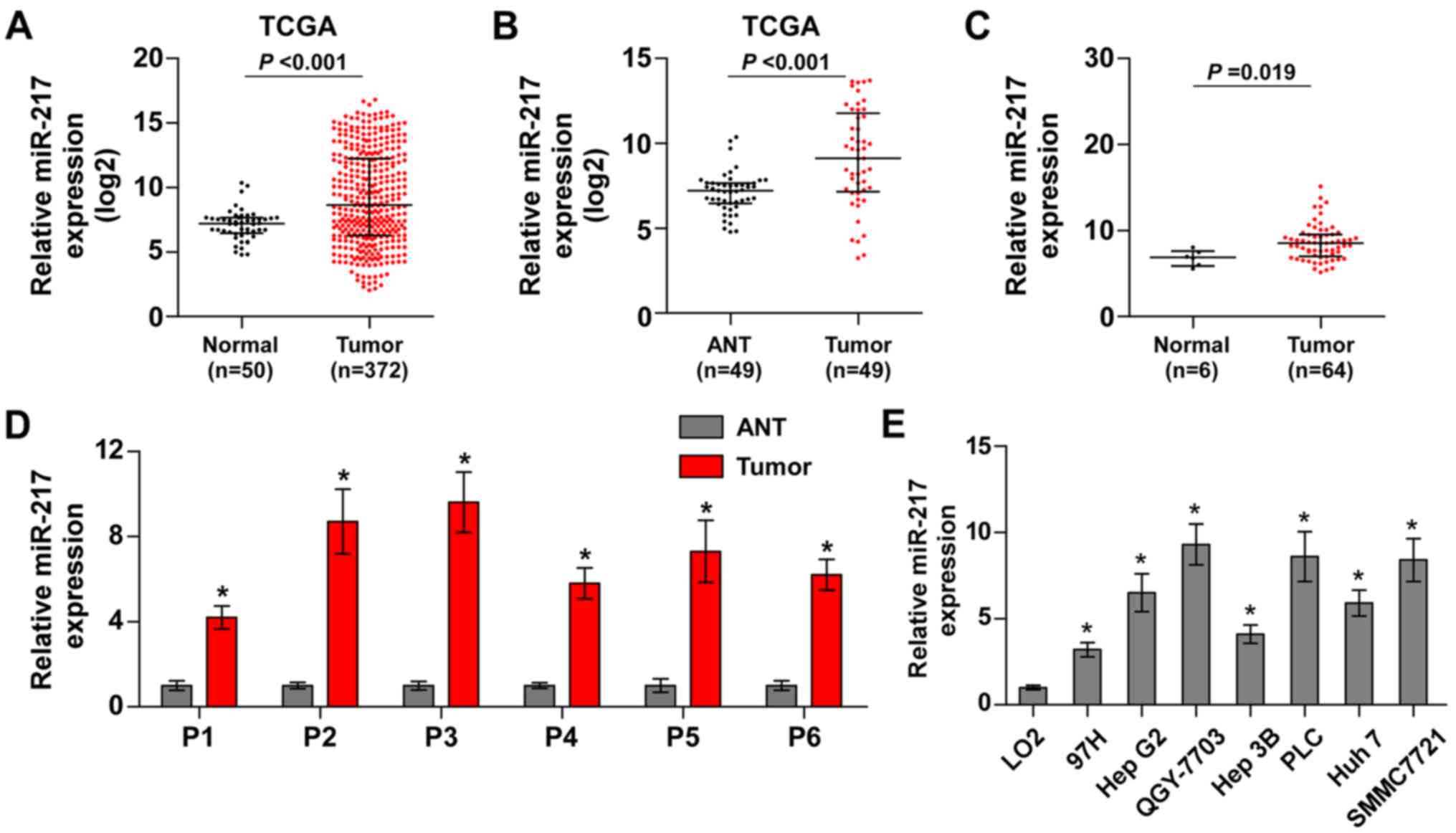
To identify the potential miRNAs in HCC, we analyzed
the HCC datasets from The Cancer Genome Atlas (TCGA). The result
revealed that miR-217 was upregulated in HCC tissues compared with
normal hepatic tissues (Fig. 1A).
We further analyzed the miR-217 expression in 49 paired HCC tissues
from TCGA and found that the expression levels of miR-217 were
upregulated in the primary HCC tissues compared with the matched
adjacent normal tissues (Fig. 1B).
Furthermore, we assessed the miR-217 expression in our own HCC
tissue samples (Table II). As
shown in Fig. 1C and D, miR-217
expression was markedly increased in HCC tissues compared with that
in 6 normal hepatic tissues, and the matched adjacent normal
tissues, respectively. We further examined the expression level of
miR-217 in HCC cells and found that compared to human liver
immortal cell line L02, the expression of miR-217 was
differentially upregulated in HCC cells. Thus, our findings
demonstrated that miR-217 is increased in HCC tissues and
cells.
 | Table II.Relationship between miR-217 and the
clinicopathological characteristics in 64 hepatocellular carcinoma
patients. |
Table II.
Relationship between miR-217 and the
clinicopathological characteristics in 64 hepatocellular carcinoma
patients.
|
|
| miR-217
expression |
|---|
|
|
|
|
|---|
| Parameters | No. of cases | High | Low | P-values |
|---|
| Gender |
|
|
| 0.448 |
|
Male | 37 | 20 | 17 |
|
|
Female | 27 | 12 | 15 |
|
| Age (years) |
|
|
| 0.211 |
|
<60 | 33 | 14 | 19 |
|
|
≥60 | 31 | 18 | 13 |
|
| AFP (ng/ml) |
|
|
| 0.003a |
|
<400 | 30 | 9 | 21 |
|
|
≥400 | 34 | 23 | 11 |
|
| Tumor size
(cm) |
|
|
|
<0.001a |
|
<5 | 38 | 11 | 27 |
|
| ≥5 | 26 | 21 | 5 |
|
|
Differentiation |
|
|
| 0.080 |
|
High | 33 | 20 | 13 |
|
|
Low | 31 | 12 | 19 |
|
| Clinical stage |
|
|
| 0.045a |
|
I–II | 30 | 11 | 19 |
|
|
III–IV | 34 | 21 | 13 |
|
| Venous
invasion |
|
|
| 0.042a |
|
Negative | 26 | 9 | 17 |
|
|
Positive | 38 | 23 | 15 |
|
| Distant
metastasis |
|
|
| 0.006a |
|
Negative | 45 | 8 | 19 |
|
|
Positive | 35 | 24 | 13 |
|
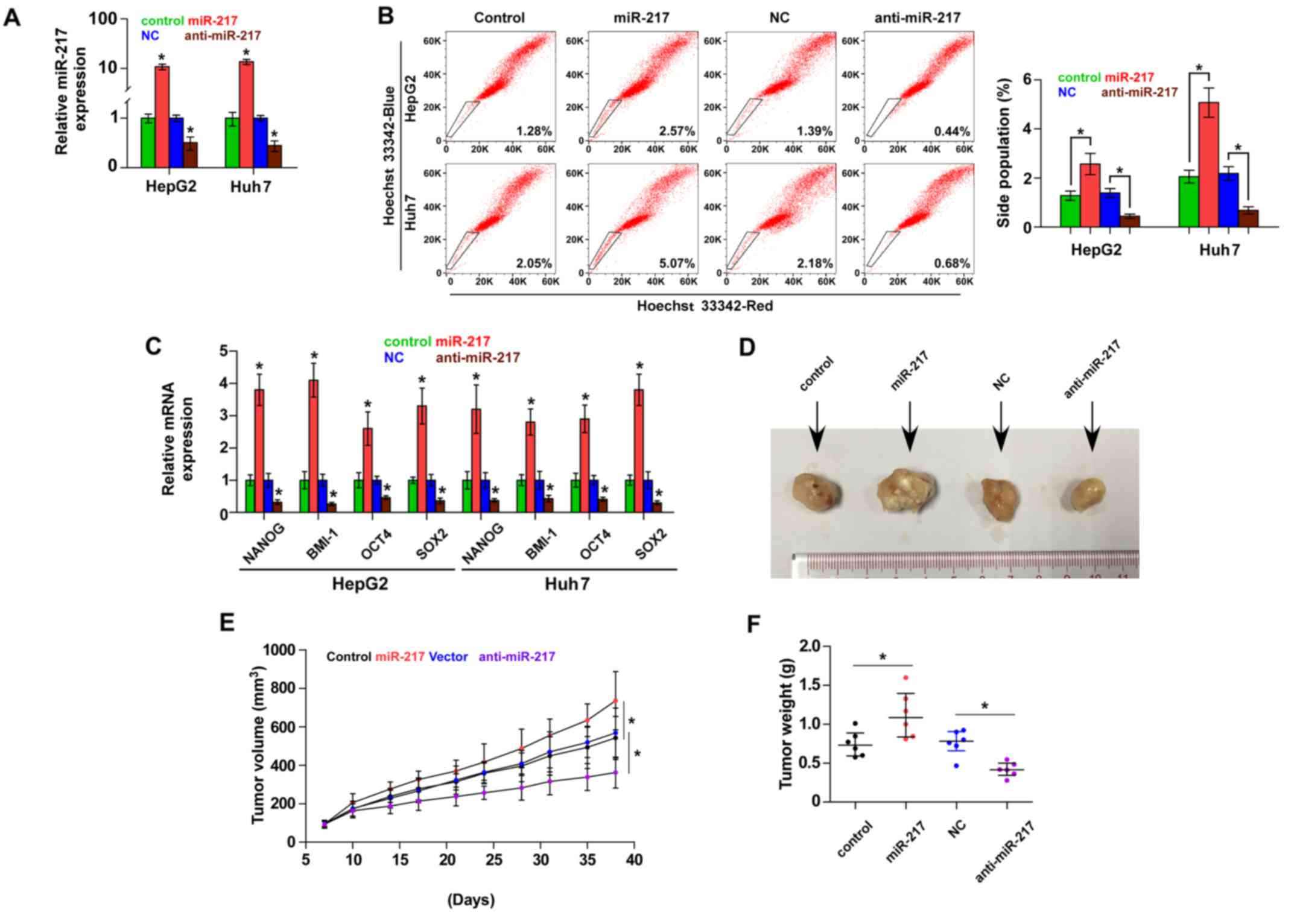
miR-217 promotes stem cell properties
and tumorigenesis in HCC cells
Accumulating studies have suggested that cancer
growth is driven by CSCs (24,25).
To further investigate the role of miR-217 in the CSC-like
phenotype of HCC cells, we first constructed miR-217-stably
expressing cells by ectopically overexpressing miR-217 and
endogenously silencing miR-217 via retrovirus infection in the
HepG2 and Huh7 cell lines (Fig.
2A). The effect of miR-217 on the SP was examined and the
result revealed that overexpression of miR-217 increased, while
silencing miR-217 decreased the fraction of SP cells in the HepG2
and Huh7 cells (Fig. 2B). We
further assessed the effect of miR-217 on stem cell markers,
including Nanog homeobox (NANOG), proto-oncogene polycomb ring
finger (BMI1), POU class 5 homeobox 1A (OCT4A) and SOX2, and found
that upregulation of miR-217 enhanced the expression of these
markers. However, knockdown of miR-217 decreased the expression of
these markers (Fig. 2C).
Collectively, these results indicated that miR-217 promotes
CSC-like phenotypes in HCC in vitro.
We next assessed the effect of miR-217 on the
tumorigenesis of HCC in vivo. Huh7 cells (1×105)
of the miR-217-overexpressing, control and anti-miR-217 cells were
inoculated into nude mice, respectively. As shown in Fig. 2D-F, tumors in the
miR-217-overexpressing cells grew more rapidly than those in the
control group. Conversely, tumors in the anti-miR-217 group were
smaller than the tumors in the negative control group. These
results revealed that miR-217 promotes the tumorigenesis of HCC
cells in vivo.
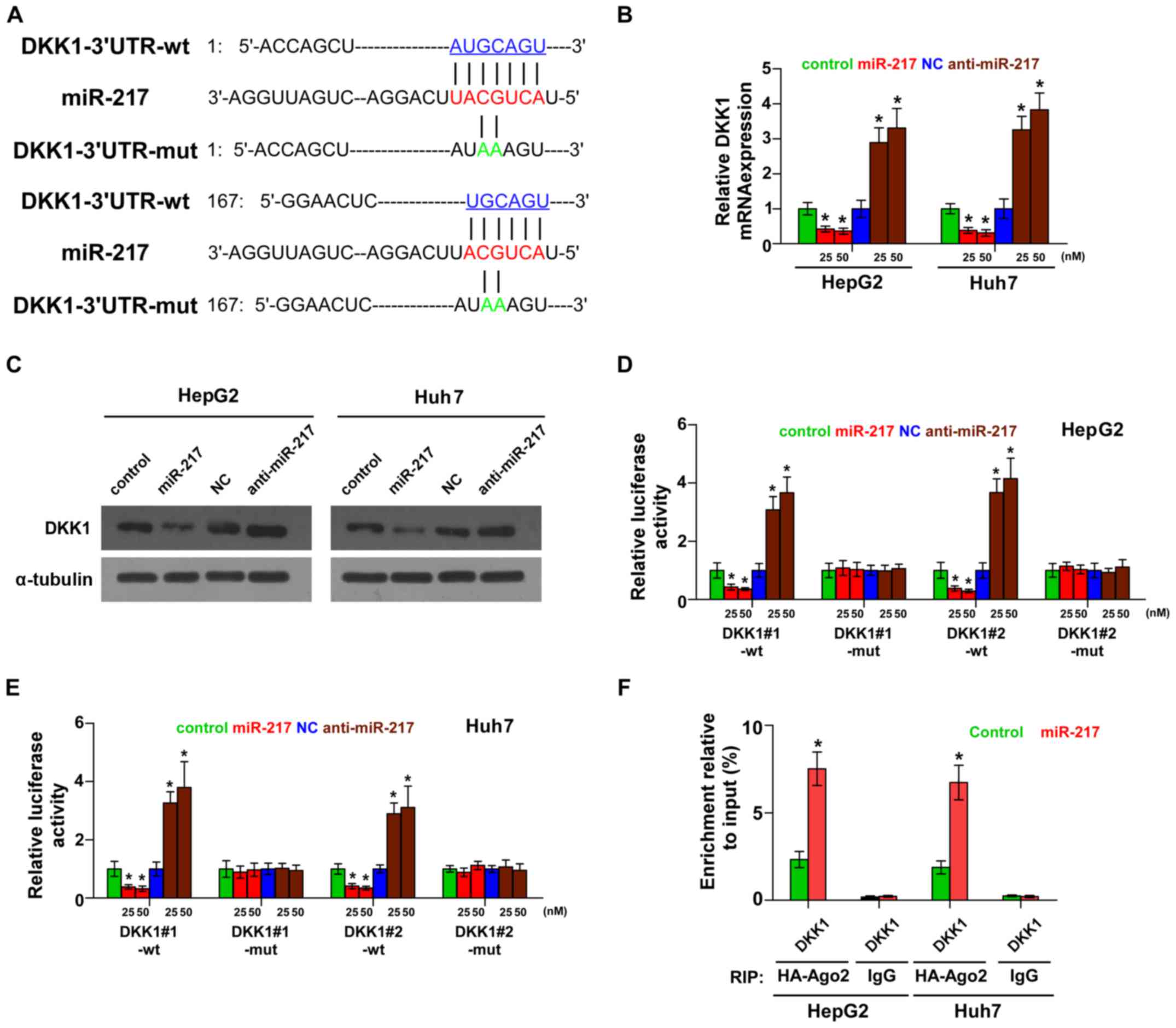
miR-217 targets a negative regulator
of the Wnt signaling pathway
By the publicly available algorithms TargetScan and
miRNA.ORG, we found that the negative regulator of Wnt
signaling, DKK1, may be a potential target of miR-217 (Fig. 3A). PCR and western blotting revealed
that miR-217 overexpression decreased the mRNA and protein
expression levels of DKK1. Conversely, silencing of miR-217
increased the expression of DKK1 (Fig.
3B and C). Luciferase assay revealed that miR-217
overexpression decreased, while silencing of miR-217 enhanced the
reporter activity driven by the 3′UTRs of DKK1 in dose-dependent
manners, but not by the mutant 3′UTRs of DKK1 within the
miR-217-binding seed regions in HCC cells (Fig. 3D and E). Furthermore,
microribonucleoprotein (miRNP) immunoprecipitation (IP) assay
demonstrated a selective association of miR-217 with DKK1
transcript (Fig. 3F), further
elucidating the direct suppressive effect of miR-217 on DKK1.
Consequently, our results indicate that DKK1 is an authentic target
of miR-217 in HCC cells.
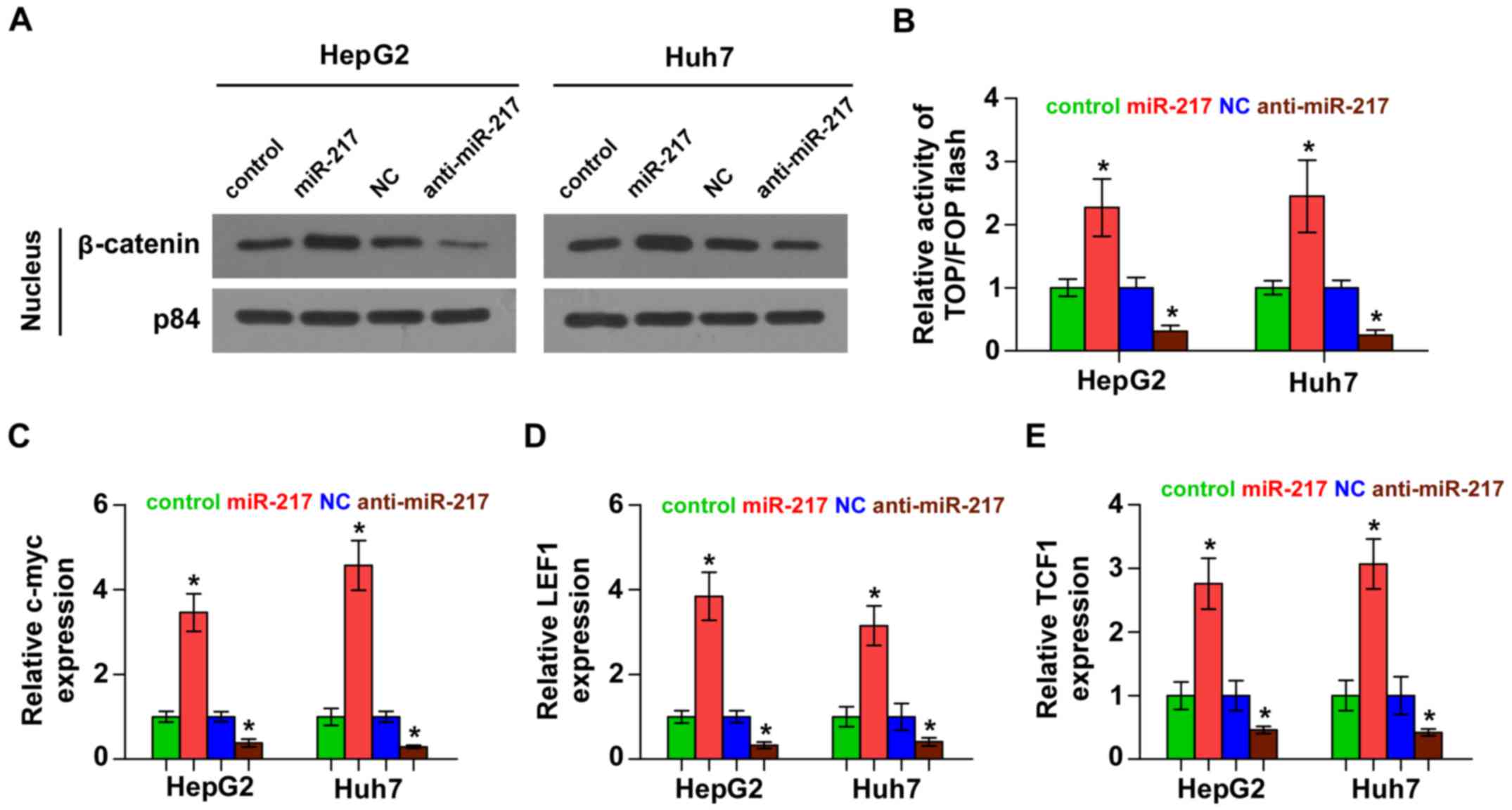
miR-217 activates Wnt signaling
pathways in HCC cells
We next investigated the effect of miR-217 on Wnt
signaling. As shown in Fig. 4A,
western blotting revealed that overexpression of miR-217 increased
nuclear accumulation of β-catenin, while silencing of miR-217
impaired the β-catenin translocation into the nucleus. Furthermore,
we found that miR-217 overexpression increased, while silencing of
miR-217 decreased, β-catenin/TCF transcriptional activity (Fig. 4B). We further examined the
expression levels of multiple downstream genes of Wnt signaling,
including c-myc, LEF1 and TCF1 and found that overexpression of
miR-217 enhanced the expression of c-myc, LEF1 and TCF1, while
silencing of miR-217 yielded the opposite effect (Fig. 4C-E). Thus, these results
demonstrated that miR-217 activates Wnt signaling pathways in HCC
cells.
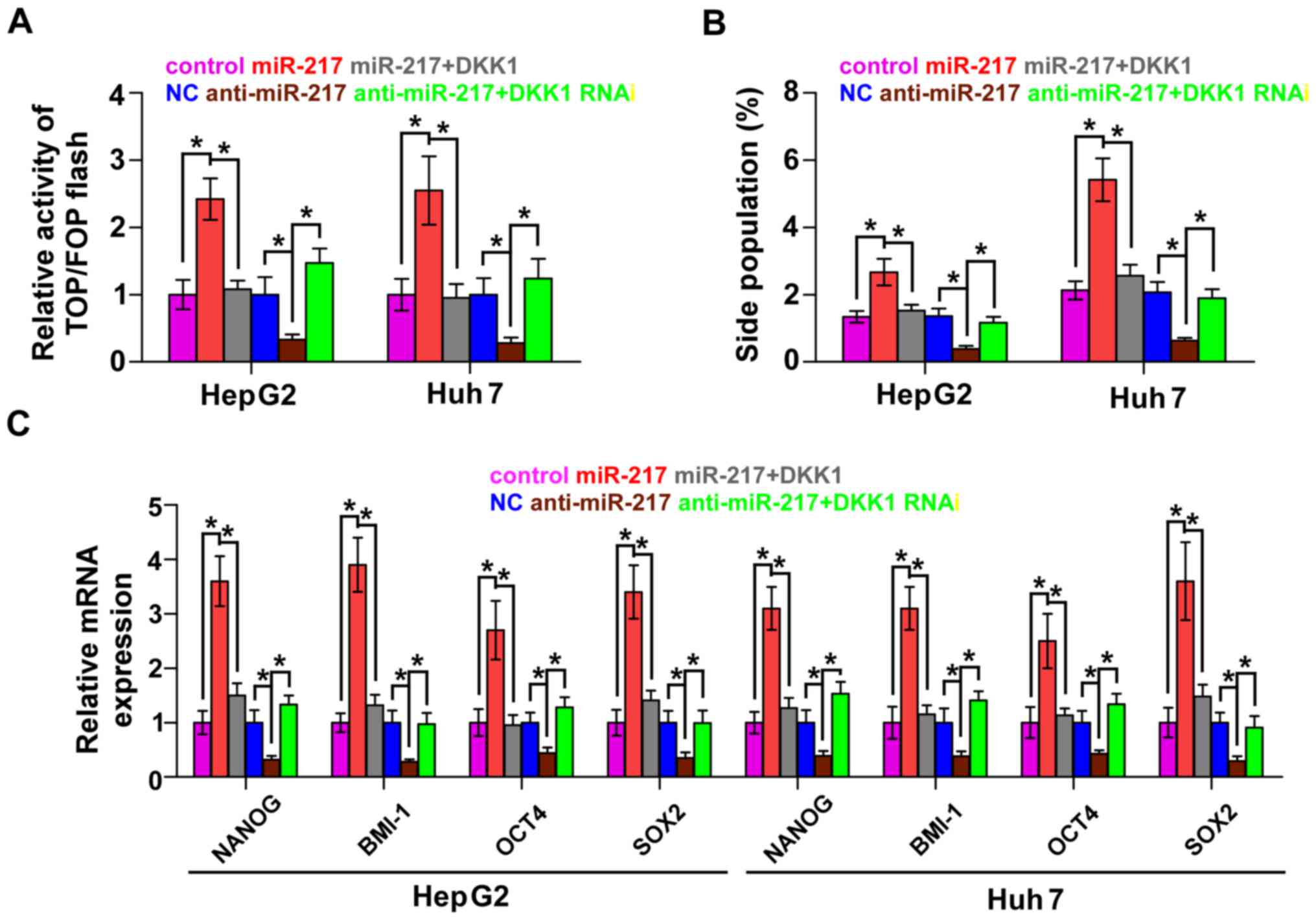
DKK1 mediates miR-217-induced CSC
phenotypes in HCC cells
To explore whether DKK1 contributed to
miR-217-induced CSCs, we transfected the DKK1 plasmid in
miR-217-overexpressing cells and RNA interference of DKK1 in
miR-217-silenced cells, respectively. β-catenin/TCF transcriptional
activity assay revealed that the stimulatory effect of miR-217 was
angatonized by the upregulation of DKK1 and the inhibitory effect
of anti-miR-217 was reversed by DKK1 RNAi (Fig. 5A). Furthermore, the upregulation of
DKK1 in miR-217-overexpressing cells significantly decreased the
SP. In contrast, knockdown of DKK1 in miR-217-silenced cells
increased the fraction of SP in HCC cells (Fig. 5B). Real-time PCR revealed that DKK1
contributed to the expression of stem cell markers regulated by
miR-217 (Fig. 5C). Collectively,
these results suggest that DKK1 mediates the regulatory role of
miR-217 in CSC-like phenotypes in HCC cells.
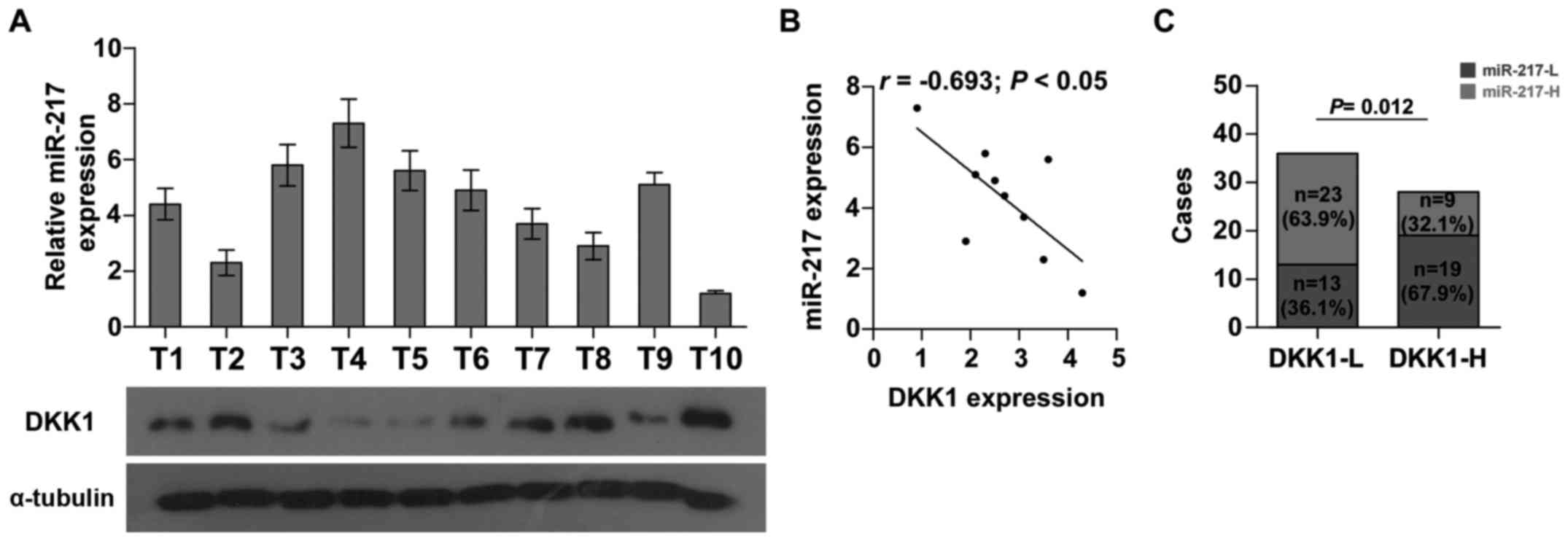
Clinical correlation of miR-217 with
DKK1 expression in HCC tissues
To further investigate whether the expression of
miR-217 was associated with DKK1 expression in HCC clinical tissue
samples, the expression levels of miR-217 were examined in 10 HCC
fresh tissues by real-time PCR. As shown in Fig. 6A and B, there was a significant
negative correlation between miR-217 and DKK1 (r=−0.693; P<0.05)
in HCC. Furthermore, we found that the expression of miR-217 was
increased in HCC tissues with low DKK1 expression when compared to
those with high DKK1 expression (Fig.
6C). Collectively, these results demonstrated that miR-217 is
negatively correlated with DKK1 expression in HCC tissues.
Discussion
In the canonical Wnt pathway, Wnt ligands bind to
frizzled receptors and lipoprotein receptor-related protein-5 or −6
(LRP5/6) co-receptors, giving rise to the accumulation and
subsequent translocation of β-catenin into the nucleus. Then,
nuclear β-catenin interacts with the T-cell factor (TCF) family of
transcription factors to initiate the transcription of downstream
target genes (26,27). Notably, several cancer stem cell
factors, including CD44, NANOG, SOX2 and OCT4, are well-known
targets of the Wnt pathway, indicating that the Wnt signaling
pathway plays crucial roles in the regulation of CSCs (28,29).
Furthermore, the constitutive activation of Wnt/β-catenin signaling
has been implicated in a variety of human cancers, including
hepatocellular carcinoma (HCC) (30,31).
Indeed, 40~70% of HCC has been reported to harbor nuclear
accumulation of the downstream effector β-catenin of Wnt signaling
(32). Wong et al reported
that constitutive activation of Wnt signaling induced the increased
translocation of β-catenin into the nucleus, which contributed to
the development of HCC (33). These
studies demonstrated that dysregulation of Wnt signaling is a
critical driver of HCC pathogenesis. In the present study, we
revealed that miR-217 was increased in HCC tissues and cells.
Overexpression of miR-217 increased, while silencing of miR-217
decreased the nuclear translocation of β-catenin, β-catenin/TCF
transcriptional activity as well as the expression levels of
downstream genes of the Wnt signaling pathway, which further
contributed to the CSC-like phenotype in HCC cells. Our results
further revealed that miR-217 activates the Wnt signaling pathway
via DKK1 targeting, an important negative regulator of Wnt
signaling. Therefore, our findings uncovered a novel mechanism of
miR-217 in the regulation of the Wnt signaling pathway.
Dickkopf-1 (DKK1) which acts as an important
negative regulator of Wnt/β-catenin signaling functions via binding
with high affinity to LRP5/6 in competition with Wnt (34). DKK1 has been reported to be
downregulated in various cancers, resulting in the constitutive
activation of the Wnt/β-catenin signaling pathway (35,36).
Furthermore, several studies have reported that the upregulation of
DKK1 inhibited sphere formation capacity and decreased the
CD24−/CD44+ cell population in breast cancer
cells (37). This evidence
indicated that DKK1 played an important role in the stemness
maintenance of CSCs. In the present study, RT-PCR and western
blotting revealed that overexpressing miR-217 decreased, while
silencing miR-217 increased the mRNA and protein levels of DKK1.
Luciferase and miRNP IP assay demonstrated the association of
miR-217 with DKK1 in HCC cells, indicating that DKK1 is a direct
target of miR-217 in HCC cells. Notably, the stimulatory effects of
miR-217 on β-catenin/TCF transcriptional activity and CSC-like
phenotype was antagonized by overexpression of DKK1 in
miR-217-overexpressing cells. Conversely, silencing of DKK1 yielded
the opposite effect in miR-217-downregulating cells. Collectively,
these findings demonstrated that miR-217 promotes a CSC-like
phenotype and activates Wnt signaling via DKK1 targeting in HCC
cells.
miR-217 has been identified to be downregulated in
multiple human cancers, and has contributed to cancer cell
proliferation, drug resistance and metastasis via varying
mechanisms (38–41). Furthermore, the expression of
miR-217 has also been found to be upregulated in gastric and breast
cancer (42,43). These findings indicated that miR-217
functions as both an oncomir and tumor-suppressive miRNA, depending
on the tumor type. Notably, several studies have reported that
miR-217 was increased in HCC (44,45),
however the specific mechanisms responsible for the progression of
HCC remain poorly elucidated. Consistent with these findings, we
revealed that miR-217 was upregulated in HCC tissues and cells.
Overexpression of miR-217 promoted, while silencing miR-217
suppressed, the CSC-like phenotypes in vitro and
tumorigenicity in vivo in HCC cells. Our findings further
demonstrated that miR-217 promotes the CSC-like phenotype via DKK1
targeting, resulting in constitutive activation of Wnt signaling.
Moreover, the stimulatory or inhibitory effects of the upregulation
or downregulation of miR-217 on stem cell properties and Wnt
signaling were antagonized by the upregulation or silencing of DKK1
in HCC cells. Notably, another study reported that miR-217
expression was much lower in highly invasive MHCC-97H HCC cells and
metastatic HCC tissues. Ectopic expression of miR-217 inhibited the
invasion of MHCC-97H cells. Inversely, inhibition of miR-217
enhanced the invasive ability of Huh7 and MHCC-97L cells (46). This evidence demonstrated that
miR-217 plays different or even contradicting roles in the
different developmental processes of HCC.
In summary, the present study demonstrated that
oncogenic miR-217 promotes CSC-like properties and tumorigenicity
by negatively regulating DKK1, leading to activation of the
Wnt/β-catenin signaling pathway in HCC. Improved understanding of
the specific role of miR-217 in the activation of the Wnt signaling
pathway and in the pathogenesis of HCC may help to develop novel
therapeutic methods in the treatment of HCC.
Acknowledgements
The present study was supported by the State Key
Program of the National Natural Science Foundation of Guangdong,
China (Program, no. 2015A030311039), the National Natural Science
Foundation of China (81272312) and the Science and Technology
Planning Project of Guangdong Province, China (no.
2014A020212390).
References
|
1
|
Fong Y, Sun RL, Jarnagin W and Blumgart
LH: An analysis of 412 cases of hepatocellular carcinoma at a
Western center. Ann Surg. 229:790–800. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gerber B, Freund M and Reimer T: Recurrent
breast cancer: Treatment strategies for maintaining and prolonging
good quality of life. Dtsch Arztebl Int. 107:85–91. 2010.PubMed/NCBI
|
|
3
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Monteiro J and Fodde R: Cancer stemness
and metastasis: Therapeutic consequences and perspectives. Eur J
Cancer. 46:1198–1203. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lapidot T, Sirard C, Vormoor J, Murdoch B,
Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA and
Dick JE: A cell initiating human acute myeloid leukaemia after
transplantation into SCID mice. Nature. 367:645–648. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee TK, Castilho A, Cheung VC, Tang KH, Ma
S and Ng IO: CD24+ liver tumor-initiating cells drive
self-renewal and tumor initiation through STAT3-mediated NANOG
regulation. Cell Stem Cell. 9:50–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao W, Wang L, Han H, Jin K, Lin N, Guo
T, Chen Y, Cheng H, Lu F, Fang W, et al: 1B50-1, a mAb raised
against recurrent tumor cells, targets liver tumor-initiating cells
by binding to the calcium channel α2δ1 subunit. Cancer Cell.
23:541–556. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren D, Wang M, Guo W, Huang S, Wang Z,
Zhao X, Du H, Song L and Peng X: Double-negative feedback loop
between ZEB2 and miR-145 regulates epithelial-mesenchymal
transition and stem cell properties in prostate cancer cells. Cell
Tissue Res. 358:763–778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ren D, Wang M, Guo W, Zhao X, Tu X, Huang
S, Zou X and Peng X: Wild-type p53 suppresses the
epithelial-mesenchymal transition and stemness in PC-3 prostate
cancer cells by modulating miR-145. Int J Oncol. 42:1473–1481.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma S, Tang KH, Chan YP, Lee TK, Kwan PS,
Castilho A, Ng I, Man K, Wong N, To KF, et al: miR-130b promotes
CD133+ liver tumor-initiating cell growth and
self-renewal via tumor protein 53-induced nuclear protein 1. Cell
Stem Cell. 7:694–707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ji J, Yamashita T, Budhu A, Forgues M, Jia
HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al: Identification
of microRNA-181 by genome-wide screening as a critical player in
EpCAM-positive hepatic cancer stem cells. Hepatology. 50:472–480.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hahn WC, Dessain SK, Brooks MW, King JE,
Elenbaas B, Sabatini DM, DeCaprio JA and Weinberg RA: Enumeration
of the simian virus 40 early region elements necessary for human
cell transformation. Mol Cell Biol. 22:2111–2123. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo W, Ren D, Chen X, Tu X, Huang S, Wang
M, Song L, Zou X and Peng X: HEF1 promotes epithelial mesenchymal
transition and bone invasion in prostate cancer under the
regulation of microRNA-145. J Cell Biochem. 114:1606–1615. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang M, Ren D, Guo W, Huang S, Wang Z, Li
Q, Du H, Song L and Peng X: N-cadherin promotes
epithelial-mesenchymal transition and cancer stem cell-like traits
via ErbB signaling in prostate cancer cells. Int J Oncol.
48:595–606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Gong LY, Song LB, Jiang LL, Liu LP,
Wu J, Yuan J, Cai JC, He M, Wang L, et al: Oncoprotein Bmi-1
renders apoptotic resistance to glioma cells through activation of
the IKK-nuclear factor-kappaB pathway. Am J Pathol. 176:699–709.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Goodell MA: Stem cell identification and
sorting using the Hoechst 33342 side population (SP). Curr Protoc
Cytom Chapter. 9:Unit9.182005.
|
|
24
|
de Sousa E, Melo F, Colak S, Buikhuisen J,
Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR,
Fessler E, van den Bergh SP, et al: Methylation of
cancer-stem-cell-associated Wnt target genes predicts poor
prognosis in colorectal cancer patients. Cell Stem Cell. 9:476–485.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Simeone DM: Pancreatic cancer stem cells:
Implications for the treatment of pancreatic cancer. Clin Cancer
Res. 14:5646–5648. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao J, Wang J, Liu B, Pan W, Farr GH III,
Flynn C, Yuan H, Takada S, Kimelman D, Li L, et al: Low-density
lipoprotein receptor-related protein-5 binds to Axin and regulates
the canonical Wnt signaling pathway. Mol Cell. 7:801–809. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gordon MD and Nusse R: Wnt signaling:
Multiple pathways, multiple receptors, and multiple transcription
factors. J Biol Chem. 281:22429–22433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barker N, van Es JH, Kuipers J, Kujala P,
van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H,
Peters PJ, et al: Identification of stem cells in small intestine
and colon by marker gene Lgr5. Nature. 449:1003–1007. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wielenga VJ, Smits R, Korinek V, Smit L,
Kielman M, Fodde R, Clevers H and Pals ST: Expression of CD44 in
Apc and Tcf mutant mice implies regulation by the WNT
pathway. Am J Pathol. 154:515–523. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gangopadhyay S, Nandy A, Hor P and
Mukhopadhyay A: Breast cancer stem cells: A novel therapeutic
target. Clin Breast Cancer. 13:7–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takebe N, Miele L, Harris PJ, Jeong W,
Bando H, Kahn M, Yang SX and Ivy SP: Targeting Notch, Hedgehog, and
Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin
Oncol. 12:445–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lachenmayer A, Alsinet C, Savic R,
Cabellos L, Toffanin S, Hoshida Y, Villanueva A, Minguez B, Newell
P, Tsai HW, et al: Wnt-pathway activation in two molecular classes
of hepatocellular carcinoma and experimental modulation by
sorafenib. Clin Cancer Res. 18:4997–5007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wong CM, Fan ST and Ng IO: beta-Catenin
mutation and overexpression in hepatocellular carcinoma:
Clinicopathologic and prognostic significance. Cancer. 92:136–145.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bafico A, Liu G, Yaniv A, Gazit A and
Aaronson SA: Novel mechanism of Wnt signalling inhibition mediated
by Dickkopf-1 interaction with LRP6/Arrow. NaT cell Biol.
3:683–686. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee J, Yoon YS and Chung JH: Epigenetic
silencing of the WNT antagonist DICKKOPF-1 in cervical
cancer cell lines. Gynecol Oncol. 109:270–274. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Na Y, Lee SM, Kim DS and Park JY: Promoter
methylation of Wnt antagonist DKK1 gene and prognostic value in
Korean patients with non-small cell lung cancers. Cancer Biomark.
12:73–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Agur Z, Kirnasovsky OU, Vasserman G,
Tencer-Hershkowicz L, Kogan Y, Harrison H, Lamb R and Clarke RB:
Dickkopf1 regulates fate decision and drives breast cancer stem
cells to differentiation: An experimentally supported mathematical
model. PLoS One. 6:e242252011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim Y, Kim H, Park D, Han M, Lee H, Lee
YS, Choe J, Kim YM and Jeoung D: miR-217 and CAGE form feedback
loop and regulates the response to anti-cancer drugs through EGFR
and HER2. Oncotarget. 7:10297–10321. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rachagani S, Macha MA, Menning MS, Dey P,
Pai P, Smith LM, Mo YY and Batra SK: Changes in microRNA (miRNA)
expression during pancreatic cancer development and progression in
a genetically engineered KrasG12D;Pdx1-Cre mouse (KC)
model. Oncotarget. 6:40295–40309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gu L, Li H, Chen L, Ma X, Gao Y, Li X,
Zhang Y, Fan Y and Zhang X: MicroRNAs as prognostic molecular
signatures in renal cell carcinoma: A systematic review and
meta-analysis. Oncotarget. 6:32545–32560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng
ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, et al:
microRNA-217 inhibits tumor progression and metastasis by
downregulating EZH2 and predicts favorable prognosis in gastric
cancer. Oncotarget. 6:10868–10879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang M, Cui G, Ding M, Yang W, Liu Y, Dai
1 and Chen L: miR-935 promotes gastric cancer cell proliferation by
targeting SOX7. Biomed Pharmacother. 79:153–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang AX, Lu FQ, Yang YP, Ren XY, Li ZF
and Zhang W: MicroRNA-217 overexpression induces drug resistance
and invasion of breast cancer cells by targeting PTEN signaling.
Cell Biol Int. Jun 24–2015.(Epub ahead of print). doi:
10.1002/cbin.10506. View Article : Google Scholar :
|
|
44
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
Identification of deregulated miRNAs and their targets in hepatitis
B virus-associated hepatocellular carcinoma. World J Gastroenterol.
18:5442–5453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang W, Zhao LJ, Tan YX, Ren H and Qi ZT:
MiR-138 induces cell cycle arrest by targeting cyclin D3 in
hepatocellular carcinoma. Carcinogenesis. 33:1113–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Su J, Wang Q, Liu Y and Zhong M: miR-217
inhibits invasion of hepatocellular carcinoma cells through direct
suppression of E2F3. Mol Cell Biochem. 392:289–296. 2014.
View Article : Google Scholar : PubMed/NCBI
|