Introduction
Microvesicles (MVs) are extracellular vesicles
released by most cells that act as mediators of intercellular
communication (1). As a package of
extracellular multi-molecular messages, MVs carry the proteins,
lipids and nucleic acids of their parental cells, providing a
potential source of biomarkers in the surveillance of disease
progression and/or relapse (2). Our
previous study demonstrated that MVs derived from K562 chronic
myeloid leukemia (CML) cells could transform mononuclear cells
(MNCs) from normal hematopoietic transplants to acute leukemia-like
cancer cells, which provided multiple implications for CML
transformation. During transformation, an increase in global DNA
methylation (GDM) in MNCs that peaked on day 3 as compared to the
control, accompanied by DNA methyltransferase (DNMT)3a and DNMT3b
mRNA and protein were consistently increased (3). DNA methylation is one of the most
frequent forms of epigenetic modification (4,5). It is
widely accepted that epigenetic and genetic alterations collaborate
in the development and maintenance of cancer. Recently, epigenetic
alterations have also been recognized as a major driving force in
the development of several cancers, including leukemia (6,7).
Although DNA hypermethylation of multiple genes (DLX4, SHP-1 and
HOXs) characterizes advanced stages of CML and/or the disease when
resistant to imatinib (8–10), the underlying epigenetic changes of
DNA methylation in CML, particularly during transformation, are not
fully implicated. In our previous transformation system, MVs lost
their transforming abilities and the mRNA and protein of these
methyltransferases decreased following RNase treatment, indicating
that RNAs in MVs were responsible for the transformation (3). The aim of the present study was to
build the connection between the miRNA inside the MVs and the
methylation of recipient cells.
Materials and methods
Cell culture and MV isolation
The human CML blast crisis cell line K562 was
purchased from the China Center for Type Culture Collection (Wuhan,
China) and cultured in RPMI-1640 medium containing 10% fetal bovine
serum (FBS) at 37°C in 5% CO2. MVs isolation was
performed using a previous protocol: cells were centrifuged at
1,000 × g for 10 min. The supernatant was centrifuged at 5,000 × g
for 20 min to remove cellular debris, and the remaining supernatant
was centrifuged at 13,000 × g for 60 min to obtain MVs. MNCs were
extracted from the peripheral blood mobilization using lymphocyte
separation medium (TBD Science, Tianjin, China), and cultured in
RPMI-1640 medium containing 15% FBS at 37°C in 5%
CO2.
Transformation of MNCs from normal
hematopoietic transplants by MVs
The retained MVs were suspended in serum-free
RPMI-1640 medium (to avoid MVs derived from the FBS). To generate
intact MVs for the cell-based assays and other experiments, the
MV-containing RPMI-1640 medium was filtered using a Millipore
Steriflip polyvinylidene difluoride filter with pore sizes of 1.2
µm (to filter cells) (Millipore, Billerica, MA, USA). MVs were
quantified according to their total RNA content and copies of
BCR-ABL1 mRNA. The MNCs were adjusted to 4×106
cells/well in 6-well plates, and 400 ng of MVs (containing RNA) was
added to the cells 3 times a day for 14–21 days. The morphology of
the transformed cells was observed using Wright-Giemsa stain. To
confirm the effect of miR-106a/b, K562 cells were transfected with
miR-106a/b mimics and inhibitors: K562 cells were seeded onto
6-well plates (2×106 cells/well) the day before
transfection. The cells were transfected with 10 µl of 20 µM
miR-106a/b inhibitors, 5 µl of 20 µM miR-106a/b mimics, 10 µl of 20
µM inhibitor negative control and 5 µl of 20 µM mimic negative
control using riboFECTTM CP reagent (both from RiboBio, Guangzhou,
China), respectively. Real-time PCR was performed to assess the
level of miR-106a/b in the cells and their MVs. The supernatant of
transfected cells was collected 72 h after transfection to isolate
MVs.
Global DNA methylation
DNA was extracted using TIANamp Genomic DNA kit
(DP304; Tiangen, Beijing, China). Total DNA methylation was
performed using MethylFlash Methylated DNA Quantification kit
(P-1035-48; Epigentek, Farmingdale, NY, USA). All procedures were
conducted in accordance with the instructions of the kits.
Real-time PCR
Total RNA was extracted using TRIzol reagent kit
(Invitrogen Life Technologies, Carlsbad, CA, USA). cDNA was
synthesized by reverse transcription of 500 ng of total RNA with
PrimeScript™ RT Reagent kit (Clontech Laboratories, Inc.; Takara
Bio USA, Inc., Mountain View, CA, USA). Real-time PCR was performed
with 5 µl of SYBR Premix Ex Taq (Clontech Laboratories, Inc.;
Takara Bio USA, Inc.), 0.8 µl of primers (Invitrogen, Carlsbad, CA,
USA), 0.2 µl of ROX reference dye, 3 µl of RNase-free
H2O and 1 µl of cDNA as a template in a final reaction
volume of 10 µl. The PCR cycling conditions were as follows:
initial melting at 95°C for 30 sec, followed by 40 cycles at 95°C
for 5 sec and 60°C for 30 sec. The fluorescence intensity was
assessed using the ABI StepOnePlus™ Real-Time PCR System. Analysis
of the melting curve for the primers was conducted to confirm the
specificity of the PCR product, and the Ct value for triplicate
reactions was averaged. The fold changes in mRNA were calculated
through relative quantification (2−ΔΔCt).
Western blotting
Cells were washed twice with phosphate-buffered
saline (PBS) and lysed in RIPA buffer [1% Nonidet™ P-40, 1 mM EDTA,
50 mM Tris (pH 7.4), 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM NaF,
1 mM sodium orthovanadate and 1 mM phenylmethylsulfonyl fluoride
(PMSF)]. The cell lysates were centrifuged at 12,000 rpm for 15 min
at 4°C, and the supernatants were collected. The protein
concentrations were determined using a BCA protein assay kit
(Pierce Biotechnology, Inc., Rockford, IL, USA). The proteins were
treated with SDS sample buffer and heated at 95°C for 10 min. The
protein samples (40 µg) for each well were separated by sodium
dodecylsulfate-polyacrylamide gel electrophoresis and transferred
to nitrocellulose membranes. The membranes were blocked with 5%
non-fat milk and incubated overnight with a monoclonal antibody
(1:500) at 4°C. The blots were developed using a horseradish
peroxidase-conjugated rabbit anti-human secondary antibody (1:500)
and a chemiluminescent detection kit (Amersham Biosciences,
Piscataway, NJ, USA).
Real-time quantitative
methylation-specific PCR
Genomic DNA was isolated with the TIANamp Genomic
DNA kit (DP304; Tiangen) according to the manufacturer's
instructions. The methylation status of p53, c-Myc and the DLX4
promoter was evaluated by methylation-specific PCR (MSP). Briefly,
genomic DNA was treated with bisulfite using the EpiTect Bisulfite
kit (Qiagen, Hilden, Germany). MSP primers designed to amplify the
methylated-MSP and unmethylated-MSP alleles were previously
described (11–13). PCR products were resolved on 4%
agarose gels, stained with ethidium bromide, and visualized under
ultraviolet illumination.
Statistical analysis
SPSS 10.0 was used for statistical analysis (SPSS,
Inc., Chicago, IL, USA). Non-parametric and unpaired t-test
comparisons were used to compare the groups; the rates between the
groups were compared by the Chi-square test. Two-sided P<0.05
was defined as being statistically significant.
Results
Relative expression of miR-106a-5p,
miR-106b-5p and lincPOU3F3 in K562 and K562-MV after
transfection
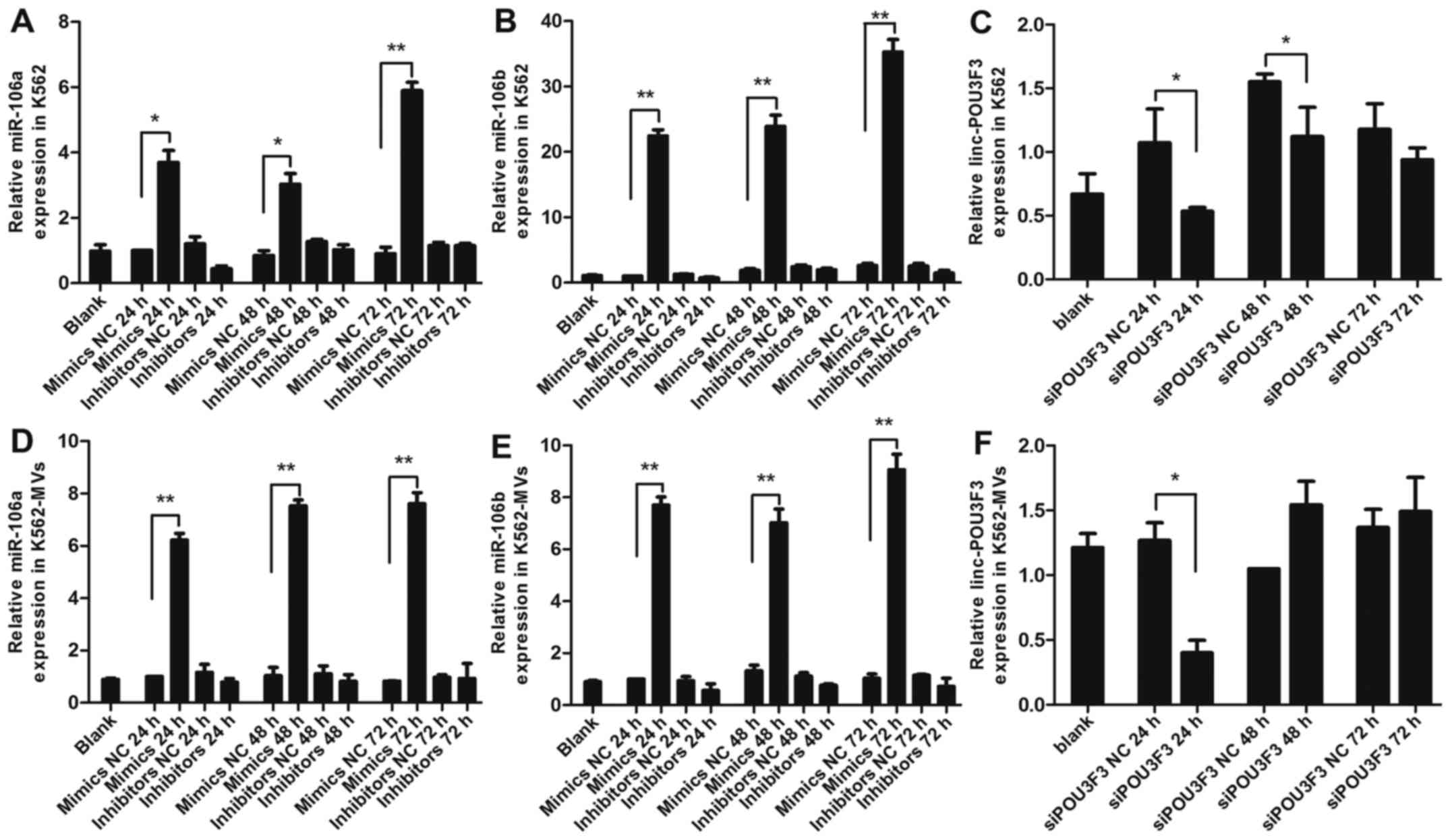
PCR analysis was used to identify the relative
expression of miRNA and lincRNA after transfection with miRNA
mimics, inhibitors, siPOU3F3 and the scrambled negative control
(NC) RNA, respectively. As expected, the expression levels of
miR-106a/b were markedly increased in the K562 and K562-MVs
transfected with the mimics, and reached a peak at 72 h (P<0.01
or P<0.05; Fig. 1A, B, D and E).
Whereas, the expression levels of miR-106a/b were relatively
decreased in cells and MVs transfected with the inhibitors.
However, compared with the control, these decreases were not
significant (P>0.05; Fig. 1A, B, D
and E). The conceivable reason for this is that inhibitors
suppress downstream pathways by binding miRNA, but do not affect
its expression. No significant differences were observed between
the mimics NC or the inhibitors NC and blank groups (P>0.05;
Fig. 1A, B, D and E). As for
lincPOU3F3, the expression in the K562 and K562-MVs was the lowest
at 24 h after transfection with siPOU3F3 (P<0.05; Fig. 1C and F). However, when detected at
48 and 72 h, there was no statistical significance between the siNC
and siPOU3F3 (both P>0.05; Fig. C
and F). According to the aforementioned results, we selected
K562-MVs transfected with miRNA at 72 h and siPOU3F3 at 24 h for
follow-up experiments.
MV-delivered miR-106a-5p, miR-106b-5p
and lincPOU3F3 accelerates the transformation process
To determine whether MV-associated miR-106a/b were
functional for the transformation, we added K562-MVs at different
levels of miR-106a/b (normal K562, miR-106a/b mimics and miR-106a/b
inhibitors) into the recipient cells, using K562-MVs as a control.
We found that a high level of miR-106a/b in K562-MVs accelerated
the transformation process (8.33±0.94 vs. 13.29±1.28 days;
P<0.001; Table I). Whereas, a
decreased level of miR-106a/b did not stop the transformation, but
resulted in a 3-day delay, leading to a transformation spanning 16
days (16±0.82 vs. 13.29±1.28 days; P<0.01; Table I). Similarly, we also found that
K562-MVs with siPOU3F3 delayed the transformation span to day 17
(17.83±0.29 vs. 13.29±1.28 days; P<0.01; Table I).
 | Table I.Roles of miR-106a/b and lincPOU3F3 in
the transformation process. |
Table I.
Roles of miR-106a/b and lincPOU3F3 in
the transformation process.
| Groups | Recipient
cells | No. of cells | Time (days) |
|---|
| K562-MVs | Mobilization |
4×106 | 13.29±1.28 |
| K562-MVs with
increased miR-106a/b | Mobilization |
4×106 |
8.33±0.94b |
| K562-MVs with
decreased miR-106a/b | Mobilization |
4×106 |
16±0.82a |
| K562-MVs with
siPOU3F3 | Mobilization |
4×106 |
17.83±0.29a |
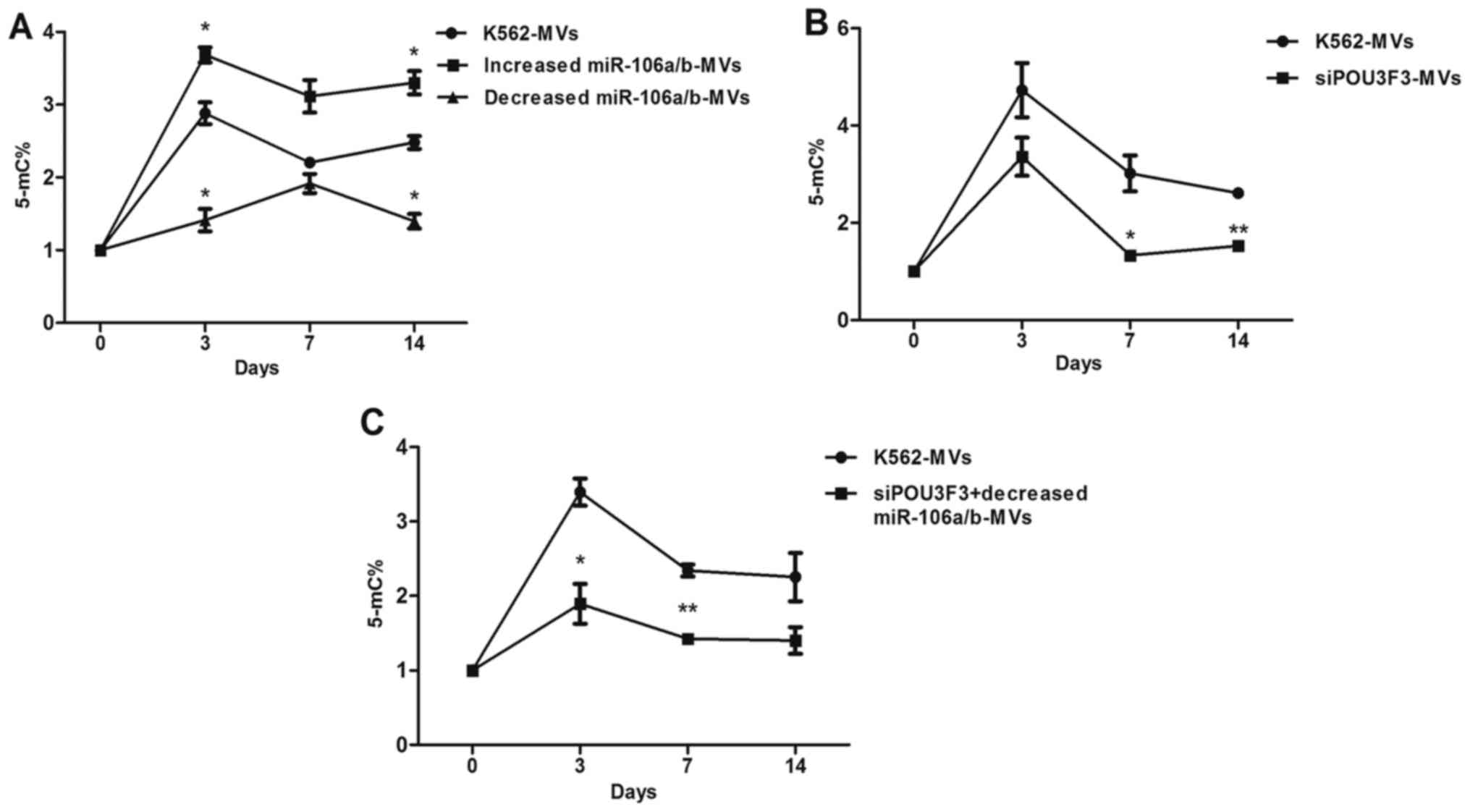
GDM is involved in transformation
We confirmed again an increase in GDM in
K562-MV-treated cells that peaked on day 3. Using K562-MV-treated
cells as a control, the GDM of the K562-MV group with increased
miR-106a/b expression was increased, particularly at days 3 and 14
(P<0.05); in contrast, decreased miR-106a/b expression in
K562-MVs decreased the GDM of recipient cells, particularly at days
3 and 14 (P<0.05), and the peak time was delayed to day 7. The
difference of GDM between the K562-MV groups with
increased/decreased miRNA and the control was not significant at
day 7 (P>0.05; Fig. 2A). In
comparison with the K562-MVs, the K562-MVs with siPOU3F3 decreased
the GDM at days 7 and 14 (P<0.05), although it made no
difference at the peak time (day 3; P>0.05; Fig. 2B). To analyze whether there is a
synergistic effect between inhibitors and siPOU3F3, we also
detected the GDM of recipient cells treated with K562-MVs
co-transfected with inhibitors and siPOU3F3. Compared with the
control, the treatment group was significantly decreased at day 3
(P<0.05) and day 7 (P<0.01), while the difference at day 14
was not significant (P>0.05; Fig.
2C).
Mechanisms of global DNA methylation
change induced by RNA within K562-MVs
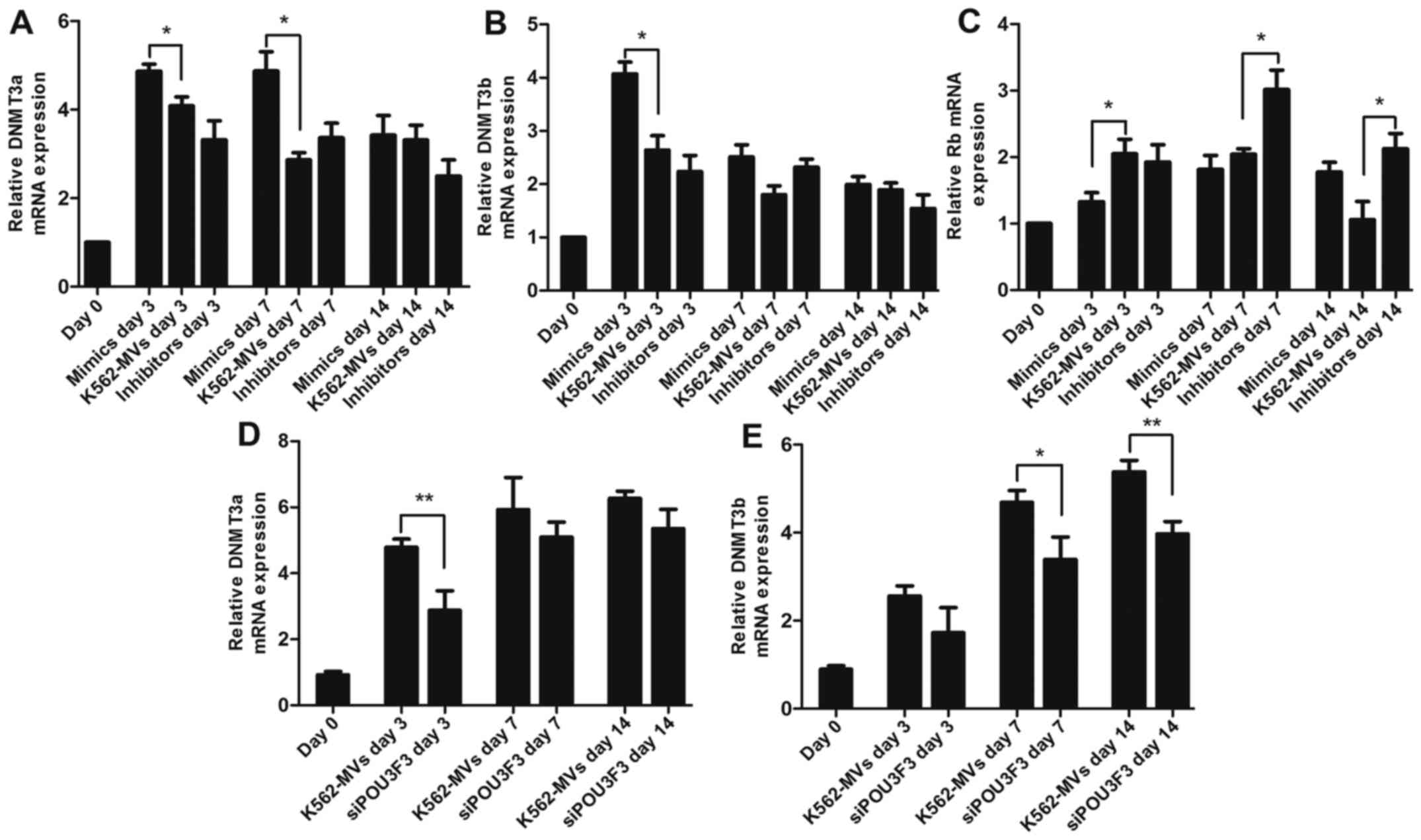
We next investigated the changes in the levels of
DNMTs by assessing the mRNA and protein levels of DNMT3a and
DNMT3b. The mRNA and protein levels of DNMT3a and DNMT3b were
generally increased during transformation induced by K562-MVs. The
mRNA of DNMT3a was consistently increased on days 3 and 7 in the
increased miR-106a/b group compared with the K562-MV group; in
contrast, the protein expression in DNMT3a in the group with
decreased miR-106a/b was suppressed (Figs. 3A, and 4A and C). miR-106a/b also increased the
expression of DNMT3b mRNA and protein (Figs. 3B, and Fig. 4A and D). Mature miRNAs lead to
translational suppression or mRNA degradation of the target
protein-coding genes (14).
According to the aforementioned data, miRNA may indirectly regulate
the expression of DNMTs. Tang et al (15) found that RB suppressed DNMT3A
promoter activity and mRNA/protein expression in lung cancer; in
contrast, we determined that RB was the target of miR-106a/b using
bioinformatics analyses (http://mirdb.org/miRDB/). Therefore, we detected the
expression of RB in recipient cells. Increased/decreased miR-106a/b
in K562-MVs led to a significant decrease/increase of Rb mRNA and
protein observed on days 3, 7 and 14 (P<0.05 or P<0.01;
Figs. 3C and 4A and E). K562-MVs with siPOU3F3
significantly decreased the expression of the mRNA of DNMT3a at day
3 and protein at day 7 (Figs. 3D
and 4B and F). In terms of DNMT3b,
its mRNA and protein were both lowest at day 14 (P<0.05 or
P<0.01, Figs. 3E and 4B and G). The mismatch between the mRNA
and protein may be explained by the fact that MVs are packages of
bioactive molecules that contain not only contributing factors but
also detractors. Collectively, we proposed a new signaling pathway
underlying the epigenetic effects of RNA in the transformation
process. The increased expression of DNMT3a and DNMT3b induced by
lincPOU3F3 may facilitate malignant transformation; whereas
miR-106a/b may indirectly regulate DNMT3a and DNMT3b through Rb,
further affecting the transformation process (Fig. 4H).
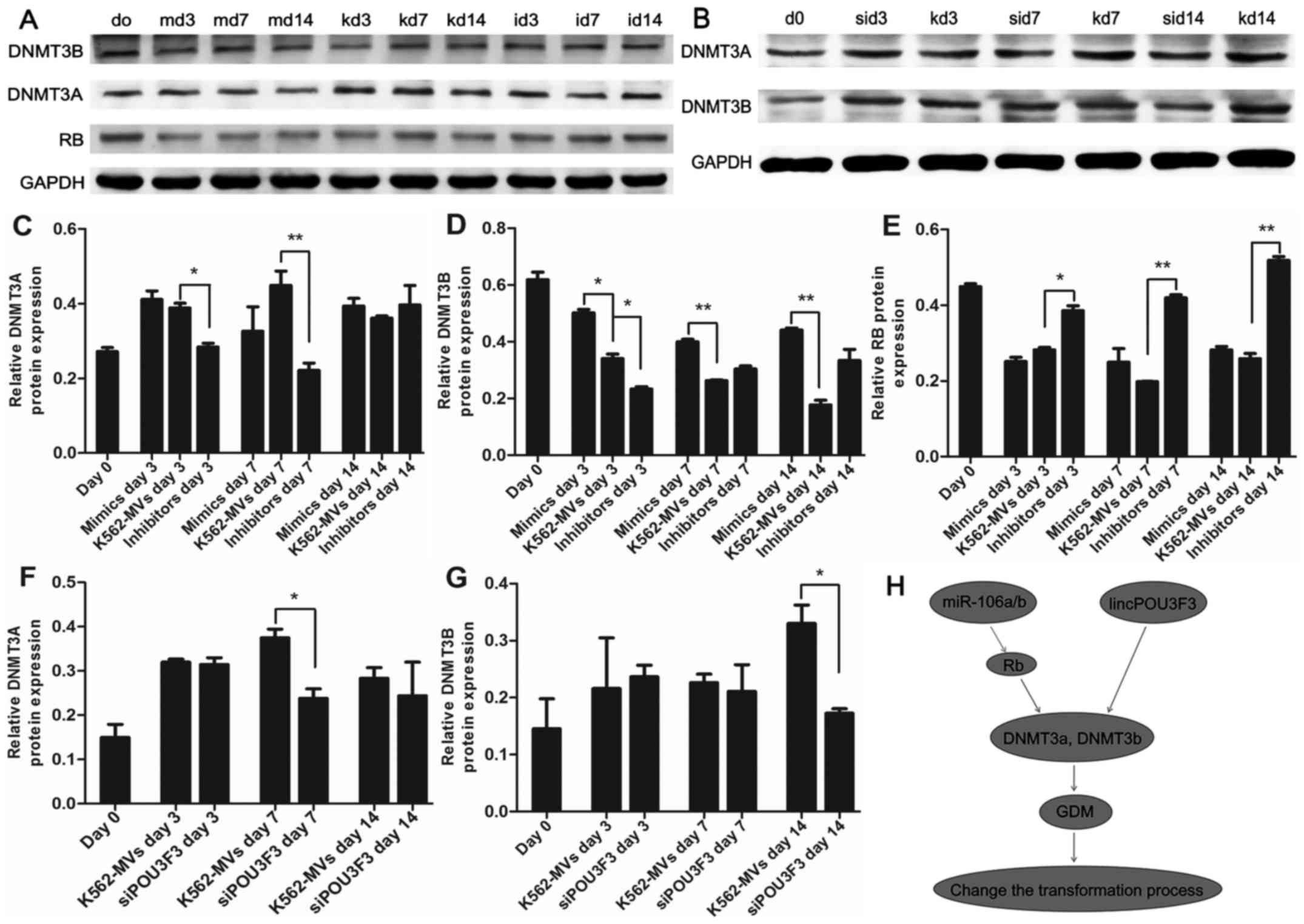 | Figure 4.Εpigenetic functions of miR-106a-5p,
miR-106b-5p and lincPOU3F3 in the transformation. (A, C, D and E)
Τhe protein levels of DNMT3A, DNMT3B and RB in recipient cells
induced by K562-MVs with regulated miR-106a/b at different
time-points. GAPDH was used as a loading control. (B, F and G) The
protein expression of DNMT3A and DNMT3B in recipient cells treated
with siPOU3F3/k562-MVs were assessed by western blotting, GAPDH was
used as a loading control. (H) Proposed signaling pathways
underlying the epigenetic effects of miR-106a-5p, miR-106b-5p and
lincPOU3F3 in the transformation process; *P<0.05 and
**P<0.01. m, mimics; k, K562-MVs; i, inhibitors; si, siPOU3F3.
MVs, microvesicles. |
The promoter methylation of p53, c-Myc
and DLX4
MSP analyses were carried out on the promoter
regions of recipient cells of p53, c-Myc and DLX4 genes at day 0, 3
and 14, respectively. Each sample was amplified by methylated
primers (M) and unmethylated primers (U). The promoters we analyzed
of the p53 and c-Myc genes were both hypomethylated in the
transformation process (day 3 and 14; Fig. 5A and B), whereas, the promoter of
the DLX4 gene was hypermethylated compared with normal peripheral
blood mobilization (day 0; Fig.
5C), which was in line with previous research which revealed
that DLX4 hypermethylation was associated with disease progression
in CML (8).
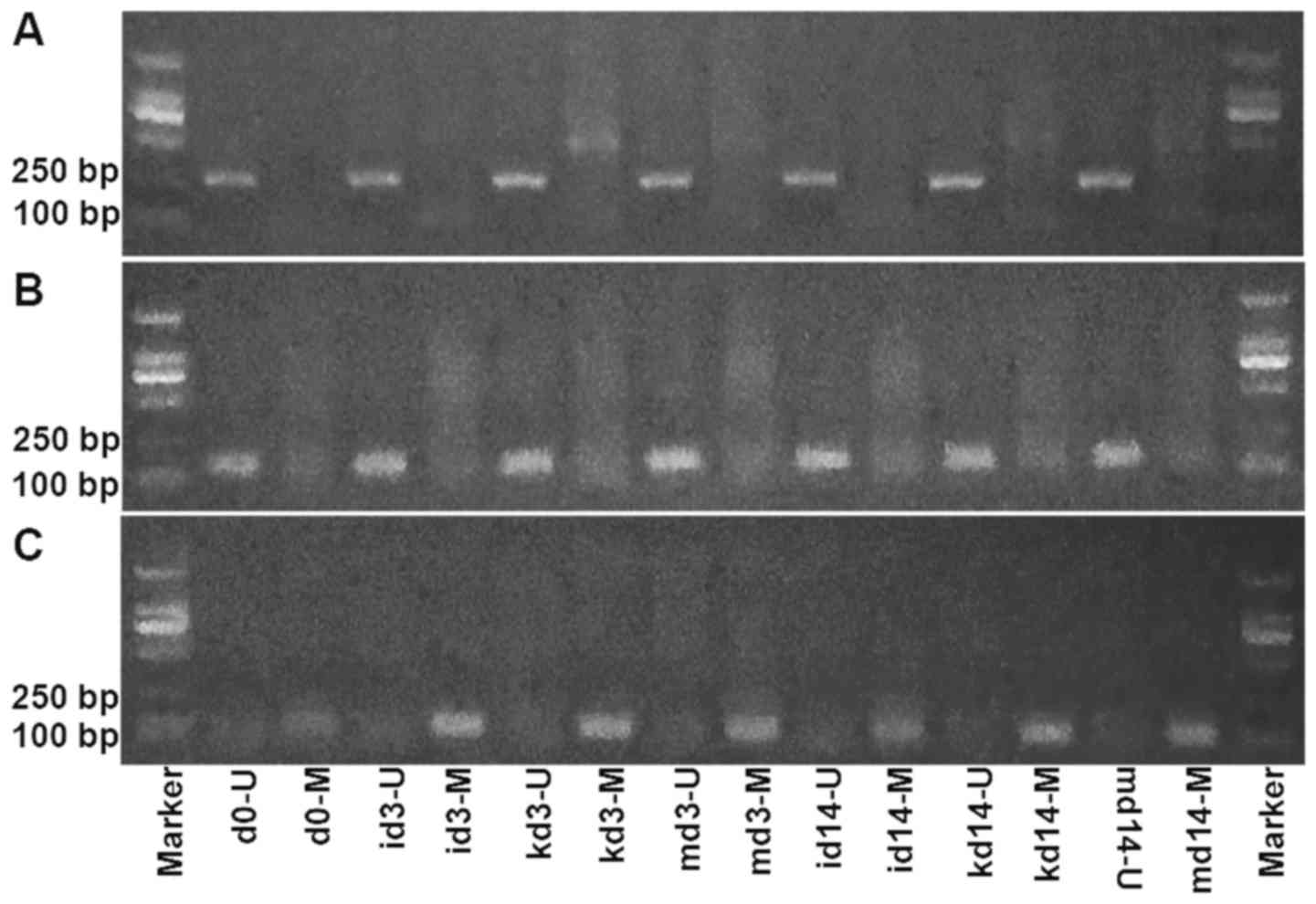 | Figure 5.The promoter methylation of p53,
c-Myc and DLX4. MSP analyses were carried out on the promoter
regions of p53, c-Myc and DLX4 of recipient cells at day 0, 3 and
14, respectively. Each sample was amplified by methylated primers
and unmethylated primers. The presence of a visible PCR product in
lane U indicated the presence of unmethylated genes, the presence
of a product in lane M indicated the presence of methylated genes.
(A) MSP of the p53 promoter region in recipient cells. (B) MSP of
the c-Myc promoter region in recipient cells. (C) MSP of the DLX4
promoter region in recipient cells. i, inhibitors; k, k562; m,
mimics; U, unmethylated; M, methylated. MSP, methylation-specific
PCR. |
Discussion
Emission of MVs represents an important emerging
element in the complexity of the cellular secretome, and likely
acts in concert with the release of ions, metabolites, hormones,
growth factors, cytokines and extracellular matrix molecules, with
which vesiculation and cell-to-cell contact likely form a
functional continuum (16). MVs
released by tumor cells are detectable in patients with cancer and
their number in the circulation correlates with poor prognosis
(17). Our previous study
demonstrated that BCR-ABL1-positive MVs initiated malignant
transformation of normal hematopoietic transplants, providing
implications for CML blast crisis (BC) and donor cell leukemia
(DCL) (3). In the present study, we
confirmed that miRNA and lincRNA within the MVs could lead to an
altered level of GDM of the recipient cells. Although demethylation
therapy has been applied in CML in some centers, little is known
concerning the impact of DNA methylation on the
evolution/progression of CML (18).
The findings of the present study helped to further our
understanding of the epigenetic change during the process of
transformation, particularly DNA methylation. Further study should
focus on the epigenetic modification induced by MVs and the
potential to translate this knowledge into innovative approaches
for monitoring and personalized therapy.
We further identified the connection between GDM and
carcinogenesis. It has been reported for years that DNA-level
epigenetic regulation plays a causative role in cancer and can thus
be targeted for treatment of the disorder (19). In CML, Heller et al observed
downregulated expression of many of these genes in BC-CML compared
with CP-CML samples, using RNA-sequencing (20). However, a single DNA methylation
change may act as a precipitating event in CML progression
(21) and may provide a useful
basis for revealing new targets of therapy in advanced CML. The
present study shed much-needed light on an unconventional and
poorly understood mechanism of exogenous RNA and its impact on GDM.
GDM contributed to carcinogenesis via epigenetic silencing of
well-known tumor-suppressor genes (TSGs) or regulators of cell
proliferation (22,23). In the present study, DLX4, a gene
correlated with disease progression of CML (8), was hypermethylated during the process,
which may be a pivotal regulator of the transformation.
One aspect merits further consideration involves the
reason why miRNA and lincRNA in MVs induced global methylation of
recipient cells. The complexity of extracellular vesicle-associated
bioactive macromolecules supports a critical role in the
modification of recipient cells (24). MVs can transfer specific proteins to
target cells for the delivery of signaling pathways (25,26).
RNA represents the main compound of cancer-derived MVs (27,28).
In cancer, miRNAs and lincRNA may act either as potent oncogenes or
TSGs and their deregulation has been associated with the etiology,
progression and prognosis (29).
Using bioinformatics analyses, we determined that lincPOU3F3
altered the expression of DNMTs. However, Tang et al
(15) revealed that RB suppressed
DNMT3A promoter activity and mRNA/protein expression in lung
cancer, leading to the decrease of methylation level globally and
TSGs specifically. RB is the target of miR-106a/b. As a result, we
confirmed that miR-106a/b and lincPOU3F3 were involved in the
transcriptional regulation of DNMT genes in our transformation
model, which may induce global DNA methylation of the recipient
cells. Thus, it could be speculated that miRNAs and lincRNAs in MVs
provided important insight into leukemogenesis and development of
CML.
Regulation of miR-106a/b and lincRNA in MVs could
alter the endpoint of the transformation. We surmised that
compounds in MVs could be regulated by two distinct pathways: via
consequent overexpression of the content, such as miRNA mimics or
inhibitors, in the parental cells; and via direct transfection of
the content with MVs (30).
Transfection into the parental cells was preferred since it could
mimic the released thus far physiological process. Notably, we
found that the expression of miRNA in K562-MVs and K562 after
transfection with parental cells was inconsistent, indicating that
MVs were shed following the selective incorporation of a host of
molecular cargo, but not randomly released. The mechanism of how
the MV content was loaded remained unclear. Several publications
considered that the RNA-induced silencing complex (RISC),
particularly the Ago2 protein, participated in the RNA transport
from plasma into MVs (31,32). It could be speculated that
modification of Ago2 and RISC could help to regulate designated
compound in MVs.
In summary, we found that horizontal transmission of
miRNA and lincRNA triggered the epigenetic modification and
downstream effect of target cells. Determinants of the cancer cell
phenotype hardwired in driver factors are superimposed with more
exogenous regulators, such as MVs, dictated by the epigenetic and
differentiation programmers. This creates several levels of
functional heterogeneity and interactive potential relevance of
therapy.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (NSFC) (nos. 81470333,
81470348 and 81500136).
Glossary
Abbreviations
Abbreviations:
|
MVs
|
microvesicles
|
|
DNMT
|
DNA methyltransferase
|
|
CML
|
chronic myeloid leukemia
|
|
MNCs
|
mononuclear cells
|
|
GDM
|
global DNA methylation
|
|
FBS
|
fetal bovine serum
|
|
MSP
|
methylation-specific PCR
|
|
NC
|
negative control
|
|
DCL
|
donor cell leukemia
|
|
BC
|
blast crisis
|
|
CP
|
chronic phase
|
|
TSGs
|
tumor suppressor genes
|
|
RISC
|
RNA-induced silencing complex
|
References
|
1
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boysen J, Nelson M, Magzoub G, Maiti GP,
Sinha S, Goswami M, Vesely SK, Shanafelt TD, Kay NE and Ghosh AK:
Dynamics of microvesicle generation in B-cell chronic lymphocytic
leukemia: Implication in disease progression. Leukemia. 31:350–360.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu X, You Y, Li Q, Zeng C, Fu F, Guo A,
Zhang H, Zou P, Zhong Z, Wang H, et al: BCR-ABL1-positive
microvesicles transform normal hematopoietic transplants through
genomic instability: Implications for donor cell leukemia.
Leukemia. 28:1666–1675. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ntziachristos P, Abdel-Wahab O and
Aifantis I: Emerging concepts of epigenetic dysregulation in
hematological malignancies. Nat Immunol. 17:1016–1024. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang LQ and Chim CS: DNA methylation of
tumor-suppressor miRNA genes in chronic lymphocytic leukemia.
Epigenomics. 7:461–473. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
San J, ose-Eneriz E, Agirre X,
Rodríguez-Otero P and Prosper F: Epigenetic regulation of cell
signaling pathways in acute lymphoblastic leukemia. Epigenomics.
5:525–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fong CY, Morison J and Dawson MA:
Epigenetics in the hematologic malignancies. Haematologica.
99:1772–1783. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou JD, Wang YX, Zhang TJ, Yang DQ, Yao
DM, Guo H, Yang L, Ma JC, Wen XM, Yang J, et al: Epigenetic
inactivation of DLX4 is associated with disease progression in
chronic myeloid leukemia. Biochem Biophys Res Commun.
463:1250–1256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esposito N, Colavita I, Quintarelli C,
Sica AR, Peluso AL, Luciano L, Picardi M, Del Vecchio L, Buonomo T,
Hughes TP, et al: SHP-1 expression accounts for resistance to
imatinib treatment in Philadelphia chromosome-positive cells
derived from patients with chronic myeloid leukemia. Blood.
118:3634–3644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Elias MH, Baba AA, Husin A, Sulong S,
Hassan R, Sim GA, Wahid SF Abdul and Ankathil R: HOXA4 gene
promoter hypermethylation as an epigenetic mechanism mediating
resistance to imatinib mesylate in chronic myeloid leukemia
patients. Biomed Res Int. 2013:1297152013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chmelarova M, Kos S, Dvorakova E, Spacek
J, Laco J, Ruszova E, Hrochova K and Palicka V: Importance of
promoter methylation of GATA4 and TP53 genes in endometrioid
carcinoma of endometrium. Clin Chem Lab Med. 52:1229–1234. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Souza CR, Leal MF, Calcagno DQ, Sozinho
EK Costa, Borges BN, Montenegro RC, Dos Santos AK, Dos Santos SE,
Ribeiro HF, Assumpção PP, et al: MYC deregulation in gastric cancer
and its clinicopathological implications. PLoS One. 8:e644202013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang TJ, Zhou JD, Yang DQ, Wang YX, Yao
DM, Ma JC, Wen XM, Guo H, Lin J and Qian J: Hypermethylation of
DLX4 predicts poor clinical outcome in patients with
myelodysplastic syndrome. Clin Chem Lab Med. 54:865–871. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang YA, Lin RK, Tsai YT, Hsu HS, Yang YC,
Chen CY and Wang YC: MDM2 overexpression deregulates the
transcriptional control of RB/E2F leading to DNA methyltransferase
3A overexpression in lung cancer. Clin Cancer Res. 18:4325–4333.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Taylor DD and Gercel-Taylor C: The origin,
function, and diagnostic potential of RNA within extracellular
vesicles present in human biological fluids. Front Genet.
4:1422013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Setti M, Osti D, Richichi C, Ortensi B,
Del Bene M, Fornasari L, Beznoussenko G, Mironov A, Rappa G, Cuomo
A, et al: Extracellular vesicle-mediated transfer of CLIC1 protein
is a novel mechanism for the regulation of glioblastoma growth.
Oncotarget. 6:31413–31427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y,
Du X, Li J, Li P, Ren R, et al: Targeting methyltransferase PRMT5
eliminates leukemia stem cells in chronic myelogenous leukemia. J
Clin Invest. 126:3961–3980. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lissa D and Robles AI: Methylation
analyses in liquid biopsy. Transl Lung Cancer Res. 5:492–504. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heller G, Topakian T, Altenberger C,
Cerny-Reiterer S, Herndlhofer S, Ziegler B, Datlinger P, Byrgazov
K, Bock C, Mannhalter C, et al: Next-generation sequencing
identifies major DNA methylation changes during progression of
Ph+ chronic myeloid leukemia. Leukemia. 30:1861–1868.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amabile G, Di Ruscio A, Müller F, Welner
RS, Yang H, Ebralidze AK, Zhang H, Levantini E, Qi L, Martinelli G,
et al: Dissecting the role of aberrant DNA methylation in human
leukaemia. Nat Commun. 6:70912015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han F, Liu W, Jiang X, Shi X, Yin L, Ao L,
Cui Z, Li Y, Huang C, Cao J, et al: SOX30, a novel epigenetic
silenced tumor suppressor, promotes tumor cell apoptosis by
transcriptional activating p53 in lung cancer. Oncogene.
34:4391–4402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kikuyama M, Takeshima H, Kinoshita T,
Okochi-Takada E, Wakabayashi M, Akashi-Tanaka S, Ogawa T, Seto Y
and Ushijima T: Development of a novel approach, the
epigenome-based outlier approach, to identify tumor-suppressor
genes silenced by aberrant DNA methylation. Cancer Lett.
322:204–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Tang J, Zhang B, Yang W, LiuGao M,
Wang R, Tan Y, Fan J, Chang Y, Fu J, et al: Epigenetic modification
of MiR-429 promotes liver tumour-initiating cell properties by
targeting Rb binding protein 4. Gut. 64:156–167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Camussi G, Deregibus MC, Bruno S, Grange
C, Fonsato V and Tetta C: Exosome/microvesicle-mediated epigenetic
reprogramming of cells. Am J Cancer Res. 1:98–110. 2011.PubMed/NCBI
|
|
26
|
Antonyak MA and Cerione RA: Microvesicles
as mediators of intercellular communication in cancer. Methods Mol
Biol. 1165:147–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ratajczak MZ, Ratajczak D and Pedziwiatr
D: Extracellular microvesicles (ExMVs) in cell to cell
communication: A role of telocytes. Adv Exp Med Biol. 913:41–49.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
ElSharawy A, Röder C, Becker T, Habermann
JK, Schreiber S, Rosenstiel P and Kalthoff H: Concentration of
circulating miRNA-containing particles in serum enhances miRNA
detection and reflects CRC tissue-related deregulations.
Oncotarget. 7:75353–75365. 2016.PubMed/NCBI
|
|
29
|
Espinosa-Parrilla Y, Muñoz X, Bonet C,
Garcia N, Venceslá A, Yiannakouris N, Naccarati A, Sieri S, Panico
S, Huerta JM, et al: Genetic association of gastric cancer with
miRNA clusters including the cancer-related genes MIR29, MIR25,
MIR93 and MIR106: Results from the EPIC-EURGAST study. Int J
Cancer. 135:2065–2076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang HM, Li Q, Zhu X, Liu W, Hu H, Liu T,
Cheng F, You Y, Zhong Z, Zou P, et al: miR-146b-5p within
BCR-ABL1-positive microvesicles promotes leukemic transformation of
hematopoietic cells. Cancer Res. 76:2901–2911. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lv Z, Wei Y, Wang D, Zhang CY, Zen K and
Li L: Argonaute 2 in cell-secreted microvesicles guides the
function of secreted miRNAs in recipient cells. PLoS One.
9:e1035992014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Turchinovich A, Weiz L and Burwinkel B:
Isolation of circulating microRNA associated with RNA-binding
protein. Methods Mol Biol. 1024:97–107. 2013. View Article : Google Scholar : PubMed/NCBI
|