Introduction
Bladder cancer is one of the most common types of
cancer worldwide. In 2015, the estimated newly diagnosed cases of
bladder cancer were 74,000 and the estimated cancer-related deaths
caused by bladder cancer were 16,000 in the United States (1). Currently, there are ~75% of cases that
present with non-muscle-invasive bladder cancer and 25% of cases
that present with muscle-invasive bladder cancer (2,3).
Although there has been some progress in the clinical treatment of
bladder cancer in the past years, the 5-year survival rate for
patients with bladder cancer remains only 50–60% (4,5). Thus,
it is urgent to reveal the potential molecular mechanisms involved
in the tumorigenesis of bladder cancer.
Leucine zipper-EF-hand containing transmembrane
protein 1 (LETM1), which was first identified in human
Wolf-Hirschhorn syndrome (6,7), is a
mitochondrial inner membrane protein which plays an important role
in mitochondrial ATP production and biogenesis by decreasing the
mitochondrial mass and expression of many mitochondrial proteins
(8). However, previous studies
revealed that the high expression levels of LETM1 have been
correlated with many human malignancies. For example, Chen et
al reported that LETM1 was highly expressed in head and neck
squamous cell carcinoma and that a high expression of LETM1
predicts poor prognosis (9).
Another study revealed that high LETM1 expression was positively
correlated with late clinical stage, poor differentiation, lymph
node metastasis, disease-free survival and 10-year overall survival
rates in triple-negative breast cancer (10). However, the role of LETM1 in human
bladder cancer have yet to be determined. In this study, we
investigated the role of LETM1 in bladder cancer. Our results
demonstrated that the expression of LETM1 was significantly
increased in bladder cancer tissues and cell lines, and that
knockdown of LETM1 markedly decreased the proliferation, migration
and invasion of bladder cancer cells. Moreover, the suppression of
LETM1 induced the accumulation of S-phase cells.
Materials and methods
Patient data
Bladder cancer tissues and their matched adjacent
normal tissues were obtained from the Shanghai Tenth People's
Hospital, Tongji University School of Medicine (Shanghai, China).
The study was approved by the Shanghai Tenth People's Hospital
Ethics Committee and written informed consents were obtained from
all patients.
Cell lines and cultures
The human bladder cancer cell lines (T24, EJ, 5637
and J82) and the human bladder epithelial immortalized SV-HUC-1
cell line were obtained from type culture collection of the Chinese
Academy of Sciences (Shanghai, China). The SV-HUC-1 cells were
cultured in F12K medium (Sigma-Aldrich, St. Louis, MO, USA). The
T24, EJ and 5637 cells were cultured in RPMI-1640 medium (Gibco,
Rockville, MD, USA) and the J82 cells were maintained in Dulbecco's
modified Eagle's medium (DMEM; Gibco). These media were
supplemented with 10% fetal bovine serum (FBS; Gibco) and 1%
penicillin/streptomycin (HyClone, Logan, UT, USA). The cells were
incubated at 37°C in a humidified atmosphere with 5%
CO2.
RNA isolation and quantitative
real-time PCR
Total RNA was extracted from the cultured cells
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer's instructions. The concentration and purity of
the RNA were determined using an ND-2000 spectrophotometer (Thermo
Fisher Scientific, Inc., Carlsbad, CA, USA). For the detection of
the LETM1 mRNA level, the cDNA was synthesized using a PrimeScript
RT Reagent kit (Takara Bio, Inc., Shiga, Japan) according to the
manufacturer's instructions. qRT-PCR was performed with the KAPA
SYBR FAST qPCR kit (Kapa Biosystems, Inc., Woburn, MA, USA). The
primers for the qRT-PCR analysis were as follows: LETM1 sense,
5′-CTCAAGGAGGAGAGGCTGAA-3′ and antisense,
5′-GAAGTTGTTGGTGCCGATG-3′; β-actin sense, 5′-CCTGGCACCCAGCACAAT-3′
and antisense, 5′-GGGCCGGACTCGTCATAC-3′. The LETM1 mRNA level was
normalized to the β-actin mRNA level. The data were analyzed using
the 2−ΔΔCt method.
Immunohistochemical (IHC)
analysis
An IHC assay was used to detect the expression
levels of LETM1 in the tissues. The paraffin-embedded tissue
samples were dewaxed and incubated with 3% hydrogen peroxide for 30
min to inhibit endogenous peroxidase activity. Then the sections
were infiltrated in citrate buffer and heated in a microwave for 10
min to carry out antigen retrieval. Subsequently the sections were
incubated with the primary antibody anti-LETM1 (1:100, sc-271234;
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) overnight at
4°C. Then the sections were washed with phosphate-buffered saline
(PBS) and the peroxidase-labeled goat anti-mouse secondary antibody
was applied at room temperature for 1 h. Next, the slides were
stained with 3,3′-diaminobenzidine tetrahydrochloride (DAB) and
hematoxylin for visualization. The intensity of LETM1 staining was
scored as: 0, none; 1, weak; and 2, strong. The proportion of
positive tumor cells was recorded as follows: 1, 1–25%; 2, 26–50%;
3, 51–75%; and 4, 76–100%. The scores were multiplied to obtain a
final score and the total expression of LETM1 was determined as
either negative/weak expression (score <4) or overexpression
(score ≥4).
Cell transfection
The small interfering RNA (siRNA) targeting human
LETM1 (si-LETM1) and the corresponding negative control (si-NC)
were purchased from GenePharma (Shanghai, China). The sequence of
LETM1 siRNA was: 5′-CCACAGAAUCGUGUCUGGAUCCACA-3′. When the cells
were grown to 50%, the aforementioned molecular products were
transfected into bladder cancer cells using Lipofectamine 2000
(Invitrogen) according to the manufacturer's instructions. The
medium containing the transfection reagents was removed 6 h after
transfection, and 48 h later, the cells were harvested for the
following assays.
Cell proliferation assay
Cell proliferation was assessed using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies Inc., Kumamoto,
Japan). Briefly, 100 µl of transfected cells were seeded into
96-well plates at a density of 1,000 cells/well, and then 10 µl of
CCK-8 was added into each well at indicated time-points (24, 48, 72
and 96 h) and incubation followed for 2 h at 37°C. The absorbance
at 450 nm was detected on a microplate spectrophotometer (BioTek
Instruments Inc., Winooski, VT, USA).
Transwell migration and invasion
assays
Transwell chambers (BD Biosciences, San Jose, CA,
USA) were used for cell migration and invasion assays. For the
invasion assay, each chamber was precoated with 25 µl of Matrigel
(BD Biosciences) at 37°C for 2 h, whereas the chambers used for the
migration assay were not precoated with Matrigel. Transfected cells
(5×104) in 200 µl serum-free medium were plated in the
upper well of the chamber, and 600 µl of RPMI-1640 medium
containing 10% FBS was added into the lower chambers as a
chemoattractant. After incubation at 37°C for 14 h, the
non-invading cells were removed with a cotton tip, and the cells
that had migrated to the lower surface of the chamber were fixed
with 95% ethanol for 20 min, stained with 0.1% crystal violet
solution for 10 min, washed for 3 times, air dried, photographed
and counted in 5 randomly selected fields for each well using a
light microscope (Olympus, Tokyo, Japan).
Cell cycle assay
For the cell cycle analysis, the transfected cells
were cultured for 48 h, digested with trypsin, washed with PBS,
centrifuged and fixed in 75% ethanol at 4°C overnight. Then the
cells were washed with PBS, centrifuged and resuspended in 1 ml of
PBS containing 1 mg/ml RNase A and 50 µg/ml propidium iodide. The
cells were then incubated for 30 min at room temperature in the
dark and analyzed immediately using a flow cytometer (BD
Biosciences).
Western blot analysis
Ice-cold RIPA buffer (Sigma-Aldrich) containing a
protease inhibitor was used to isolate protein from cells or
tissues. A BCA protein assay kit was used to determine the
concentration of total cellular protein according to the
manufacturer's instructions. Then equal amounts of protein were
loaded onto 10% SDS-PAGE and were transferred onto nitrocellulose
membranes. The membranes were blocked in 5% non-fat milk for 1 h
and then incubated with the following primary antibodies: mouse
anti-LETM1 (sc-271234) and mouse anti-β-actin (sc-130300) (both
from Santa Cruz Biotechnology, Inc.), rabbit anti-β-catenin (no.
8480; Cell Signaling Technology, Danvers, MA, USA), rabbit
anti-cyclin D1 (ab134175; Abcam, Cambridge, UK), and rabbit
anti-c-Myc (no. 5605; Cell Signaling Technology), overnight at 4°C.
After being washed with PBST three times, the membranes were
incubated with the corresponding secondary antibodies at room
temperature for 1 h. The protein bands were visualized using the
Odyssey scanner (LI-COR Biosciences, Lincoln, NE, USA).
Statistical analysis
SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) was
used for statistical analysis. All the data are presented as the
means ± standard deviation (SD) from at least three independent
experiments. The immunohistochemical staining results were
evaluated using Pearson's Chi-square test and the other
experimental results were calculated using Student's t-test. A
P-value <0.05 was considered to indicate a statistically
significant result. The diagrams were drawn using GraphPad Prism 5
software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
LETM1 is overexpressed in human
bladder cancer cell lines and tissues
To explore the expression differences of LETM1
between the bladder cancer cells and the normal human bladder
transitional cell line SV-HUC-1, western blotting and qRT-PCR were
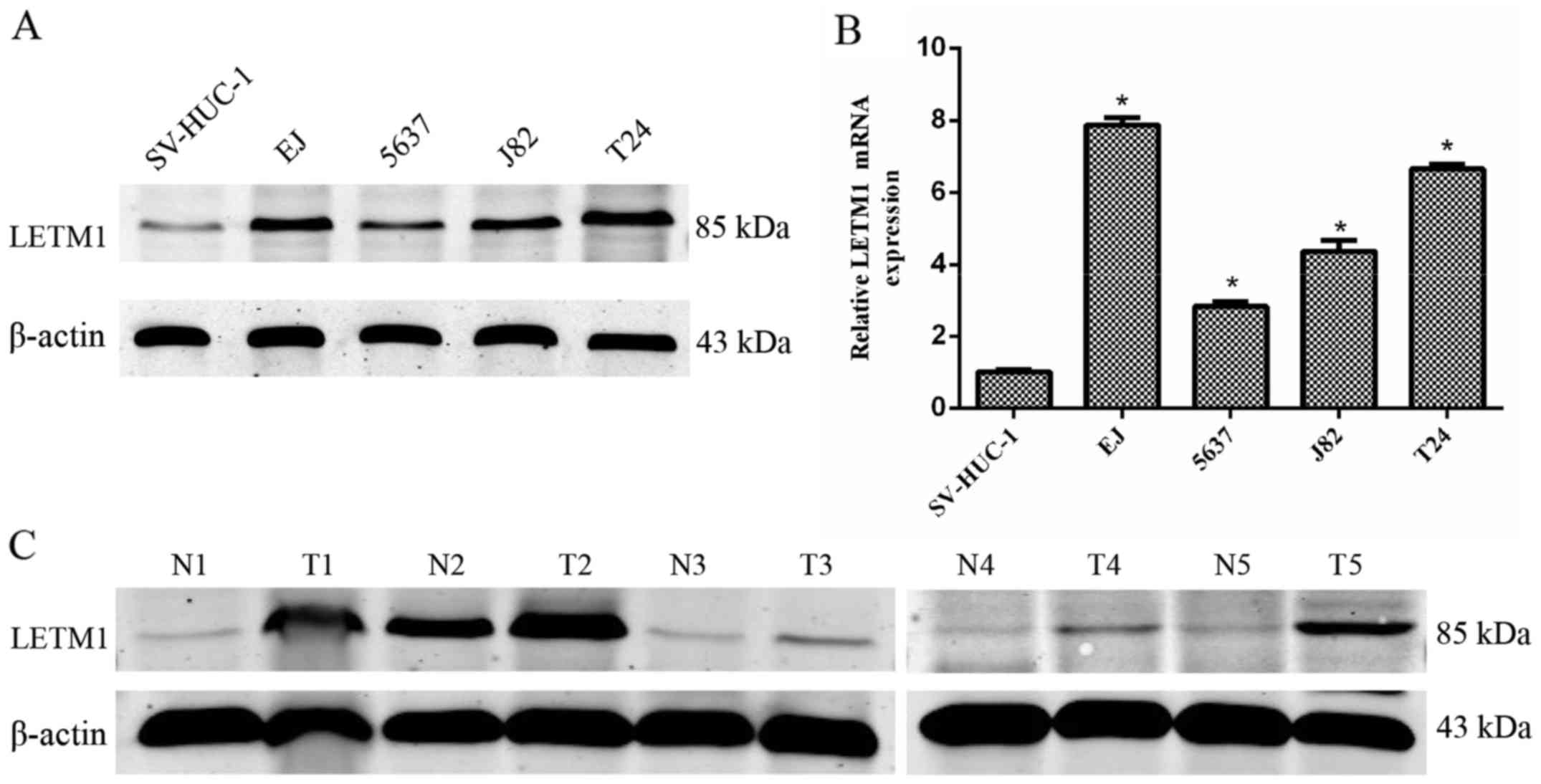
performed. As shown in Fig. 1A, the
protein level of LETM1 was significantly higher in the bladder
cancer cell lines, compared to that in the normal human bladder
transitional cells. Moreover, LETM1 mRNA expression was further
confirmed to be abundant in bladder cancer cell lines (Fig. 1B), which was consistent with the
western blot analysis. Furthermore, we detected the protein
expression of LETM1 in 5 pairs of tumor tissues and matched
adjacent normal tissues by western blot analysis. As shown in
Fig. 1C, the protein expression of
LETM1 was higher in the bladder cancer tissues than that in the
adjacent normal tissues.
To explore the correlation between the expression of
LETM1 and the clinicopathological parameters of bladder cancer,
immunohistochemistry was applied to examine the LETM1 protein
expression level in 86 cases of bladder cancer tissues and 5
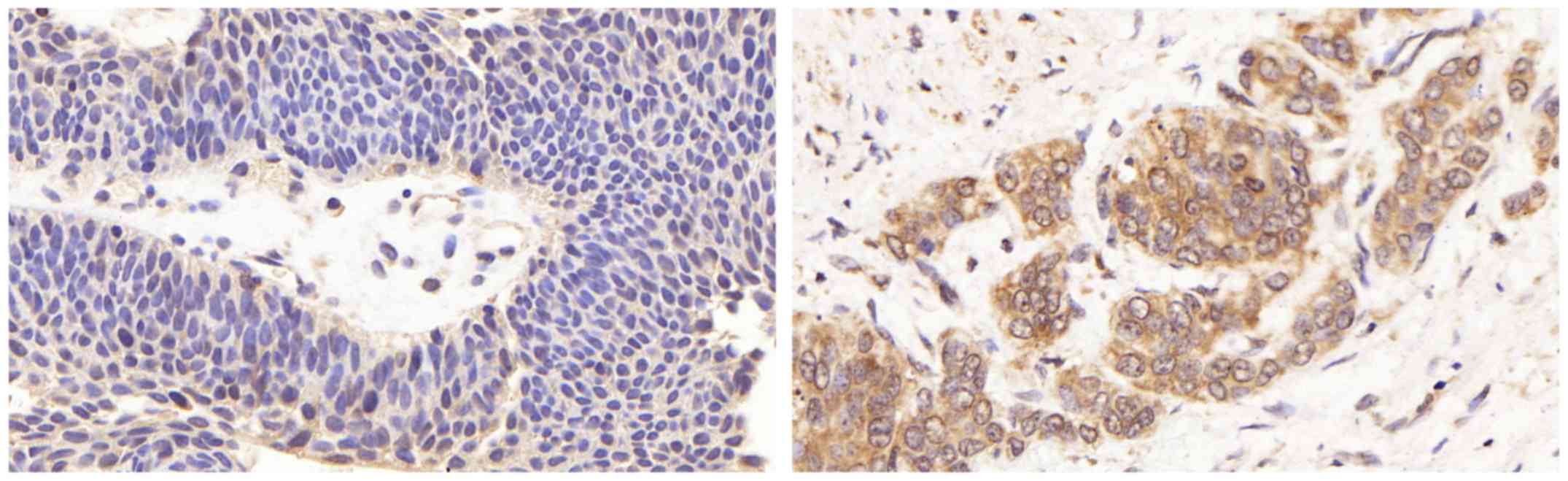
adjacent normal tissues. We found that negative LETM1 staining was
observed in normal bladder tissues, and positive LETM1 staining was
observed in bladder cancer tissues (Fig. 2). Moreover the expression of LETM1
in bladder cancer was associated with the pathological stage, lymph
node metastasis and recurrence of bladder cancer (p<0.05)
(Table I). These results indicated
that LETM1 was upregulated in bladder cancer cell lines and
tissues, therefore we assumed that LETM1 may act as an oncogene in
bladder cancer.
 | Table I.Relationship between the expression of
LETM1 and the clinicopathological parameters in bladder cancer
patients. |
Table I.
Relationship between the expression of
LETM1 and the clinicopathological parameters in bladder cancer
patients.
| Variables | No. of cases | LETM1 overexpression
cases (%) | χ2 | P-valuea |
|---|
| Sex |
|
| 0.167 | 0.683 |
| Male | 57 | 36 (63.2) |
|
|
|
Female | 29 | 17 (58.6) |
|
|
| Age (years) |
|
| 3.230 | 0.072 |
|
<60 | 39 | 20 (51.3) |
|
|
| ≥60 | 47 | 33 (70.2) |
|
|
| Pathological |
|
| 9.466 | 0.009 |
|
stage |
|
|
|
|
| Ta | 19 | 6 (31.6) |
|
|
| T1 | 41 | 28 (68.3) |
|
|
| ≥T2 | 26 | 19 (73.1) |
|
|
| Grade |
|
| 0.614 | 0.433 |
| Low | 27 | 15 (55.6) |
|
|
| High | 59 | 38 (64.4) |
|
|
| Lymph node
metastasis |
|
| 10.185 | 0.001 |
| No | 58 | 29 (50.0) |
|
|
| Yes | 28 | 24 (85.7) |
|
|
| Recurrence |
|
| 5.029 | 0.025 |
| No | 39 | 19 (53.8) |
|
|
| Yes | 47 | 34 (68.1) |
|
|
Knockdown of LETM1 inhibits bladder
cancer cell proliferation
To further explore the role of LETM1 in bladder
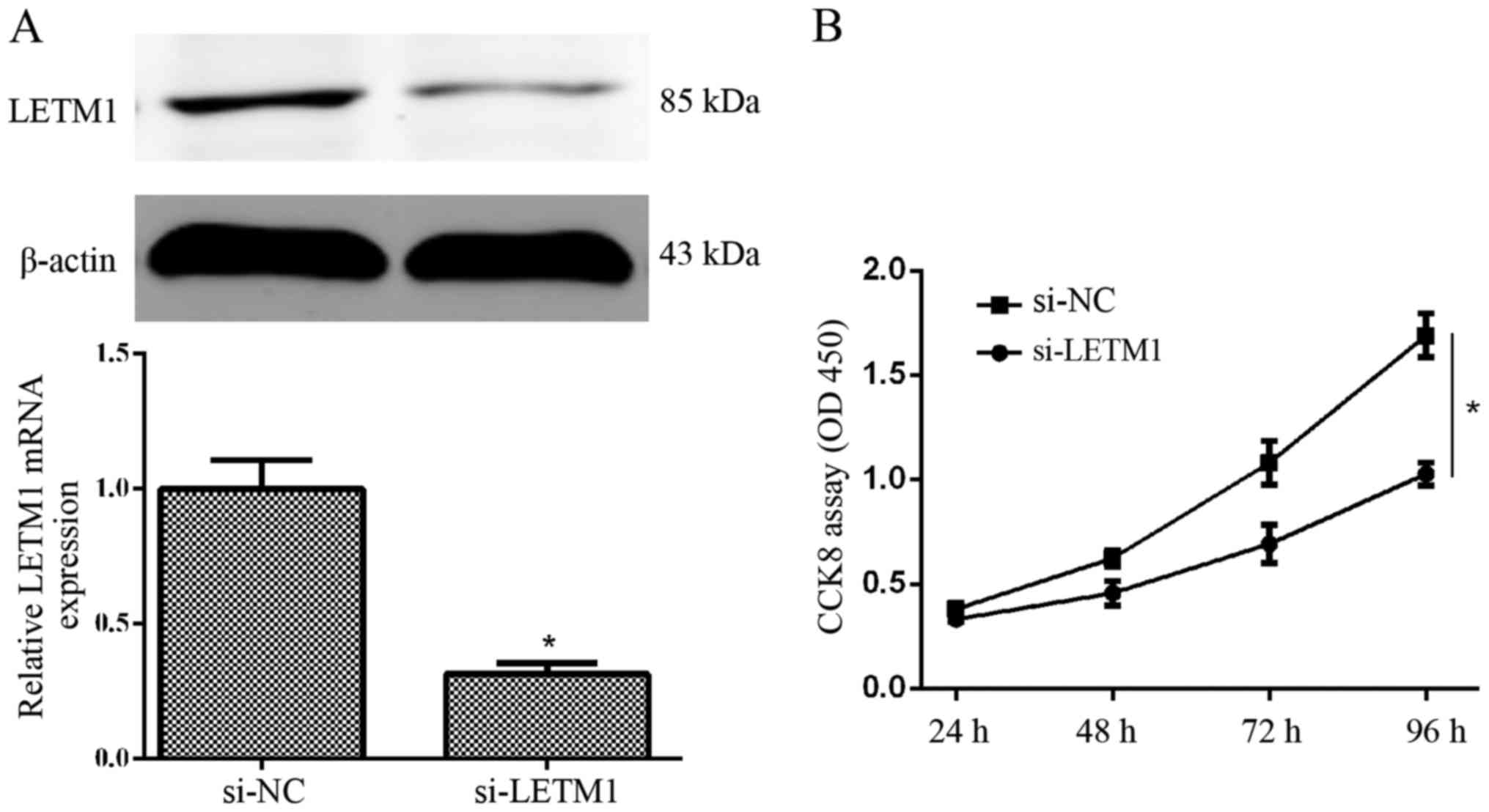
cancer cells, T24 cells were transfected with si-LETM1 or si-NC,
and then the expression of LETM1 at the mRNA and protein level was
detected at 48 and 72 h after transfection, respectively. We found
that knockdown of LETM1 by si-LETM1 significantly decreased the
expression of LETM1 at the mRNA and protein level in T24 cells
(Fig. 3A). A CCK-8 proliferation
assay revealed that knockdown of LETM1 markedly decreased the
proliferation of T24 cells, when compared with the respective
control (Fig. 3B). Collectively,
knockdown of LETM1 markedly inhibited bladder cancer cell
proliferation.
LETM1 knockdown induces accumulation
of S-phase cells
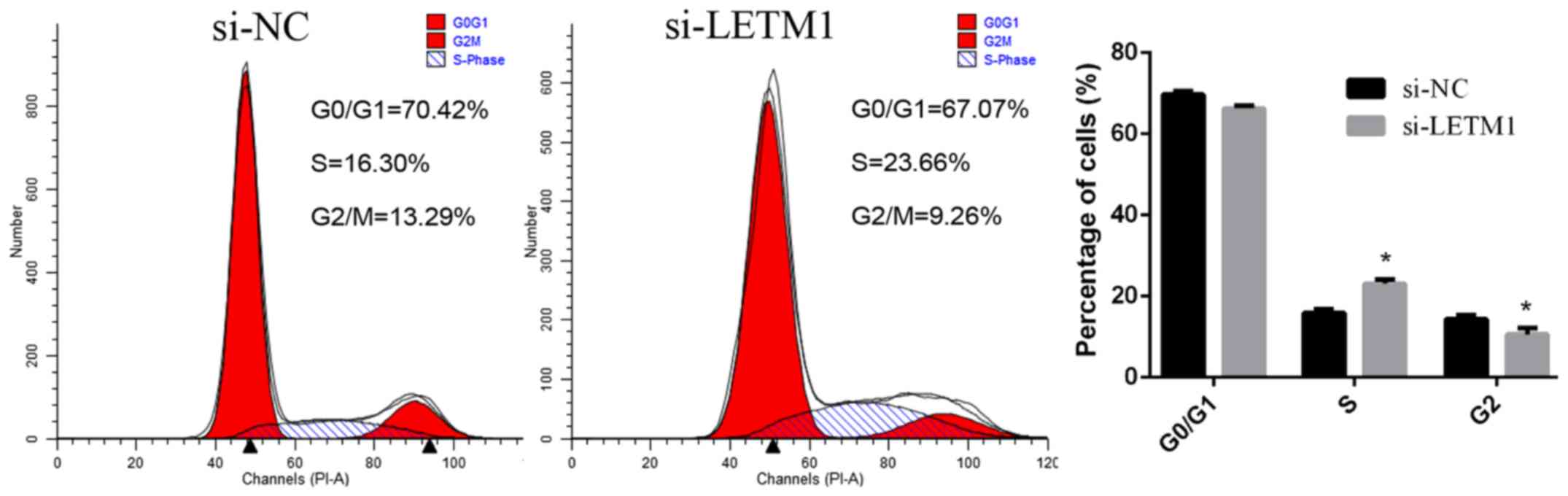
To further investigate the possible mechanism
underlying the cell growth inhibition effect by suppression of the
expression of LETM1 in bladder cancer cells, the cell cycle was
analyzed using a flow cytometer. The percentage of cells at
different phases revealed that LETM1 knockdown induced bladder
cancer cell accumulation at the S phase (Fig. 4).
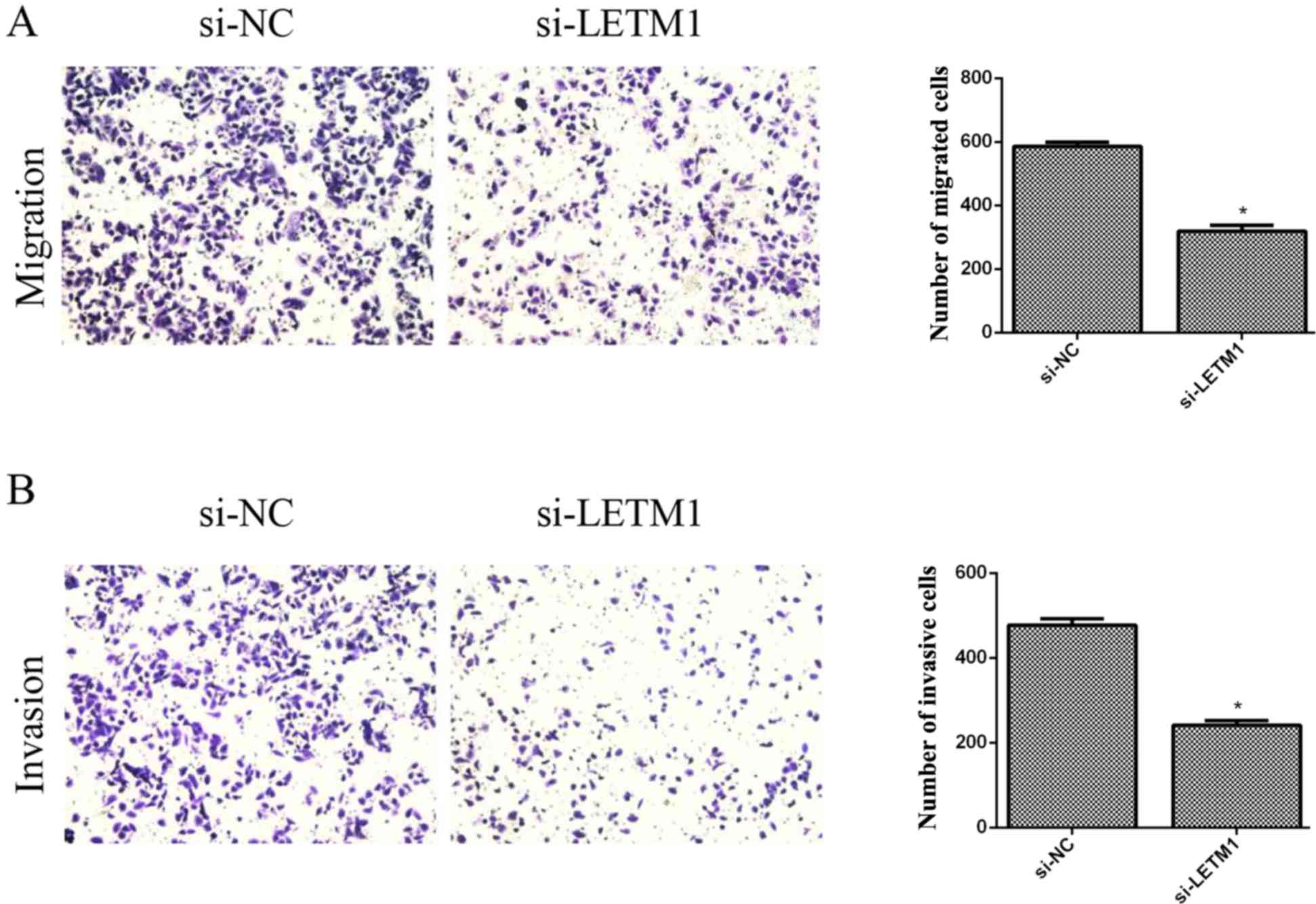
Knockdown of LETM1 inhibits bladder
cancer cell migration and invasion
We then investigated the effects of LETM1 on the
cell migration of bladder cancer cells using Transwell chambers
without Matrigel. The results revealed that knockdown of LETM1
markedly inhibited the migration of T24 cells, when compared with
the respective control (Fig. 5A).
In addition, we explored the role of LETM1 on cell invasion of
bladder cancer cells using Transwell chambers with Matrigel. As
revealed in Fig. 5B, knockdown of
LETM1 significantly suppressed the number of T24 cells that invaded
through the Matrigel, as compared with the respective control
group.
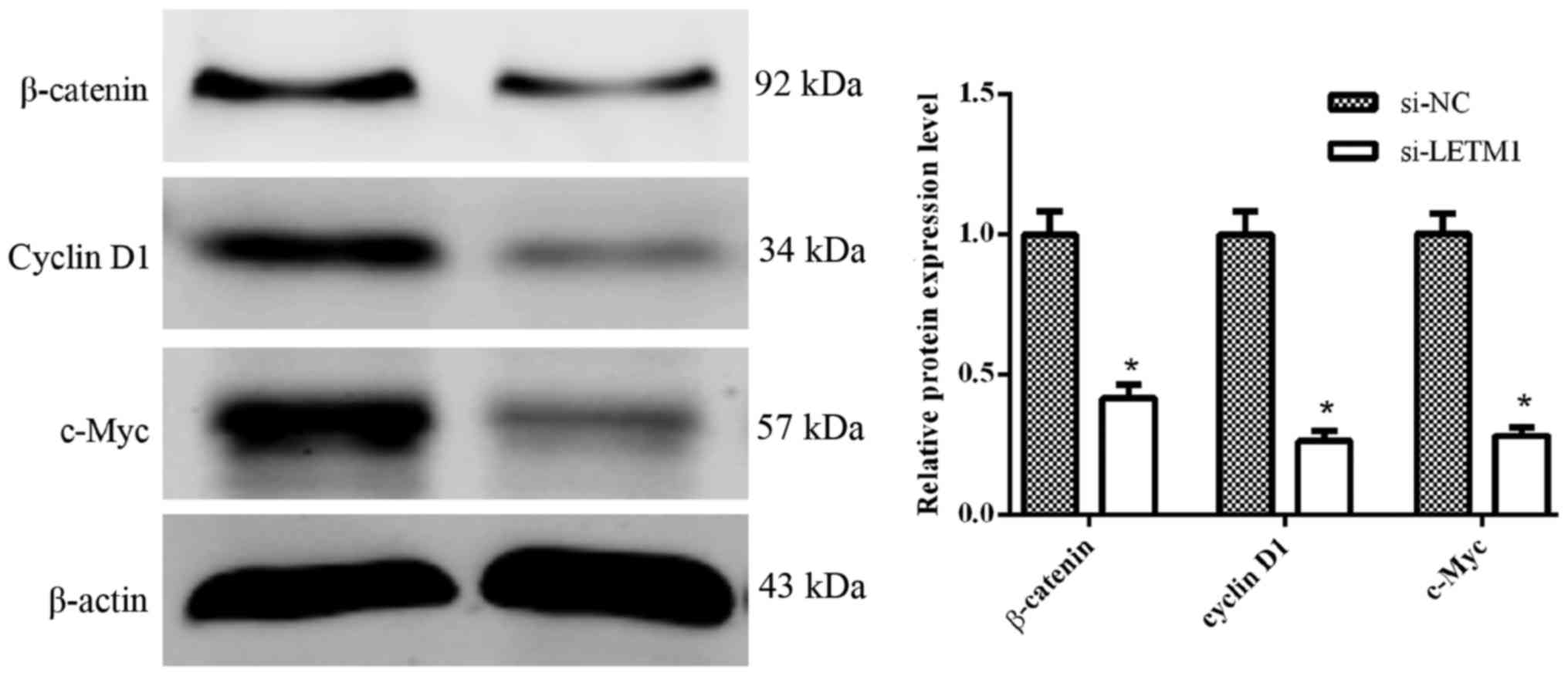
Knockdown of LETM1 inhibits the
activation of the Wnt/β-catenin pathway
Some evidence has indicated that the Wnt/β-catenin
pathway plays a critical role in the tumor progression and
metastasis of human bladder cancer cells (11,12).
To further explore the effects of si-LETM1 on the Wnt/β-catenin
signaling pathway in human bladder cancer cells, we determined the
protein expression of β-catenin, cyclin D1 and c-Myc when knocking
down LETM1. As illustrated in Fig.
6, knockdown of LETM1 significantly suppressed the protein
expression of β-catenin, cyclin D1 and c-Myc in T24 cells.
Discussion
Emerging evidence has indicated that many oncogene
and anti-oncogene alterations are involved in the tumorigenesis of
bladder cancer (13). Although
great progress in diagnosis and treatment has been achieved in the
past decades, the underlying molecular mechanism of bladder
tumorigenesis still remains to be elucidated and targets for gene
therapy are limited. In the present study, we demonstrated that the
suppression of LETM1 significantly suppressed cell proliferation,
migration and invasion and disrupted the cell cycle
distribution.
Recent studies revealed that LETM1 is highly
expressed in many types of human cancer and predicts poor prognosis
(8–10,14).
This is consistent with our results which revealed that the
expression of LETM1 was increased in bladder cancer and that
upregulated LETM1 expression was related with pathological stage,
lymph node metastasis and recurrence of bladder cancer. As for the
molecular function of LETM1, Doonan et al found that
knockdown of LETM1 caused accumulation of S-phase cells and the
re-expression of LETM1 could reverse S-phase accumulation (15), which was consistent with our
results, indicating that the suppression of LETM1 inhibited cell
proliferation possibly by disrupting the cell cycle distribution.
Piao et al found that the overexpression of LETM1 could
induce necrotic cell death in HeLa cells by decreasing
mitochondrial biogenesis and ATP production (8). However, Dimmer et al found that
downregulation of LETM1 also caused necrotic cell death (16). It is still unclear how both the gain
and loss of LETM1 causes similar phenotypes in cells.
The Wnt/β-catenin pathway is an important signaling
pathway involved in the malignant progression of various tumors,
and regulates the proliferation, migration and invasion of cancer
cells (17–19). When Wnt is activated, β-catenin
translocates from the cytoplasm to the nucleus and stimulates
proto-oncogene cyclin D1 and c-Myc transcription (20,21).
Cyclin D1 promotes cell proliferation by regulating the G1 phase
progression of the cell cycle (22)
and c-Myc encodes a transcription factor, that triggers selective
gene expression amplification and promotes cell proliferation by
genetic and epigenetic elimination of checkpoints (23). In our study, we found that knockdown
of LETM1 inhibited the expression of β-catenin, cyclin D1 and c-Myc
in bladder cancer cells. These results indicated that silencing of
LETM1-inhibited cell proliferation may partially be epigenetically
inhibiting β-catenin, cyclin D1 and c-Myc expression. However, more
research is warranted in order to explore the underlying mechanism
of how LETM1 affects the Wnt/β-catenin pathway. The expression of
some specific indicators of Wnt/β-catenin signaling and the nuclear
localization of β-catenin need to be analyzed after knockdown of
LETM1.
Collectively, our results revealed that the
knockdown of LETM1 suppresses proliferation, migration and invasion
of bladder cancer cells possibly via the suppression of the
Wnt/β-catenin signaling pathway. Therefore, our results indicate
that LETM1 may be a novel target for bladder cancer therapy.
Acknowledgements
This study was supported by a research grant from
the National Natural Science Foundation of China (no.
81370699).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Babjuk M, Böhle A, Burger M, Capoun O,
Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M,
et al: EAU guidelines on non-muscle-invasive urothelial carcinoma
of the bladder: Update 2016. Eur Urol. 3:447–461. 2017. View Article : Google Scholar
|
|
3
|
Kamat AM, Hahn NM, Efstathiou JA, Lerner
SP, Malmström PU, Choi W, Guo CC, Lotan Y and Kassouf W: Bladder
cancer. Lancet. 388:2796–2810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chester JD, Hall GD, Forster M and
Protheroe AS: Systemic chemotherapy for patients with bladder
cancer - current controversies and future directions. Cancer Treat
Rev. 30:343–358. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Budman LI, Kassouf W and Steinberg JR:
Biomarkers for detection and surveillance of bladder cancer. Can
Urol Assoc J. 2:212–221. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hart L, Rauch A, Carr AM, Vermeesch JR and
O'Driscoll M: LETM1 haploinsufficiency causes mitochondrial defects
in cells from humans with Wolf-Hirschhorn syndrome: Implications
for dissecting the underlying pathomechanisms in this condition.
Dis Model Mech. 7:535–545. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Endele S, Fuhry M, Pak SJ, Zabel BU and
Winterpacht A: LETM1, a novel gene encoding a putative EF-hand
Ca2+−-binding protein, flanks the Wolf-Hirschhorn
syndrome (WHS) critical region and is deleted in most WHS patients.
Genomics. 60:218–225. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Piao L, Li Y, Kim SJ, Byun HS, Huang SM,
Hwang SK, Yang KJ, Park KA, Won M, Hong J, et al: Association of
LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP
production and necrotic cell death. Cancer Res. 69:3397–3404. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Yang Y, Liu S, Piao L, Zhang Y,
Lin Z and Li Z: High expression of leucine zipper-EF-hand
containing transmembrane protein 1 predicts poor prognosis in head
and neck squamous cell carcinoma. Biomed Res Int.
2014:8503162014.PubMed/NCBI
|
|
10
|
Wang CA, Liu Q, Chen Y, Liu S, Xu J, Cui
X, Zhang Y and Piao L: Clinical implication of leucine zipper/EF
hand-containing transmembrane-1 overexpression in the prognosis of
triple-negative breast cancer. Exp Mol Pathol. 98:254–259. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahmad I: The role of WNT signalling in
urothelial cell carcinoma. Ann R Coll Surg Engl. 97:481–486. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pierzynski JA, Hildebrandt MA, Kamat AM,
Lin J, Ye Y, Dinney CP and Wu X: Genetic variants in the
Wnt/β-catenin signaling pathway as indicators of bladder cancer
risk. J Urol. 194:1771–1776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Costello JC and Theodorescu D: Decade in
review-bladder cancer: International progress: From cytology to
genomics. Nat Rev Urol. 11:609–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li N, Zheng Y, Xuan C, Lin Z, Piao L and
Liu S: LETM1 overexpression is correlated with the clinical
features and survival outcome of breast cancer. Int J Clin Exp
Pathol. 8:12893–12900. 2015.PubMed/NCBI
|
|
15
|
Doonan PJ, Chandramoorthy HC, Hoffman NE,
Zhang X, Cárdenas C, Shanmughapriya S, Rajan S, Vallem S, Chen X,
Foskett JK, et al: LETM1-dependent mitochondrial Ca2+
flux modulates cellular bioenergetics and proliferation. FASEB J.
28:4936–4949. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dimmer KS, Navoni F, Casarin A, Trevisson
E, Endele S, Winterpacht A, Salviati L and Scorrano L: LETM1,
deleted in Wolf-Hirschhorn syndrome is required for normal
mitochondrial morphology and cellular viability. Hum Mol Genet.
17:201–214. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang HS, Nie X, Wu RB, Yuan HW, Ma YH, Liu
XL, Zhang JY, Deng XL, Na Q, Jin HY, et al: Downregulation of human
Wnt3 in gastric cancer suppresses cell proliferation and induces
apoptosis. Onco Targets Ther. 9:3849–3860. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li K, Zhou ZY, Ji PP and Luo HS: Knockdown
of β-catenin by siRNA influences proliferation, apoptosis and
invasion of the colon cancer cell line SW480. Oncol Lett.
11:3896–3900. 2016.PubMed/NCBI
|
|
19
|
Wu D, Li L and Yan W: Knockdown of TC-1
enhances radiosensitivity of non-small cell lung cancer via the
Wnt/β-catenin pathway. Biol Open. 5:492–498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Klein EA and Assoian RK: Transcriptional
regulation of the cyclin D1 gene at a glance. J Cell Sci.
121:3853–3857. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qin X, Zhang H, Zhou X, Wang C, Zhang H,
Zhang X and Ye L: Proliferation and migration mediated by
Dkk-1/Wnt/β-catenin cascade in a model of hepatocellular carcinoma
cells. Transl Res. 150:281–294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cicatiello L, Addeo R, Sasso A, Altucci L,
Petrizzi VB, Borgo R, Cancemi M, Caporali S, Caristi S, Scafoglio
C, et al: Estrogens and progesterone promote persistent CCND1 gene
activation during G1 by inducing transcriptional derepression via
c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex
assembly to a distal regulatory element and recruitment of cyclin
D1 to its own gene promoter. Mol Cell Biol. 24:7260–7274. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|