Introduction
CRC is the most common digestive system cancer and
is the third most common cancer in the world. Early stage CRC does
not have obvious symptoms and is often diagnosed in advanced stage.
CRC patients without metastases can be cured by surgery, but
patients with advanced CRC are mainly treated with chemotherapy
(1). Therefore, early diagnosis and
treatment of CRC is needed.
Genetic changes have been investigated in CRC. Some
significant oncogenes and tumor suppressor genes such as APC, KRAS
and p53 are mutated in a considerable part of CRCs (2). This could allow screening and
diagnosis of early-stage CRC, which may lead to substantial
decreases in the mortality rate of CRC (3).
miRNAs are a series of small, non-coding regulatory
RNAs. By repressing their target mRNAs, miRNAs regulate many
cellular pathways, such as proliferation, apoptosis and
differentiation. This feature of miRNAs provides a novel approach
for cancer therapy (4). miRNAs such
as miRNA-92a and miRNA-21 have shown significant connection with
CRC, and can serve as biomarkers (5). Ng et al (6) reported plasma miR-92a can effectively
discriminate CRC from control subjects. It has been verified that
miR-21 can repress the tumor suppressor gene and induce cell
invasion (7).
Microarray is a multiple lab-on-a-chip. To identify
the biomarkers of CRC, we downloaded the gene and miRNA expression
profiles of CRC from GEO database. Expression differences were
compared between CRC tissues and normal colorectal tissues. By
analyzing GO (8) and pathway
enrichment (9) and constructing PPI
network (10) and miRNA-gene
network, we aimed to find key genes and miRNAs which play
significant roles in the occurrence and development of CRC and
discover new biomarkers for diagnosis and therapy.
Materials and methods
Microarray data
Three gene expression profiles (GSE21815, GSE32323
and GSE44076) and two miRNA expression profiles (GSE39845 and
GSE53592) were obtained from GEO database (http://www.ncbi.nlm.nih.gov/geo/) (11). The GSE21815 datasets contained 132
CRC samples and 9 normal samples. GSE32323 included 17 CRC samples
and 17 normal samples. GSE44076 consisted of 98 CRC samples and 98
normal samples. The miRNA expression profile of GSE39845 contained
3 CRC samples and 3 normal tissue samples. GSE53592 included 3 CRC
samples and 3 normal samples.
Identification of DEGs and DE
miRNAs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/info/geo2r.html) is an
interactive web tool for comparing two or more groups of samples in
a GEO series to identify DEGs or DE miRNAs across experimental
conditions. We used GEO2R to identify DEGs and DE miRNAs. The
P-value <0.05 and |logFC|>1 were chosen as cut-off
criteria.
Functional enrichment analysis of
DEGs
KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/) is a latest updated web
server for gene/protein functional annotation and functional sets
enrichment of gene (12). The GO
enrichment and KEGG (13) pathway
analysis were performed by KOBAS 3.0 online tool. In addition,
P<0.05 was set as the cut-off criterion.
PPI network construction and module
analysis
To explore the interactive relationships among the
DEGs, PPI network was constructed by the Search Tool for the
Retrieval of Interacting Genes (STRING, version 10.0, http://string.embl.de/) and combined score >0.4 was
set as the cut-off criterion. Then, PPI network was visualized by
Cytoscape software (14). The
Molecular Complex Detection (MCODE) app was performed to analyze
modules of PPI network. MCODE scores >3 and the number of nodes
>4 were set as cut-off criteria. The pathway enrichment analysis
of genes in the modules was performed by KOBAS 3.0. P<0.05 and
input number >3 were considered to be significant.
miRNA-gene network construction
The target genes of DE miRNAs were predicted by five
established miRNA target prediction programs (miRanda, MirTarget2,
PicTar, PITA and TargetScan). The genes predicted by at least three
programs were chosen as the targets of DE miRNAs. Then, miRNA-gene
network was constructed. To identify the key gene biomarkers, we
combined both PPI and miRNA-gene network. Genes with degree ≥20 in
PPI network and degree ≥3 in miRNA-gene network were selected as
gene biomarkers.
Cell culture
The human CRC cell lines HCT116 and HT-29 were
obtained from the American Type Culture Collection Cell Center and
cultured in RPMI-1640 medium (HyClone Laboratories, Inc., Logan UT,
USA) supplemented with 10% fetal bovine serum (FBS; PAN-Biotech,
Aidenbach, Germany) and 1% penicillin-streptomycin (Beyotime
Institute of Biotechnology, Haimen, China) at 37°C in 5%
CO2.
Patient samples
CRC tissue samples and normal colorectal tissue
samples were collected from patients with CRC in The First
Affiliated Hospital of Chongqing Medical University (Chongqing,
China). Ethics approval was obtained from the Clinical Research
Ethics Committee of The First Affiliated Hospital of Chongqing
Medical University. Informed consents were signed by all patients
for the acquisition and use of tissue samples.
Quantitative real-time PCR
Total RNA was extracted from cultured cells (HCT116
and HT-29) by RNAiso Plus (Takara Bio, Dalian, China) and tissues
by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA (1 µg) was
reverse transcribed by the GoScript™ Reverse Transcription system
(Promega, Madison, WI, USA). Real-time PCR was performed by the
GoTaq qPCR Master Mix (Promega). All q-PCR values of each gene were
normalized against ACTB. The relative expressions of eight DEGs
were calculated by the 2−∆∆Ct method (15).
Statistical analysis
The final data were presented as mean ± SD. The
results were analyzed by the Students t-test, and P<0.05 was
considered to show statistical difference.
Results
Identification of DEGs
A total of 8027, 1328 and 1230 upregulated DEGs were
identified with the 653, 1231 and 1422 downregulated DEGs from
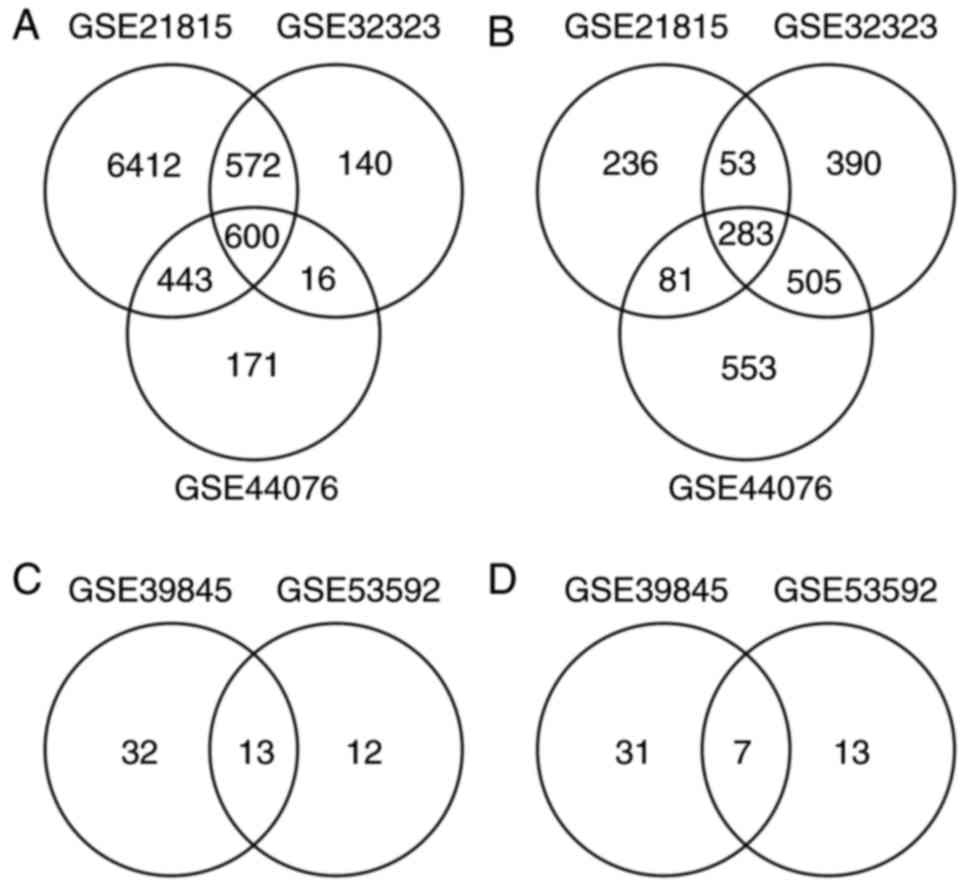
GSE21815, GSE32323 and GSE44076, respectively. There were 600
upregulated DEGs (Fig. 1A) and 283
downregulated DEGs (Fig. 1B) in the
overlap of the three datasets.
Functional enrichment analysis of
DEGs
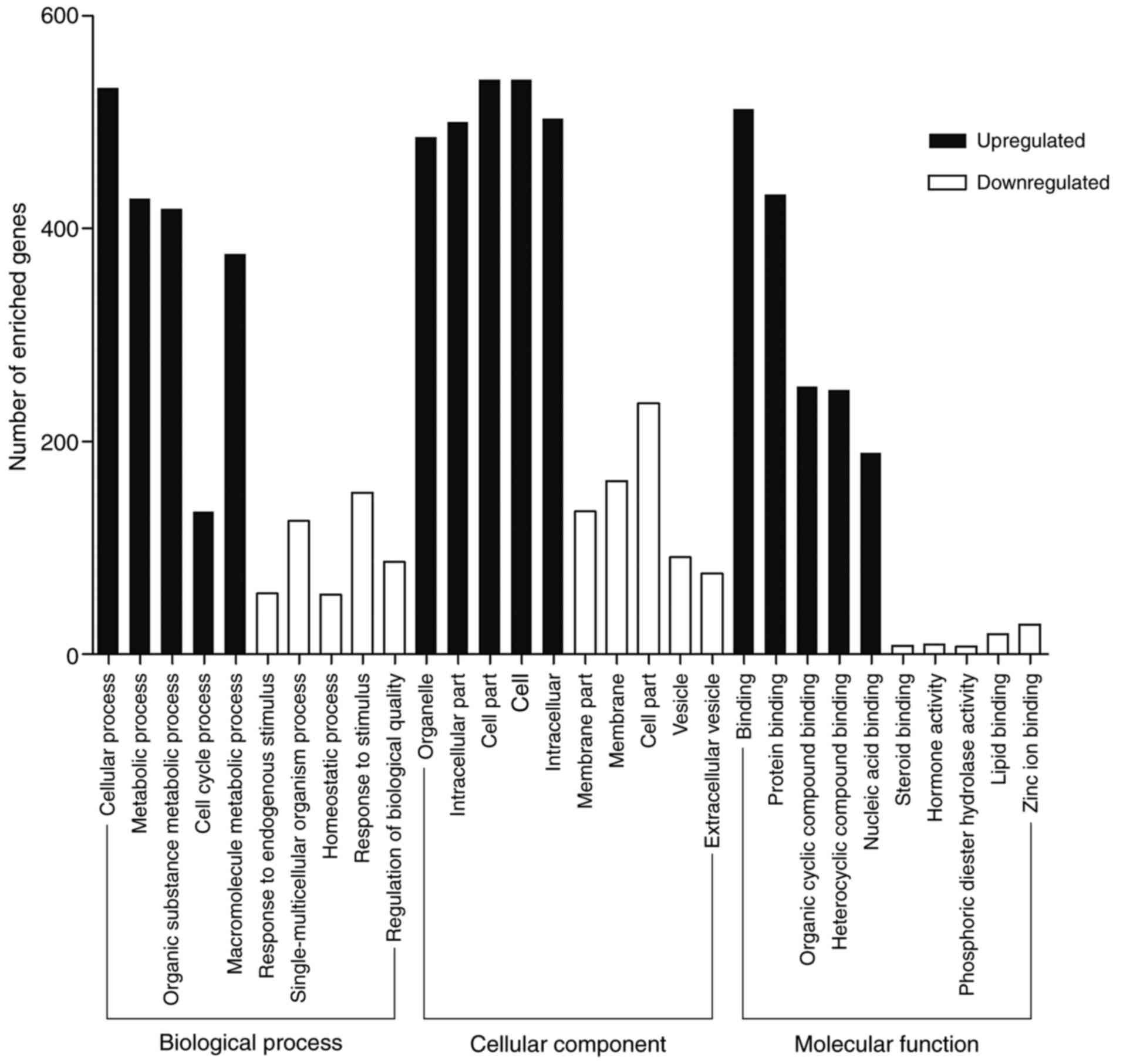
KOBAS 3.0 was performed to analyze the functional
and pathway enrichment of identified DEGs. GO analysis results
showed that upregulated DEGs were significantly enriched in
binding, protein binding and organic cyclic compound binding at MF
level; organelle, intracellular part and cell part at CC level and
cellular process, metabolic process and organic substance metabolic
process at BP level (Fig. 2).
Downregulated DEGs were enriched in binding, receptor binding and
protein binding at MF level; extracellular region, cell periphery
and plasma membrane at CC level and chemical homeostasis, cellular
response to chemical stimulus and single-organism process at BP
level (Fig. 2). Most GO terms
enriched in the regulation processes. Binding is a basic step of
regulation process. The GO terms mostly enriched in binding which
is consistent with the knowledge that tumor development and
progression is regulated by multiple molecules (16–18).
The GO analysis supported the correlation between the identified
DEGs and CRC.
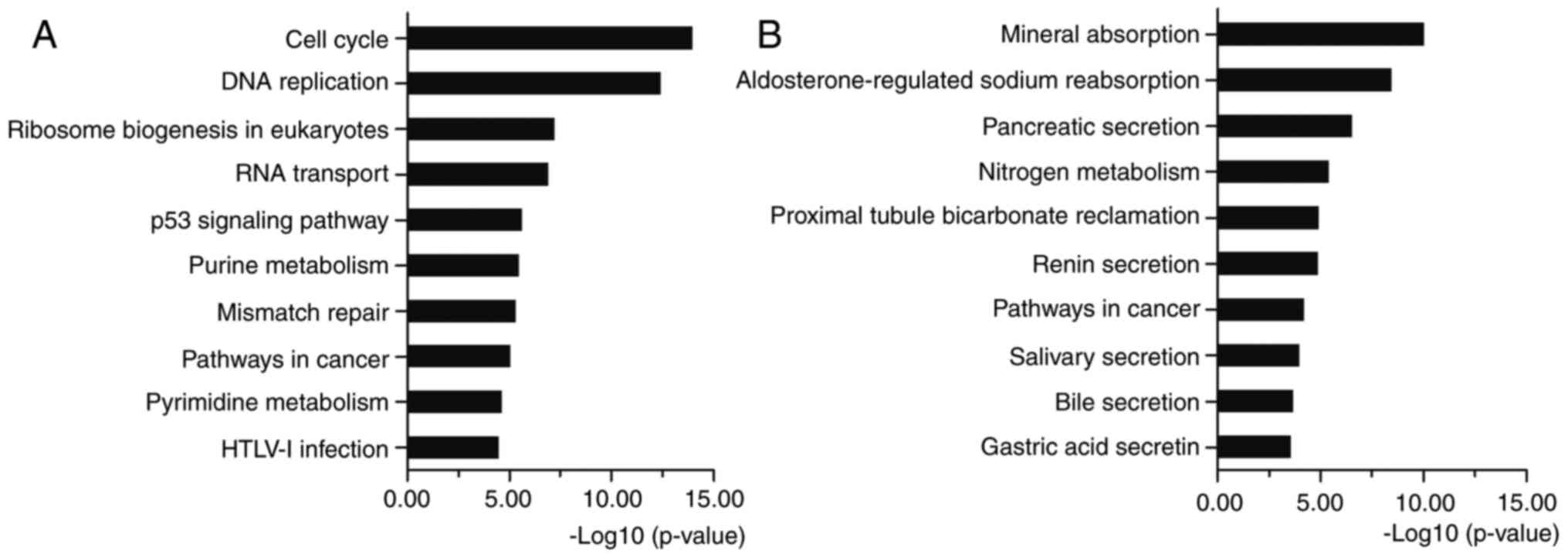
KEGG analysis showed that the upregulated DEGs were
mostly enriched in cell cycle, DNA replication, p53 signaling
pathway and pathways in cancer (Fig.
3A). The downregulated DEGs were enriched in metabolic pathways
and pathways in cancer (Fig. 3B).
Cell cycle is the series of events that take place in a cell
leading to its division and DNA replication to produce two new
cells. Abnormal cell cycle regulation will lead to the occurrence
of cancer. Many molecules participate in cancers by regulating the
cell cycle (19,20). Multiple genes can disturb DNA
replication and cause DNA replication stress and genome instability
(21). Oncogenes increase DNA
replicative stress and induce a DNA damage response early in
tumorigenesis (22). Activated by
various factors, p53 signaling pathway represses cancer progression
by arresting growth, or by promoting cellular death programs
(23). Various molecules regulate
the cell cycle and apoptosis though activating or repressing p53
signaling pathway (24–26). Pathways in cancer consist of many
classical signaling pathways, such as Wnt, PI3K-Akt, MAPK, VEGF,
PPAR, TGF-β and p53 signaling pathways, which play significant
roles in regulating cell proliferation, apoptosis, invasion,
metastasis and adhesion. The KEGG pathway enrichment proved the
relationship between the identified DEGs and CRC. All the above
analyses supported the reliability of the results in the present
study.
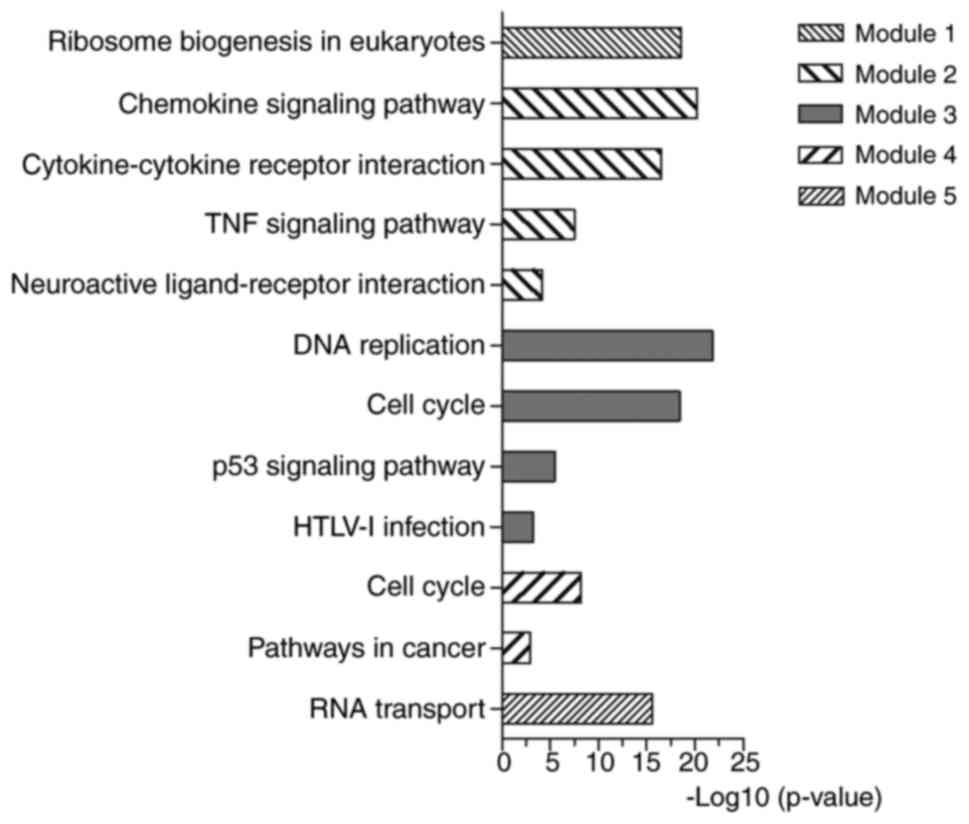
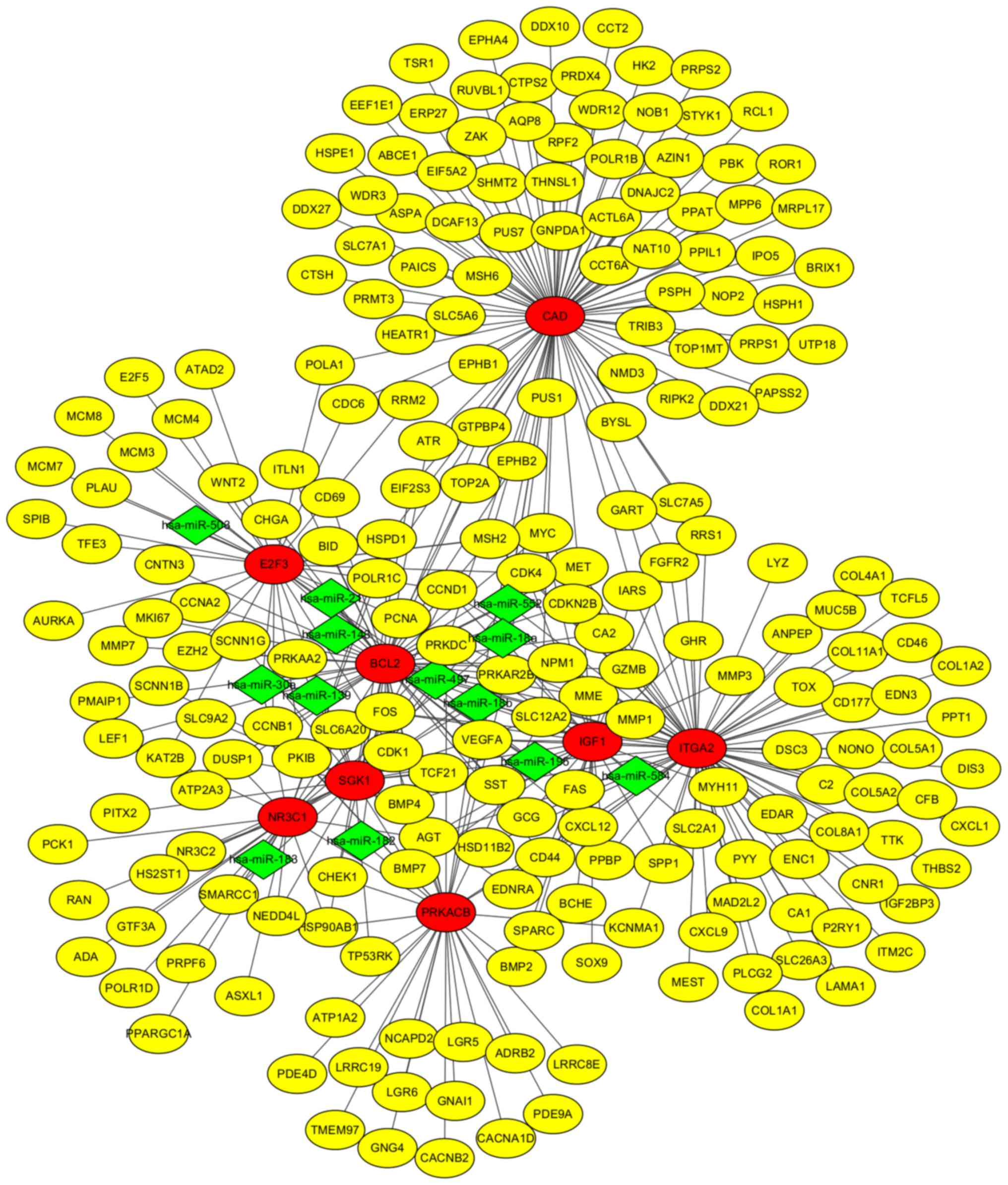
PPI network construction and analysis
of modules
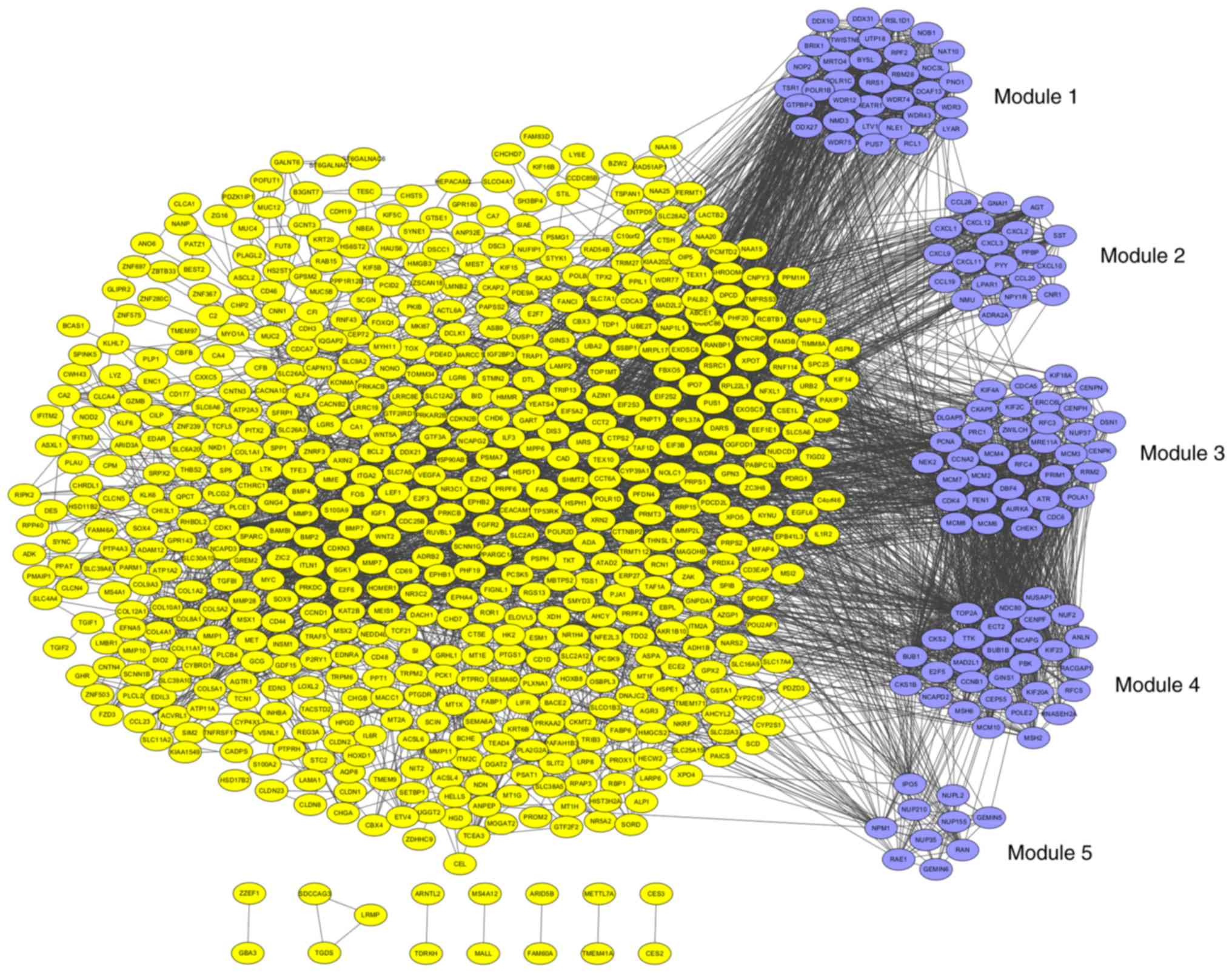
The PPI network was constructed by STRING and
visualized by Cytoscape (version 3.4.0) (Fig. 4). Degree ≥20 was set as the cut-off
criterion. A total of 168 DEGs were chosen as hub genes, such as
CAD, ITGA2, BCL2, PRKACB, IGF1, SGK1, E2F3 and NR3C1. Moreover, the
whole PPI network was analyzed by MCODE. The top five modules were
selected, and the KEGG pathway enrichment analysis of the genes
involved in modules were performed by KOBAS 3.0. The pathway
enrichment analysis revealed that DEGs in these modules were mostly
enriched in TNF signaling pathway, DNA replication, cell cycle, p53
signaling pathway and pathways in cancer (Fig. 5).
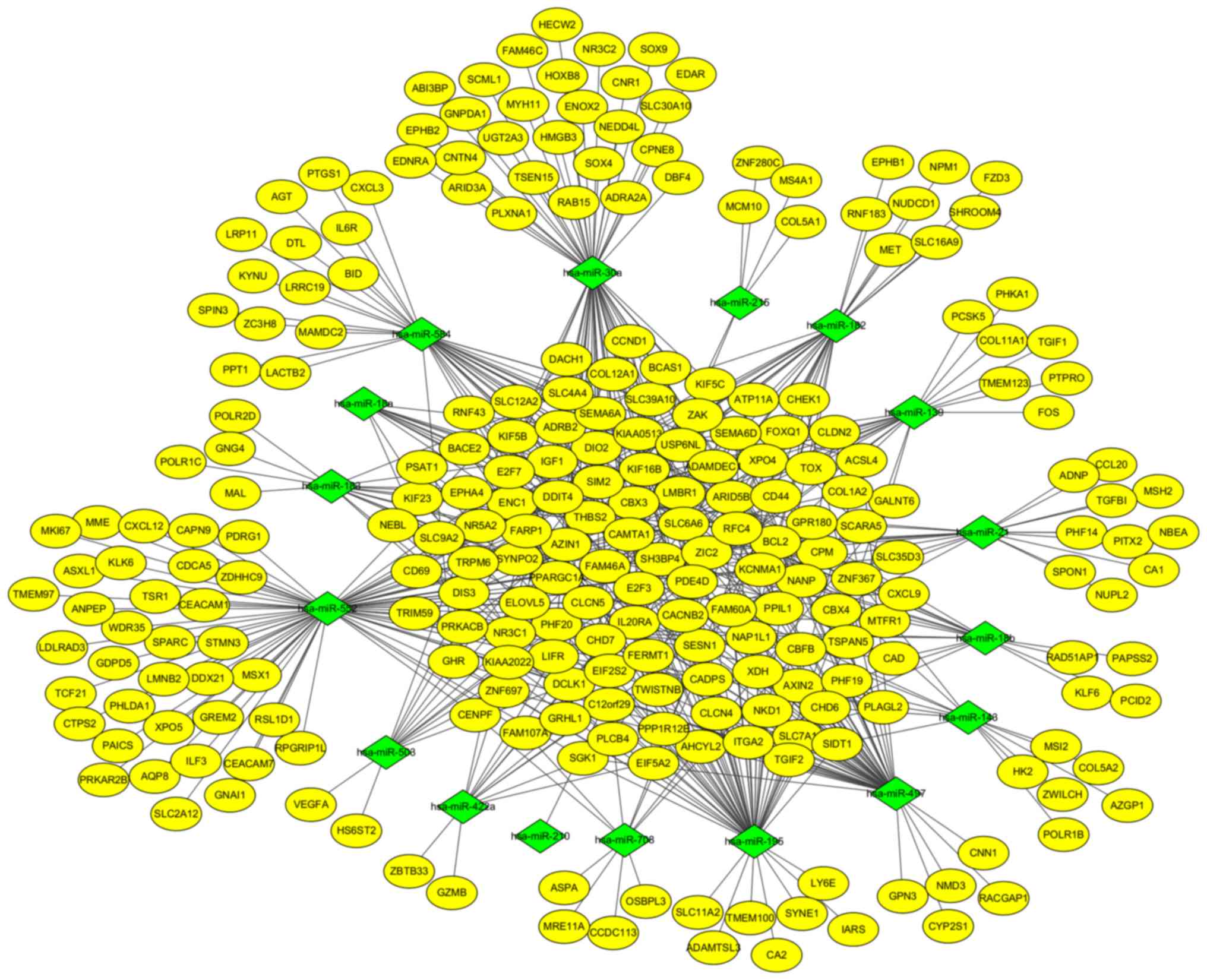
miRNA-gene network
To further understand the regulatory relationship
between identified DEGs and miRNAs, two miRNA profiles were
analyzed. A total of 45 and 25 upregulated DE miRNAs (Fig. 1C) were identified with 38 and 20
downregulated DE miRNAs (Fig. 1D)
from GSE39845 and GSE53592, respectively. Among them, 13
upregulated DE miRNAs and seven downregulated DE miRNAs were
screened out in both datasets. The targeted genes were predicted by
the five programs. Besides, to identify the reliable target genes,
we compared the targets with DEGs. Only the overlapping genes were
selected. Following, miRNA-gene network was constructed (Fig. 6). By analyzing PPI and miRNA-gene
network, a total of eight genes were chosen, consisting of 3
upregulated DEGs (CAD, ITGA2 and E2F3) and 5 downregulated DEGs
(BCL2, PRKACB, IGF1, SGK1, and NR3C1) (Fig. 7).
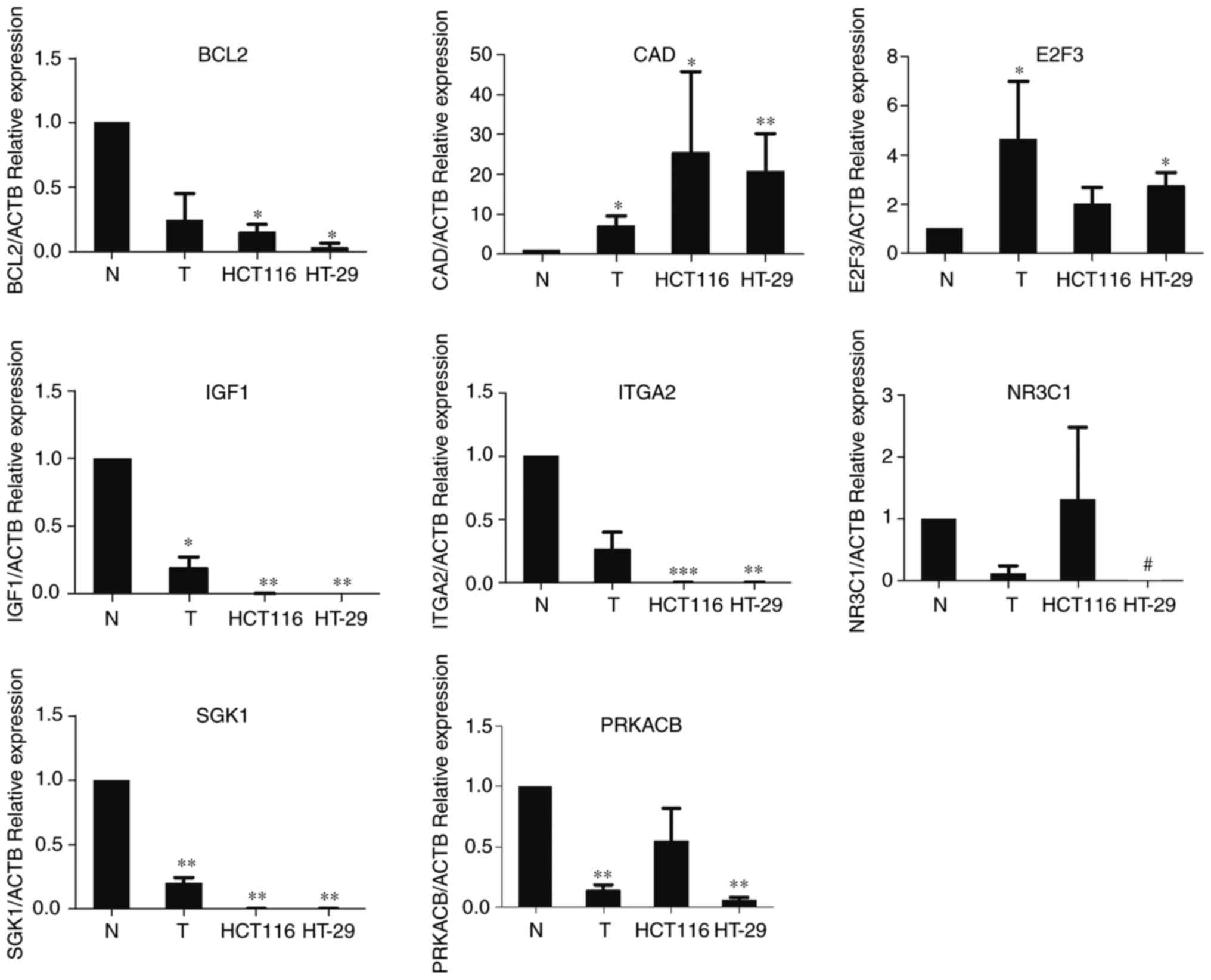
Experimental validation of the
identified biomarker genes
To verify the bioinformatics analysis for predicting
the potential biomarkers in CRC, the eight candidate genes were
selected for further quantitative real-time PCR. The expression of
eight genes was detected in both tissue samples and CRC cell lines
(HCT116 and HT-29). As shown in Fig.
8, seven of the eight identified genes were confirmed to have
the same expression trend as predicted by bioinformatics analysis.
CAD and E2F3 were overexpressed in tumor tissue samples and CRC
cell lines, while the expression of BCL2, PRKACB, IGF1, SGK1 and
NR3C1 was reduced in CRC tissue samples and cell lines. However,
the expression of ITGA2 in tumor tissue samples and cell lines was
lower than that in normal tissue samples. Besides, there was no
significant difference in the expression of NR3C1 and PRKACB in
HCT116 cell line and normal tissues.
Discussion
Recently, microarray and bioinformatics analysis
have been widely used in identifying potential targets for
diagnosis and therapy of different cancers. In the present study, a
total of 883 DEGs were screened, including 600 upregulated DEGs and
283 downregulated DEGs. We then performed functional enrichment
analysis on the DEGs. For the upregulated DEGs, GO terms were
significantly enriched in binding at MF level, organelle at CC
level and cellular process at BP level. For the downregulated DEGs,
most enriched GO terms were binding at MF level, extracellular
region part at CC level and chemical homeostasis at BP level. Most
of these GO terms were basic regulatory concepts in cell. Pathway
enrichment analysis revealed that cell cycle, p53 signaling pathway
and pathways in cancer were significantly enriched, which were
associated with the occurrence of cancers. In addition, we
constructed the PPI network and analyzed the top five modules. KEGG
pathway analysis revealed that the DEGs in these modules were also
enriched in cell cycle, p53 signaling pathway and pathways in
cancer, which confirmed the findings above. These enriched pathways
revealed insight into the molecular mechanisms of CRC, which could
provide effective therapeutic strategies for CRC.
miRNAs have been reported to participate in the
occurrence and progress of cancer. In the present study, we
identified 13 upregulated miRNAs and seven downregulated miRNAs. We
then constructed the miRNA-gene network. To identify the key
biomarkers of CRC, we combined the two networks. A gene with higher
degree in PPI network and regulated by more miRNAs in miRNA-gene
network was considered to play a more important role in CRC. A
total of eight genes were identified, consisting of three
upregulated DEGs (CAD, ITGA2 and E2F3) and five downregulated DEGs
(BCL2, PRKACB, IGF1, SGK1 and NR3C1). Besides, hsa-miR-552 and
hsa-miR-30a represented the more important role in regulating
target genes.
Experimental validation is necessary to confirm our
results predicted by bioinformatics analysis. We found that seven
of the eight candidate genes have the same expression trend as
predicted by qPCR (P<0.05). Although ITGA2 did not overexpress
in CRC tissue as predicted, it has been reported to be
downregulated in prostate cancer (27). In addition, Ferraro et al
(28) proved that depressed ITGA2
could reduce cell migration in colon cancer. As for NR3C1, its
expression in HCT116 cell line was significantly higher than that
in tumor tissues, but could not be detected in HT-29 cell line.
Lind et al (29) reported
that hypermethylation was significantly associated with the absence
or reduced expression of NR3C1, which leads to different expression
levels of NR3C1 in different tissues and cells.
The pathogenesis of cancer is a complex process
driven by many factors, including genetic and epigenetic
alterations. There was no report on CAD in CRC. However, increased
expression of CAD was associated with local tumor extension and
cancer recurrence in prostate cancer (30). It may become a novel biomarker of
CRC. Downregulation of E2F3 could inhibit cell proliferation and
induce apoptosis in CRC. Besides, E2F3 is a target gene of several
miRNAs (31,32). BCL2 is an apoptosis regulator. It
has been identified as a cause of several cancers, including CRC,
melanoma, breast and lung cancer (33–36).
PRKACB is a member of the serine/threonine protein kinase family
and encodes a catalytic subunit of cAMP-dependent protein kinase.
Kvissel et al (37) reported
that PRKACB isoforms play different roles in proliferation and
differentiation in prostate cancer. IGF1 polymorphisms could
influence the risk of CRC (38).
For example, the SNP rs6214 of IGF1 could increase the CRC risk
(39). Besides, the expression
level of IGF1 correlates significantly with tumor size (P=0.0024)
and depth of invasion (P=0.0147) in CRC (40). SGK1 promotes survival, invasiveness,
and metastasis of CRC cells (41).
It can also enhance the growth and migration of NSCLC cells by
activating β-catenin/TCF signaling pathway (42). SGK1 expression is also increased in
endometrial cancer. Suppression of the expression of SGK1 can
induce autophagy and apoptosis (43). hsa-miR-552 and hsa-miR-30a were
identified as key miRNAs in the miRNA-gene network. Downregulation
of miR-552 reduced cell proliferation, migration and clonogenicity
in CRC (44). Conversely,
inhibition of miR-30a promoted cell proliferation, migration and
invasion in CRC (45).
Cancer is a complex disease caused by multiple
factors. A single reason is not sufficient to explain the mechanism
of cancer. Combination of mRNAs, miRNAs and interaction networks
can help us to investigate and explain the molecular mechanism of
cancer (46). It is an effective
approach to identify biomarkers of various diseases.
In the present study, we applied bioinformatics
analysis to identify the biomarkers of CRC and performed
experimental validation. But there were still some limitations in
this study. First, the amount of CRC tissue samples was not enough.
Because of the difficulty to obtain clinical samples and
information. Second, we only checked two cell lines, which cannot
fully reveal the difference in expression of biomarkers in CRC.
In conclusion, eight genes and two miRNAs were
identified as the biomarkers of CRC. Besides, this study also
provided a series of significant pathways and mechanisms for
diagnosis and therapy. Eight genes were verified by experiments,
including CAD, ITGA2, E2F3, BCL2, PRKACB, IGF1, SGK1 and NR3C1,
which might become the new stars in research of CRC.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
DEGs
|
differentially expressed genes
|
|
DE miRNAs
|
differentially expressed genes
miRNAs
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
|
STRING
|
Search Tool for the Retrieval of
Interacting Genes
|
|
MCODE
|
molecular complex detection
|
|
qPCR
|
quantitative real-time PCR
|
References
|
1
|
Haraldsdottir S, Einarsdottir HM,
Smaradottir A, Gunnlaugsson A and Halfdanarson TR: Colorectal
cancer - review. Laeknabladid. 100:75–82. 2014.(in Icelandic).
PubMed/NCBI
|
|
2
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Simon K: Colorectal cancer development and
advances in screening. Clin Interv Aging. 11:967–976. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thomas J, Ohtsuka M, Pichler M and Ling H:
MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci.
16:28063–28076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren A, Dong Y, Tsoi H and Yu J: Detection
of miRNA as non-invasive biomarkers of colorectal cancer. Int J Mol
Sci. 16:2810–2823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of patients with colorectal cancer: A potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thomas PD: The Gene Ontology and the
meaning of biological function. Methods Mol Biol. 1446:15–24. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xing Z, Chu C, Chen L and Kong X: The use
of Gene Ontology terms and KEGG pathways for analysis and
prediction of oncogenes. Biochim Biophys Acta. 1860:2725–2734.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khunlertgit N and Yoon BJ: Incorporating
topological information for predicting robust cancer subnetwork
markers in human protein-protein interaction network. BMC
Bioinformatics. 17 Suppl 13:3512016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clough E and Barrett T: The Gene
Expression Omnibus Database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:(Web Server issue). W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44(D1): D457–D462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Killcoyne S, Carter GW, Smith J and Boyle
J: Cytoscape: A community-based framework for network modeling.
Methods Mol Biol. 563:219–239. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dashwood MR and Loizidou M: Determination
of cell-specific receptor binding using a combination of
immunohistochemistry and in vitro autoradiography: Relevance to
therapeutic receptor targeting in cancer. Methods Mol Biol.
878:137–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Lacerda TC, Costa-Silva B, Giudice FS,
Dias MV, de Oliveira GP, Teixeira BL, Dos Santos TG and Martins VR:
Prion protein binding to HOP modulates the migration and invasion
of colorectal cancer cells. Clin Exp Metastasis. 33:441–451. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsujitani S, Saito H, Honboh T, Ataka M,
Tanida T, Makino M and Ikeguchi M: Prognostic significance of
receptor-binding cancer antigen expressed on SiSo cells (RCAS1)
expression in relation to cadherin expression in patients with
colorectal carcinoma. Dis Colon Rectum. 50:1241–1249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang B, Liu Z, Cao Y, Zhu C, Zuo Y, Huang
L, Wen G, Shang N, Chen Y, Yue X, et al: MC37, a new mono-carbonyl
curcumin analog, induces G2/M cell cycle arrest and
mitochondria-mediated apoptosis in human colorectal cancer cells.
Eur J Pharmacol. 796:139–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Wu X, Ouyang W, Gu M, Gao Z, Song
M, Chen Y, Lin Y, Cao Y and Xiao H: Novel ent-Kaurane
diterpenoid from Rubus corchorifolius L. f. inhibits human
colon cancer cell growth via inducing cell cycle arrest and
apoptosis. J Agric Food Chem. 65:1566–1573. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Boyer AS, Walter D and Sørensen CS: DNA
replication and cancer: From dysfunctional replication origin
activities to therapeutic opportunities. Semin Cancer Biol.
37–38:16–25. 2016. View Article : Google Scholar
|
|
22
|
Hills SA and Diffley JF: DNA replication
and oncogene-induced replicative stress. Curr Biol. 24:R435–R444.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stegh AH: Targeting the p53 signaling
pathway in cancer therapy - the promises, challenges and perils.
Expert Opin Ther Targets. 16:67–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu YH, Liu GH, Mei JJ and Wang J: The
preventive effects of hyperoside on lung cancer in vitro by
inducing apoptosis and inhibiting proliferation through caspase-3
and P53 signaling pathway. Biomed Pharmacother. 83:381–391. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen J, Wei Y, Feng Q, Ren L, He G, Chang
W, Zhu D, Yi T, Lin Q, Tang W, et al: Ribosomal protein S15A
promotes malignant transformation and predicts poor outcome in
colorectal cancer through misregulation of p53 signaling pathway.
Int J Oncol. 48:1628–1638. 2016.PubMed/NCBI
|
|
26
|
Xiao S, Zhou Y, Yi W, Luo G, Jiang B, Tian
Q, Li Y and Xue M: Fra-1 is downregulated in cervical cancer
tissues and promotes cervical cancer cell apoptosis by p53
signaling pathway in vitro. Int J Oncol. 46:1677–1684. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shaikhibrahim Z, Lindstrot A, Buettner R
and Wernert N: Analysis of laser-microdissected prostate cancer
tissues reveals potential tumor markers. Int J Mol Med. 28:605–611.
2011.PubMed/NCBI
|
|
28
|
Ferraro A, Boni T and Pintzas A: EZH2
regulates cofilin activity and colon cancer cell migration by
targeting ITGA2 gene. PLoS One. 9:e1152762014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lind GE, Kleivi K, Meling GI, Teixeira MR,
Thiis-Evensen E, Rognum TO and Lothe RA: ADAMTS1, CRABP1, and NR3C1
identified as epigenetically deregulated genes in colorectal
tumorigenesis. Cell Oncol. 28:259–272. 2006.PubMed/NCBI
|
|
30
|
Morin A, Fritsch L, Mathieu JR, Gilbert C,
Guarmit B, Firlej V, Gallou-Kabani C, Vieillefond A, Delongchamps
NB and Cabon F: Identification of CAD as an androgen receptor
interactant and an early marker of prostate tumor recurrence. FASEB
J. 26:460–467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang SW, Yue J, Wang BC and Zhang XL:
miR-503 inhibits cell proliferation and induces apoptosis in
colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol.
8:12853–12860. 2015.PubMed/NCBI
|
|
32
|
Fang Y, Gu X, Li Z, Xiang J and Chen Z:
miR-449b inhibits the proliferation of SW1116 colon cancer stem
cells through downregulation of CCND1 and E2F3 expression. Oncol
Rep. 30:399–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tong Z, Liu N, Lin L, Guo X, Yang D and
Zhang Q: miR-125a-5p inhibits cell proliferation and induces
apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1.
Biomed Pharmacother. 75:129–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaluzki I, Hrgovic I, Hailemariam-Jahn T,
Doll M, Kleemann J, Valesky EM, Kippenberger S, Kaufmann R, Zoeller
N and Meissner M: Dimethylfumarate inhibits melanoma cell
proliferation via p21 and p53 induction and bcl-2 and cyclin B1
downregulation. Tumour Biol. 37:13627–13635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meka Bhushann P, Jarjapu S, Vishwakarma
SK, Nanchari SR, Cingeetham A, Annamaneni S, Mukta S, Triveni B and
Satti V: Influence of BCL2-938 C>A promoter polymorphism and
BCL2 gene expression on the progression of breast cancer. Tumour
Biol. 37:6905–6912. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang X, Gao F, Ma F, Ren Y, Chen H, Liang
X, Han S, Xiong X, Pan W, Zhou C, et al: Association of the
functional BCL-2 rs2279115 genetic variant and small cell lung
cancer. Tumour Biol. 37:1693–1698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kvissel AK, Ramberg H, Eide T, Svindland
A, Skålhegg BS and Taskén KA: Androgen dependent regulation of
protein kinase A subunits in prostate cancer cells. Cell Signal.
19:401–409. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wong HL, Koh WP, Probst-Hensch NM, Van den
Berg D, Yu MC and Ingles SA: Insulin-like growth factor-1 promoter
polymorphisms and colorectal cancer: A functional genomics
approach. Gut. 57:1090–1096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feik E, Baierl A, Hieger B, Führlinger G,
Pentz A, Stättner S, Weiss W, Pulgram T, Leeb G, Mach K, et al:
Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps
and colorectal cancer risk. Cancer Causes Control. 21:91–97. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shiratsuchi I, Akagi Y, Kawahara A,
Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M and
Shirouzu K: Expression of IGF-1 and IGF-1R and their relation to
clinicopathological factors in colorectal cancer. Anticancer Res.
31:2541–2545. 2011.PubMed/NCBI
|
|
41
|
Lang F, Perrotti N and Stournaras C:
Colorectal carcinoma cells -regulation of survival and growth by
SGK1. Int J Biochem Cell Biol. 42:1571–1575. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiaobo Y, Qiang L, Xiong Q, Zheng R,
Jianhua Z, Zhifeng L, Yijiang S and Zheng J: Serum and
glucocorticoid kinase 1 promoted the growth and migration of
non-small cell lung cancer cells. Gene. 576:339–346. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Conza D, Mirra P, Calì G, Tortora T,
Insabato L, Fiory F, Schenone S, Amato R, Beguinot F, Perrotti N,
et al: The SGK1 inhibitor SI113 induces autophagy, apoptosis, and
endoplasmic reticulum stress in endometrial cancer cells. J Cell
Physiol. Feb 8–2017.(Epub ahead of print). https://doi.org/10.1002/jcp.25850 View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang J, Li H, Wang Y, Wang L, Yan X, Zhang
D, Ma X, Du Y, Liu X and Yang Y: MicroRNA-552 enhances metastatic
capacity of colorectal cancer cells by targeting a disintegrin and
metalloprotease 28. Oncotarget. 7:70194–70210. 2016.PubMed/NCBI
|
|
45
|
Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv
G, Zou Z, Jiang XC, Zou C, Liu W, et al: Role of microRNA 30a
targeting insulin receptor substrate 2 in colorectal tumorigenesis.
Mol Cell Biol. 35:988–1000. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zeng T, Sun SY, Wang Y, Zhu H and Chen L:
Network biomarkers reveal dysfunctional gene regulations during
disease progression. FEBS J. 280:5682–5695. 2013. View Article : Google Scholar : PubMed/NCBI
|