Introduction
Medulloblastoma (MB) accounts for approximately
one-fifth of all pediatric brain tumors and is one of the most
common malignant brain tumors in children (1–3). Due
to the progress in radiation therapy and chemotherapy during the
past decades, the survival of MB patients has considerably improved
(4,5). Nevertheless, in WHO grade IV central
nervous system tumors, the prognosis of MB remains dismal due to
rapid recurrence or metastasis in a considerable proportion of the
tumors (6). Therefore, it is
necessary to gain more insights in the carcinogenesis of MB and
explore molecular targets potentially useful for treating the
disease.
Ubiquitin-like with PHD and RING finger domains 1
(UHRF1) is comprised of an N-terminal ubiquitylation-like domain, a
plant homeodomain (PHD) domain, a RING finger, and a SET- and
RING-associated (SRA) domain, which plays a major role in DNA
methylation and cell proliferation (7,8). The
SRA domain of UHRF1 recognizes hemi-methylated DNA, recruits DNA
methyltransferase 1 (DNMT1) and maintains DNA-methylation patterns
(9). In the past few years, UHRF1
was revealed to be aberrantly upregulated and functioned as an
oncogene in various types of cancers, including laryngeal squamous
cell carcinoma (10), non-small
cell lung (11), breast (12) and gastric cancer (13), hepatocellular carcinoma (14), and colorectal (15), bladder (16) and prostate cancer (17). Most recently, we demonstrated that
UHRF1 was overexpressed in MB tissues and was an independent
prognostic factor. We further revealed that UHRF1 promoted the
proliferation and progression of MB cells, indicating that UHRF1
could be a potential therapeutic target for MB (18).
MicroRNAs (miRNAs), a class of ~19–25 nt non-coding
RNAs, are evolutionarily conserved, and serve as endogenous
regulators of gene expression. They also play a vital role in
carcinogenesis. miRNAs could cause either mRNA degradation or
inhibition of protein translation via direct interaction with the
3′-UTR of target mRNA (19,20). A growing body of compelling data has
revealed that the malfunction of certain miRNAs is involved in a
series of tumorigenic processes including cell proliferation,
apoptosis, metastasis and angiogenesis (21). Recent studies revealed that
dysregulation of miR-378 was shown related to tumorigenesis and
cancer progression. Notably, miR-378 was reported as an onco-miR in
nasopharyngeal carcinoma (22), but
as a tumor suppressor microRNA in colorectal (23) and prostate cancer (24), and glioma (25). Nevertheless, to the best of our
knowledge, the molecular mechanism of miR-378 in the regulation of
MB is unknown.
Playing a vital part in connection to DNA
methylation, UHRF1 was implicated in MB carcinogenesis in our
previous study (18). However,
little is known about the factors that modulated UHRF1 expression.
In the present study, we found that miR-378 was downregulated in MB
specimens and was inversely associated with UHRF1 expression. We
further predicted and confirmed that miR-378 modulated the
expression of UHRF1, and exerted a tumor-suppressive function in MB
proliferation.
Materials and methods
Patients, tissue specimens
Fresh frozen tissues of 19 patients with MB and 9
cases of normal cerebellum (collected in brain trauma surgery) were
gathered at The First Affiliated Hospital of Zhengzhou University
between January 2015 and September 2016. Informed consent was
obtained from all individual participants included in the study and
all procedures performed involving human participants were in
accordance with the ethical standards of the Institutional Research
Committee of The First Affiliated Hospital of Zhengzhou
University.
Cell culture and transfection
DAOY and HEK 293T cell lines were purchased from
American Type Culture Collection (ATCC, Manassas, VA, USA). ONS-76
cell line was obtained from Japanese Collection of Research
Bioresources Cell Bank (JCRB Cell Bank, Osaka, Japan). ONS-76 cells
were cultured in RPMI-1640 medium (Thermo Scientific HyClone,
Beijing, China) and DAOY cells were maintained in MEM-α medium
(Life Technologies, Carlsbad, CA, USA). The transfection was
performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad,
CA, USA) according to the manufacturer's instructions. miR-378 was
cloned into the CMV-miRNA-PGK-puromycin vector (Genomeditech,
Shanghai, China). UHRF1 cDNA was cloned into
pLVX-Neo-IRES-ZsGreen1 (Genomeditech). Puromycin was used to
establish the miR-378 stably transfected cell lines. Fluorescence
was detected for selection of clones with stable UHRF1 expression.
Stably transfected clones were validated by quantitative real-time
polymerase chain reaction (qRT-PCR) and immunoblotting. Transfected
cells were then used for Cell Counting Kit-8 (CCK-8) and clonogenic
assays, and flow cytometry.
Real-time quantitative PCR
Small RNAs were isolated from tissue samples with
the mirVana miRNA Isolation kit (Ambion, Austin, TX, USA) following
the manufacturer's instructions. The total RNA of the fresh
tissues, ONS-76 and DAOY cells was extracted using TRIzol reagent
(Invitrogen) and reversely transcribed with RT Primer Mix and
PrimeScript RT Enzyme Mix 1 (Takara, Shiga, Japan). The mRNA level
was quantified with SYBR Premix Ex Taq (Takara). Human-U6 RNA was
chosen as an endogenous control. The PCR reaction was conducted
with a real-time PCR system (Roche Diagnostics, Penzberg, Germany).
Conditions for amplification were 94°C for 2 min followed by 40
cycles of 94°C for 25 sec, 61°C for 31 sec. The fold change was
calculated using the 2−ΔΔCt method. The levels of
miR-378 in normal brain tissues were normalized to an arbitrary
value of 1. The primers used are listed in Table I.
 | Table I.Primers used in quantitative real-time
PCR. |
Table I.
Primers used in quantitative real-time
PCR.
| Primer name | Sequence (5′-3′) |
|---|
| Pre-miR-378 | F
CGACGCGTCGGGCTGCGAGGAGTGAGCG |
|
| R
CCATCGATGGGAGTTCAAATGGCTTGCTCC |
| U6 | F
TGCGGGTGCTCGCTTCGGCAGC |
|
| R
CCAGTGCAGGGTCCGAGGT |
| UHRF1 | F
CCACATCGTCCTCACAGC |
|
| R
GGTCCACATCATCCTCATAGC |
| GAPDH | F
TGATGACATCAAGAAGGTGGTGAAG |
|
| R
TCCTTGGAGGCCATGTGGGCCAT |
Western blot analysis
Total protein was extracted from fresh tissues,
ONS-76 and DAOY cells using cell lysates. Gene expression was
normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Fifty micrograms of samples were first electrophoresed on a 10%
SDS-polyacrylamide gels, transferred to nitrocellulose filter
membranes, and incubated in blocking solution consisting of 5%
non-fat milk in Tris-buffered saline with Tween-20 (TBST) at room
temperature for 1 h. Subsequently, the membranes were immunoblotted
with UHRF1 (1:250 dilution; BD Biosciences, San Jose, CA, USA),
cleaved PARP (1:1,000 dilution), cleaved caspase-3 (1:1,000
dilution) and GAPDH antibodies (1:5,000 dilution) [all from Cell
Signaling Technology (CST) Beverly, MA, USA], overnight at 4°C.
Subsequently, the membranes were washed and incubated with
appropriate HRP-conjugated secondary IgG antibodies (Dako,
Glostrup, Denmark). The protein bands were visualized using ECL
Western Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Target prediction and dual-luciferase
reporter assay
Three miRNA target prediction algorithms were used
(miRWalk 2.0, TargetScan and microrna.org).
For the dual-luciferase report assay, the wild-type (WT) reporter
construct pmirGLO/UHRF1-3′-UTR and the mutant (MUT) reporter
construct pmirGLO/UHRF1-3′-UTR MUT were cloned. The site of perfect
complementarity to miR-378 was mutated using site-directed
mutagenesis PCR. Subsequently, 293 cells were co-transfected with
miR-378 in a 48-well plate followed by the pmirGLO/UHRF1-3′-UTR
reporter vector or the pmirGLO/UHRF1-3′-UTR MUT vector. Firefly
luciferase activity for each sample was normalized to
Renilla luciferase activity and was assessed at 48 h after
transfection.
CCK-8 assay
Cell viability was assessed using a CCK-8 assay.
Stably transfected ONS-76 and DAOY cells were seeded at a density
of 103 cells/well into 96-well culture plates and
incubated for 24, 48, 72 and 96 h. For the CCK-8 assay, the cells
were further incubated with 10 µl CCK-8 (Sigma, Santa Clara, CA,
USA) for 4 h. The absorbance was assessed at 450 nm.
Clonogenic assay
A flat plate clone formation assay was used to
investigate cell clonogenic ability. Cells in the logarithmic
growth phase were digested into a single-cell suspension using a
trypsin-EDTA solution, and 2 ml of cell suspension was then seeded
onto a 6-well cell culture plate at a density of 1,000 cells/ml.
The cells were cultured at 37°C in a well-humidified 95% air/5%
CO2 incubator for 2 weeks, and the colony formation was
photographed and counted.
Flow cytometry for cell apoptosis
The apoptotic ratios of cells were determined using
the Annexin V/7-ADD apoptosis detection kit (Roche, Basel,
Switzerland). Forty-eight hours after transfection, the cells were
harvested and washed twice with phosphate-buffered saline (PBS)
buffer, suspended with binding buffer, incubated with Annexin
V-R-PE for 20 min in a dark ice bath, and then with 7-AAD before
analysis on a BD FACSCalibur (BD Biosciences).
Statistical analysis
All in vitro experiments were repeated at
least 3 times. The numerical data were documented as the mean ±
standard deviation (SD). The Student's t-test or one-way analyses
of variance were employed to compare the differences in numerical
variables between different groups. Statistical significance was
defined as a P-value <0.05. Data statistics were performed using
software GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA,
USA) and IBM SPSS Statistics 19 (IBM Corp., Armonk, NY, USA).
Results
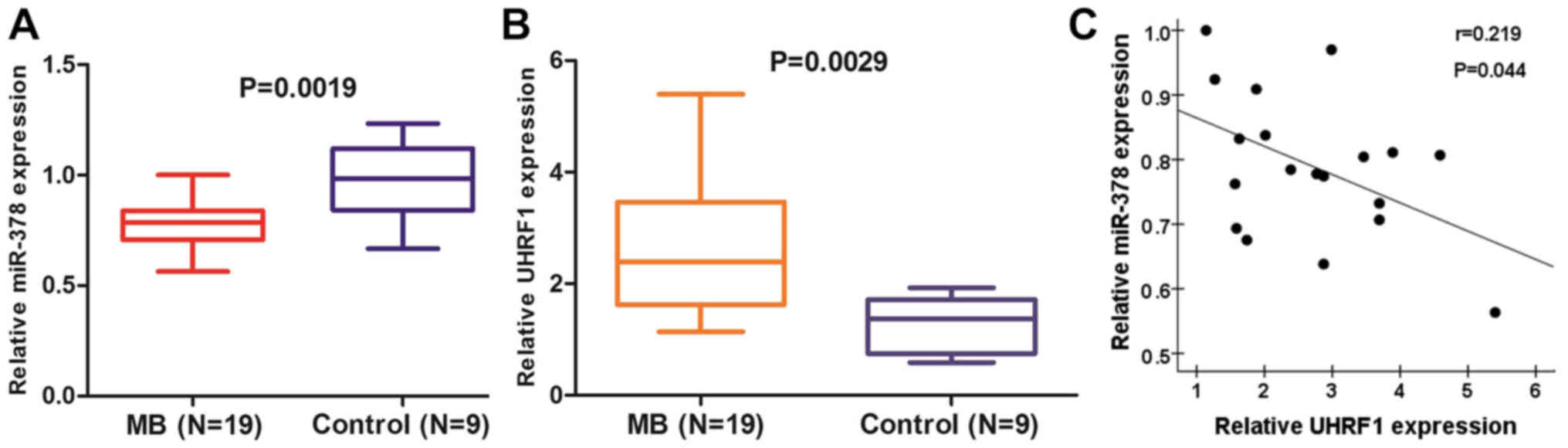
miR-378 is downregulated in MB tissues
and inversely correlated with UHRF1
To evaluate the expression of miR-378 in MB, we
first quantitatively detected the levels of reverse
transcription-PCR in 19 MB and 9 normal cerebellum tissues. The
results demonstrated that miR-378 was considerably decreased in MB
compared with normal cerebellum (Fig.
1A). Meanwhile, the expression of UHRF1 was revealed to be
significantly upregulated in MB tissues compared with normal
cerebellum tissues (Fig. 1B). In
combination with the two data, an inverse correlation between the
mRNA levels of miR-378 and UHRF1 in MB tissues was observed
(Fig. 1C).
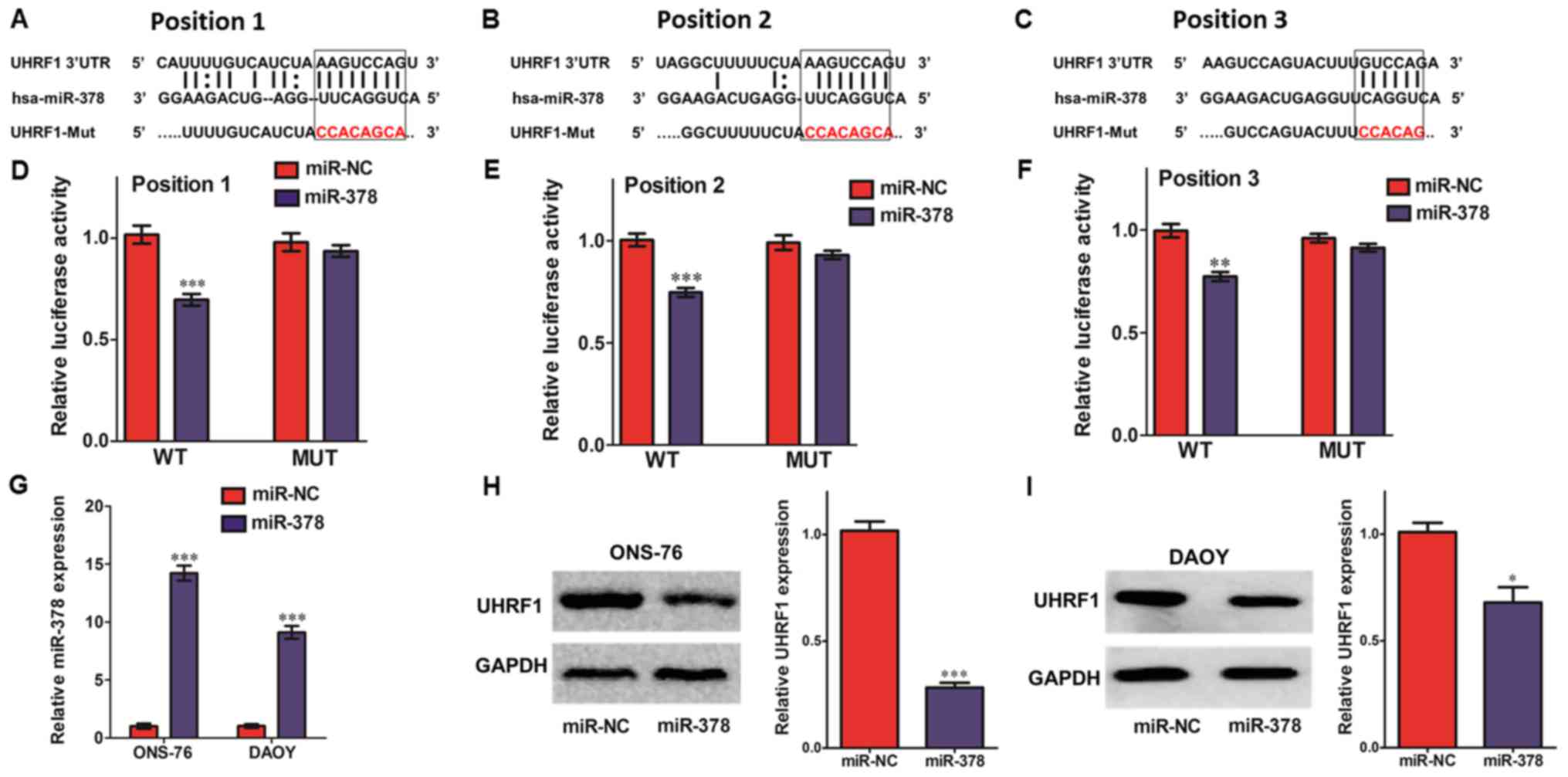
miR-378 is a potent miRNA regulator of
UHRF1 expression
Three miRNA target prediction algorithms including
miRWalk 2.0, Targetscan and microrna.org
were used to search for potential miRNA binding sites with the
3′-UTR region of UHRF1. Due to the higher prediction score (data
not shown), we chose miR-378 for further study. As shown in
Fig. 2A-C, the 3′-UTR of UHRF1 mRNA
contained a putative binding site for miR-378. We then cloned the
human UHRF1 3′-UTR sequence and inserted it into a luciferase
reporter plasmid and co-transfected it with miR-378 into 293 cells.
The luciferase reporter assays demonstrated that miR-378 markedly
inhibited the activity of firefly luciferase that contained the WT
but not the MUT 3′-UTR of UHRF1 (Fig.
2D-F). Moreover, we further investigated whether miR-378 could
modulate the endogenous expression of UHRF1 in MB cells. After
transfection of mature miR-378 by ectopic plasmids in MB cells, the
expression levels of miR-378 were substantially increased, ~14-fold
in ONS-76 cells and 9-fold in DAOY cells, respectively, compared to
the miR-NC control (Fig. 2G).
Consistently with previous luciferase reporter assays,
overexpression of miR-378 significantly inhibited UHRF1 expression
in ONS-76 and DAOY cells (Fig. 2H and
I). These data indicated that miR-378 negatively regulated the
expression of UHRF1 by directly binding to its 3′-UTR.
Overexpression of miR-378 suppresses
the proliferation of MB cells and restoration of UHRF1 counteracts
the effect of miR-378 in part
To better understand how miR-378 influences the
proliferation ability of MB cells, CCK-8 and colony formation
assays were conducted in ONS-76 and DAOY cells. In the meantime, to
confirm whether the effect of miR-378 on MB cells is mediated by
its regulatory role on UHRF1 expression, a rescue assay was
performed. Forced overexpression of miR-378 resulted in a marked
decrease in the expression of UHRF1, and this decrease was rescued
by transfection of UHRF1 (Fig. 3A and
E). A CCK-8 assay revealed that upregulation of miR-378
substantially suppressed the viability of ONS-76 and DAOY cells,
and ectopic expression of UHRF1 partly rescued the inhibition of
viability (ONS-76 MB cells, Fig.
3B; DAOY MB cells, Fig. 3F).
Additionally, the plate colony formation assay revealed that the
colony formation ability was markedly decreased in miR-378
overexpressed MB cells, and restoration of UHRF1 partly increased
the formation ability (ONS-76 MB cells, Fig. 3C and D; DAOY MB cells, Fig. 3G and H).
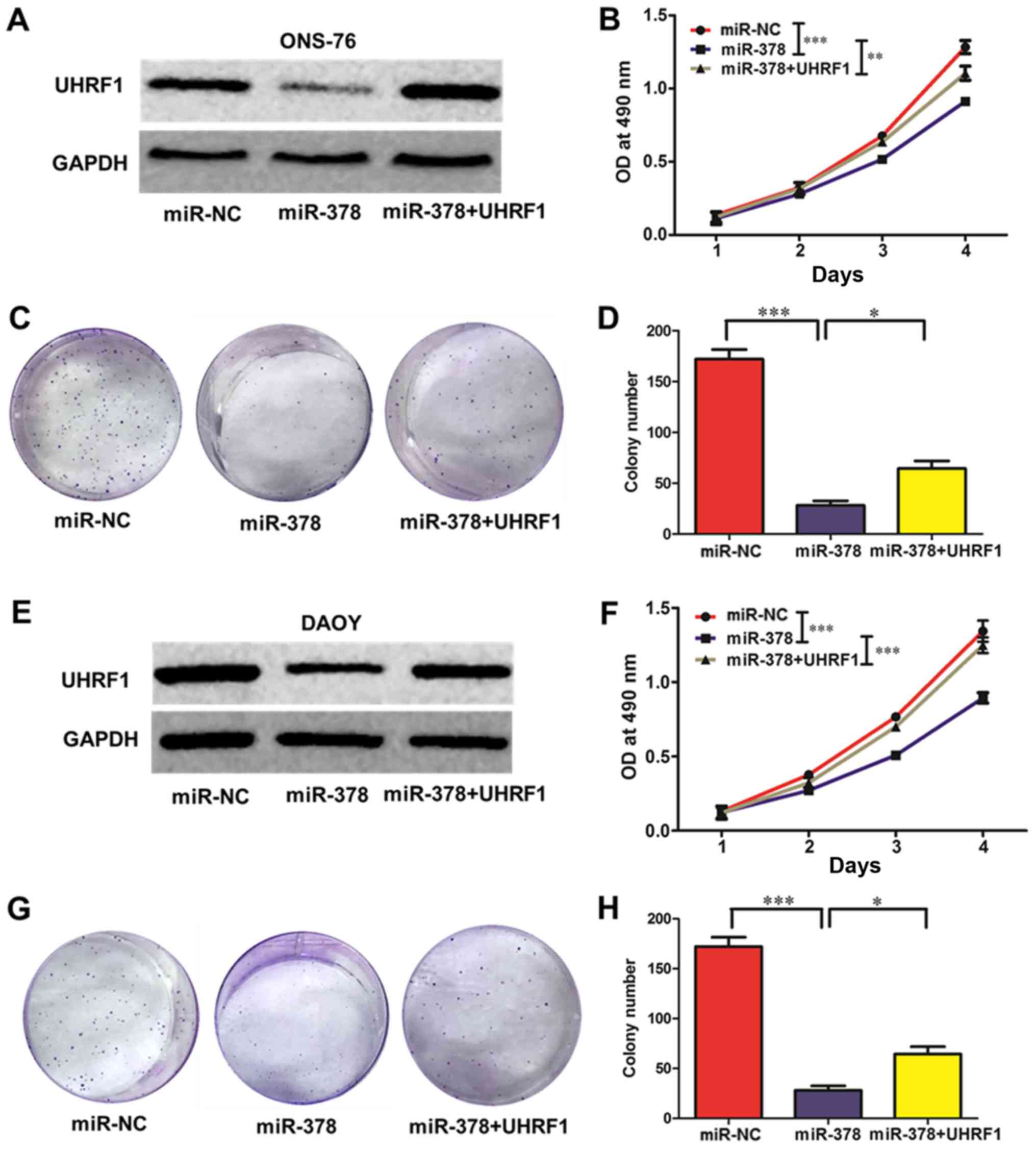 | Figure 3.miR-378 regulates ONS-76 and DAOY cell
viability and clonogenic ability. (A and E) The protein expression
of UHRF1 was assessed by western blot analysis. ONS-76 and DAOY
cells were transfected with control (miR-NC), miR-378, and miR-378
along with UHRF1. miR-378 suppressed UHRF1 protein expression
compared with the control group, while UHRF1 rescued the protein
level of UHRF1 compared with that of the miR-378 group. (B and F)
Cell viability was detected by CCK-8 assays. After transfection
with miR-NC, miR-378 and miR-378 along with UHRF1 in (B) ONS-76 and
(F) DAOY cells, the CCK-8 assay was used to determine the relative
cell viability at 1, 2, 3 and 4 days. (C, D, G and H) Cell
clonogenic ability was evaluated by colony formation assay. (C and
D) ONS-76 and (G and H) DAOY cells that were transfected with
miR-NC, miR-378 and miR-378 along with UHRF1 were seeded into
12-well plates. On the 12th day after seeding, the number of
colonies was counted; *P<0.05, **P<0.01, ***P<0.001. |
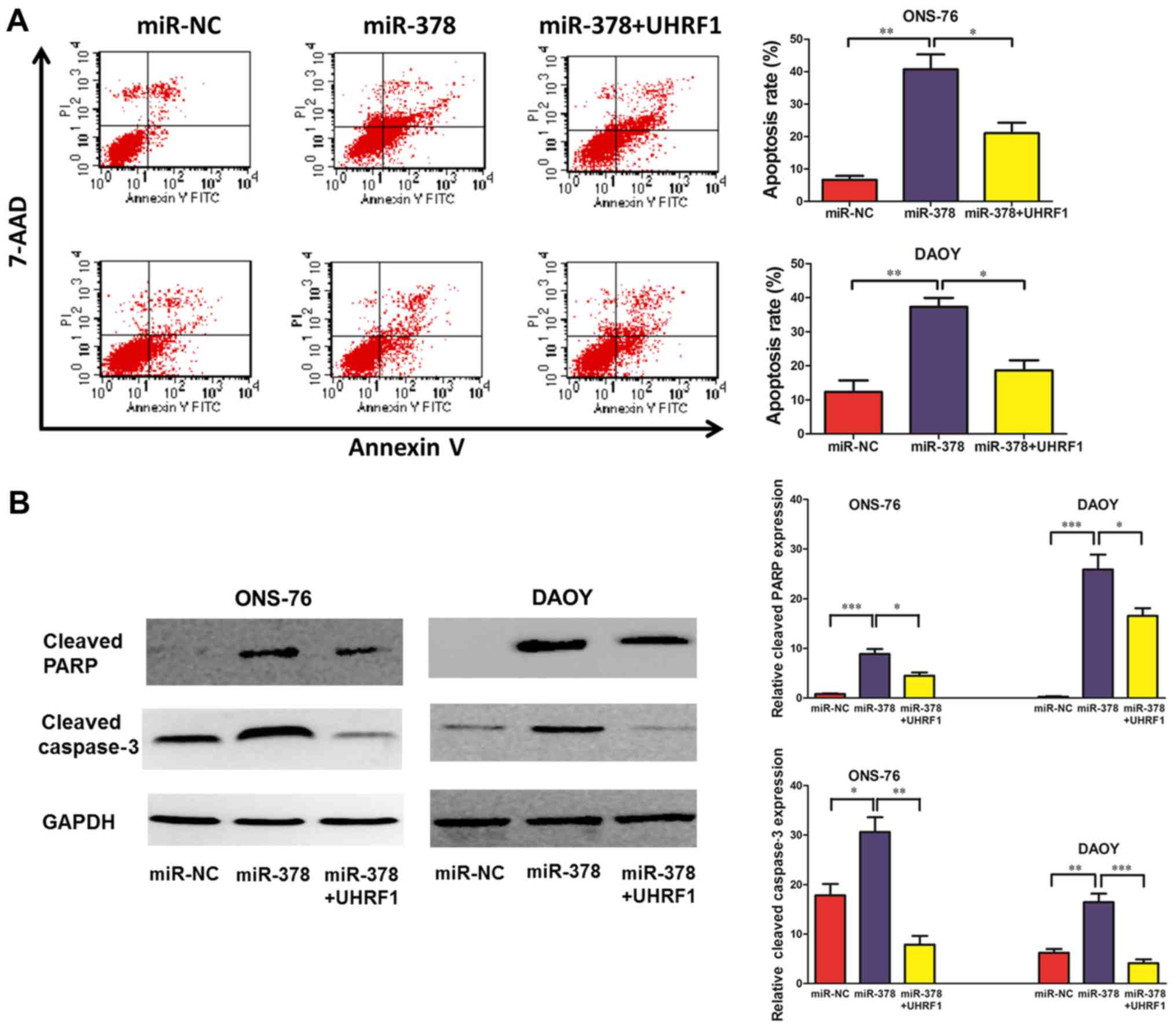
Overexpression of miR-378 promotes
apoptosis and restoration of UHRF1 counteracts the effect of
miR-378 in part
Fluorescence-activated cell sorting (FACS) analysis
was used to investigate the effects of miR-378 on MB cell
apoptosis. The results demonstrated that enforced expression of
miR-378 led to significant cancer cell apoptosis. The percentage of
total apoptotic cells was markedly increased in response to miR-378
upregulation compared with the control group in ONS-76 and DAOY
cells. Moreover, ectopic overexpression of UHRF1 reversed this
tendency (Fig. 4A). We subsequently
investigated the expression of two putative apoptosis-related
proteins PARP and activated caspase-3 in ONS-76 and DAOY cells.
Western blotting revealed that upregulation of miR-378 markedly
increased the expression of cleaved PARP and cleaved caspase-3 in
ONS-76 and DAOY cells, and ectopic expression of UHRF1 partly
inhibited the expression of the two apoptosis-related proteins
(Fig. 4B).
Discussion
As a putative oncogene, UHRF1 has been demonstrated
to be involved in the process of cancer cell proliferation,
invasion and apoptosis. UHRF1 has also been revealed to be
upregulated in various types of cancer, and its expression was
associated with poor clinical outcomes (9–17). In
our previous study (18), UHRF1
expression was not detected in normal cerebellum tissues but was
detected in the majority of MB tissues. In univariate and
multivariate survival analysis, the expression level of UHRF1 was
identified as an independent prognostic factor influencing overall
survival (OS) and progression-free survival (PFS) of MB patients.
Furthermore, downregulation of UHRF1 by RNAi inhibited the
proliferation and clonogenic ability of MB cell lines with cell
cycle arrest at the G1/G2 phase. These data for the first time
revealed the important roles of UHRF1 in MB tumorigenesis and
progression and suggested that UHRF1 may be a potential therapeutic
target for MB.
Recently, studies have implicated miRNAs as
important modulators in MB cell proliferation and migration. For
instance, miR-218 functions as a tumor suppressor by inhibiting
several phenotype-associated genes in MB (26). miR-124, miR-219 and miR-383 were
reported to suppress cell proliferation, migration and invasion of
MB cells (27–29). miR-378 was reported to function as a
tumor suppressor microRNA in several types of cancer including
glioma (23–25). Nonetheless, whether and how miR-378
plays a role in MB development and progression remains
unexplored.
Based on our previous study, we used various miRNA
target prediction algorithms and predicted miR-378 as one of the
most possible miRNAs that interact with the 3′-UTR of UHRF1.
Several previous studies had revealed that the expression of
miRNA-378 was decreased in certain tumors (30,31).
It has been reported that miR-378 was significantly downregulated
in colorectal cancer tissues and forced expression of miR-378
inhibited tumor cell viability and invasion (30). Fei and Wu demonstrated that the
expression of miR-378 was decreased in gastric cancer tissues and
cell lines, and delivery of miR-378 could hinder cell proliferation
and induce cell apoptosis (31). In
the present study, we used a luciferase reporter assay to validate
the interaction between miRNA-378 and UHRF1, and the results
revealed that miR-378 negatively regulated the expression of UHRF1.
Subsequent transfection of miR-378 in MB cell lines resulted in the
inhibition of proliferation, colony formation ability and promotion
of cell apoptosis. A restoration assay demonstrated that
transfection of UHRF1 could partly counteract the effect of
miR-378, indicating that miR-378 mediated the biological behavior
of MB cells, at least partly via regulation of UHRF1.
Even though the present study first demonstrated
that miR-378 negatively regulates UHRF1, it has certain
limitations. For instance, due to the limited sample size of MB and
normal cerebellum tissues, the association between the expression
of miR-378 in tumors and controls was preliminary, and the sample
size needs to be increased for more compelling evidence.
Additionally, extensive biological and functional characterization
of miR-378 and UHRF1 are needed to better clarify the regulation of
miR-378 on UHRF1 in MB carcinogenesis and progression in
vitro and in vivo.
In conclusion, our findings revealed that UHRF1, a
cancer gene in MB demonstrated by our previous study, is in a
modulation axis downstream of miR-378. In addition, our results
shed light on the roles of miR-378, which negatively regulated
UHRF1, in proliferation and apoptosis of MB cells. Although more
detailed experimental and clinical investigations to better clarify
the mechanism of miR-378 and its effects on MB patients are
warranted, the present study shed light on the promotion of a novel
targeted therapy for MB treatment.
Acknowledgements
The authors are thankful for the financial support
of the Youth Innovation Fund of The First Affiliated Hospital of
Zhengzhou University to Y.-M.W. and to Y.-H.B., the Science and
Technology Program of Henan Province to J.-Q.Z. (172102310648), and
the Medical Science and Technology Program of Henan Province to
J.-Q.Z. (201602058).
References
|
1
|
Gottardo NG, Hansford JR, McGlade JP,
Alvaro F, Ashley DM, Bailey S, Baker DL, Bourdeaut F, Cho YJ, Clay
M, et al: Medulloblastoma Down Under 2013: A report from the third
annual meeting of the International Medulloblastoma Working Group.
Acta Neuropathol. 127:189–201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kool M, Korshunov A, Remke M, Jones DT,
Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van
Meeteren A, van Vuurden D, et al: Molecular subgroups of
medulloblastoma: An international meta-analysis of transcriptome,
genetic aberrations, and clinical data of WNT, SHH, Group 3, and
Group 4 medulloblastomas. Acta Neuropathol. 123:473–484. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Packer RJ, Cogen P, Vezina G and Rorke LB:
Medulloblastoma: Clinical and biologic aspects. Neuro-oncol.
1:232–250. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Packer RJ, Sutton LN, Goldwein JW,
Perilongo G, Bunin G, Ryan J, Cohen BH, D'Angio G, Kramer ED,
Zimmerman RA, et al: Improved survival with the use of adjuvant
chemotherapy in the treatment of medulloblastoma. J Neurosurg.
74:433–440. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
del Charco JO, Bolek TW, McCollough WM,
Maria BL, Kedar A, Braylan RC, Mickle JP, Buatti JM, Mendenhall NP
and Marcus RB Jr: Medulloblastoma: Time-dose relationship based on
a 30-year review. Int J Radiat Oncol Biol Phys. 42:147–154. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Northcott PA, Korshunov A, Witt H,
Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins
CE, French P, et al: Medulloblastoma comprises four distinct
molecular variants. J Clin Oncol. 29:1408–1414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bronner C, Achour M, Arima Y, Chataigneau
T, Saya H and Schini-Kerth VB: The UHRF family: Oncogenes that are
drugable targets for cancer therapy in the near future? Pharmacol
Ther. 115:419–434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Muto M, Kanari Y, Kubo E, Takabe T,
Kurihara T, Fujimori A and Tatsumi K: Targeted disruption of Np95
gene renders murine embryonic stem cells hypersensitive to DNA
damaging agents and DNA replication blocks. J Biol Chem.
277:34549–34555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mudbhary R, Hoshida Y, Chernyavskaya Y,
Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson
RT, et al: UHRF1 overexpression drives DNA hypomethylation and
hepatocellular carcinoma. Cancer Cell. 25:196–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pi JT, Lin Y, Quan Q, Chen LL, Jiang LZ,
Chi W and Chen HY: Overexpression of UHRF1 is significantly
associated with poor prognosis in laryngeal squamous cell
carcinoma. Med Oncol. 30:6132013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Daskalos A, Oleksiewicz U, Filia A,
Nikolaidis G, Xinarianos G, Gosney JR, Malliri A, Field JK and
Liloglou T: UHRF1-mediated tumor suppressor gene inactivation in
nonsmall cell lung cancer. Cancer. 117:1027–1037. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Geng Y, Gao Y, Ju H and Yan F: Diagnostic
and prognostic value of plasma and tissue ubiquitin-like,
containing PHD and RING finger domains 1 in breast cancer patients.
Cancer Sci. 104:194–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou L, Zhao X, Han Y, Lu Y, Shang Y, Liu
C, Li T, Jin Z, Fan D and Wu K: Regulation of UHRF1 by miR-146a/b
modulates gastric cancer invasion and metastasis. FASEB J.
27:4929–4939. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang D, Xue H, Yu Y, Lv F, You W and
Zhang B: Elevated expression of UHRF1 predicts unfavorable
prognosis for patients with hepatocellular carcinoma. Int J Clin
Exp Pathol. 8:9416–9421. 2015.PubMed/NCBI
|
|
15
|
Wang F, Yang YZ, Shi CZ, Zhang P, Moyer
MP, Zhang HZ, Zou Y and Qin HL: UHRF1 promotes cell growth and
metastasis through repression of p16ink4a in colorectal
cancer. Ann Surg Oncol. 19:2753–2762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang GL, Zhang LH, Bo JJ, Chen HG, Cao M,
Liu DM and Huang YR: UHRF1 is associated with tumor recurrence in
non-muscle-invasive bladder cancer. Med Oncol. 29:842–847. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jazirehi AR, Arle D and Wenn PB: UHRF1: A
master regulator in prostate cancer. Epigenomics. 4:251–252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang ZY, Cai JJ, Hong J, Li KK, Ping Z,
Wang Y, Ng HK, Yao Y and Mao Y: Clinicopathological analysis of
UHRF1 expression in medulloblastoma tissues and its regulation on
tumor cell proliferation. Med Oncol. 33:992016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
21
|
Sato F, Tsuchiya S, Meltzer SJ and Shimizu
K: MicroRNAs and epigenetics. FEBS J. 278:1598–1609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang
L, Li G, Chen HH and Li XP: MicroRNA-378 functions as an onco-miR
in nasopharyngeal carcinoma by repressing TOB2 expression. Int J
Oncol. 44:1215–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang GJ, Zhou H, Xiao HX, Li Y and Zhou
T: MiR-378 is an independent prognostic factor and inhibits cell
growth and invasion in colorectal cancer. BMC Cancer. 14:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen QG, Zhou W, Han T, Du SQ, Li ZH,
Zhang Z, Shan GY and Kong CZ: MiR-378 suppresses prostate cancer
cell growth through downregulation of MAPK1 in vitro and in vivo.
Tumour Biol. 37:2095–2103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li B, Wang Y, Li S, He H, Sun F, Wang C,
Lu Y, Wang X and Tao B: Decreased expression of miR-378 correlates
with tumor invasiveness and poor prognosis of patients with glioma.
Int J Clin Exp Pathol. 8:7016–7021. 2015.PubMed/NCBI
|
|
26
|
Venkataraman S, Birks DK, Balakrishnan I,
Alimova I, Harris PS, Patel PR, Handler MH, Dubuc A, Taylor MD,
Foreman NK, et al: MicroRNA 218 acts as a tumor suppressor by
targeting multiple cancer phenotype-associated genes in
medulloblastoma. J Biol Chem. 288:1918–1928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li KK, Pang JC, Lau KM, Zhou L, Mao Y,
Wang Y, Poon WS and Ng HK: MiR-383 is downregulated in
medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol.
23:413–425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi JA, Lu DL, Huang X and Tan W: miR-219
inhibits the proliferation, migration and invasion of
medulloblastoma cells by targeting CD164. Int J Mol Med.
34:237–243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Silber J, Hashizume R, Felix T, Hariono S,
Yu M, Berger MS, Huse JT, VandenBerg SR, James CD, Hodgson JG, et
al: Expression of miR-124 inhibits growth of medulloblastoma cells.
Neuro Oncol. 15:83–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang GJ, Zhou H, Xiao HX, Li Y and Zhou
T: MiR-378 is an independent prognostic factor and inhibits cell
growth and invasion in colorectal cancer. BMC Cancer. 14:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fei B and Wu H: MiR-378 inhibits
progression of human gastric cancer MGC-803 cells by targeting
MAPK1 in vitro. Oncol Res. 20:557–564. 2012. View Article : Google Scholar : PubMed/NCBI
|