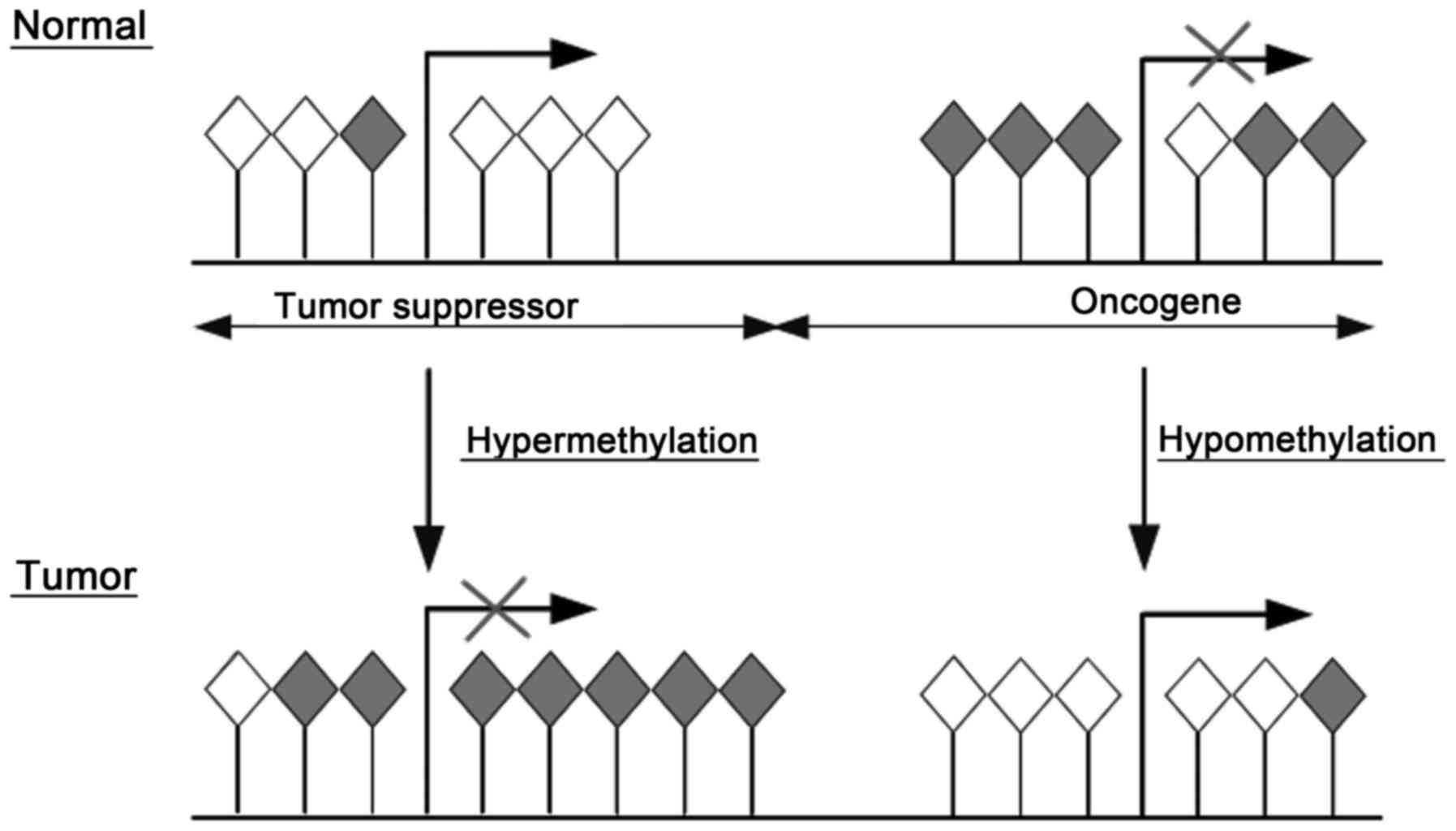
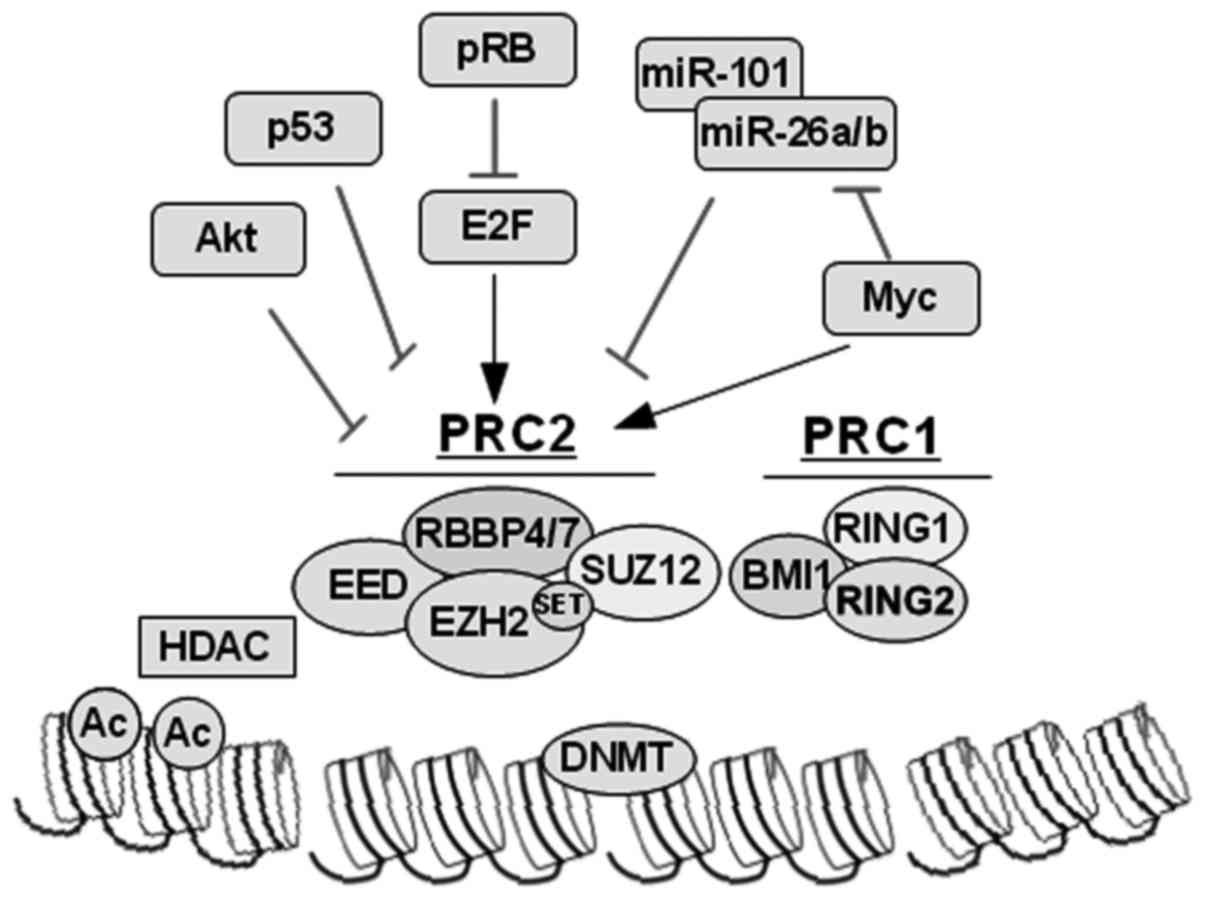
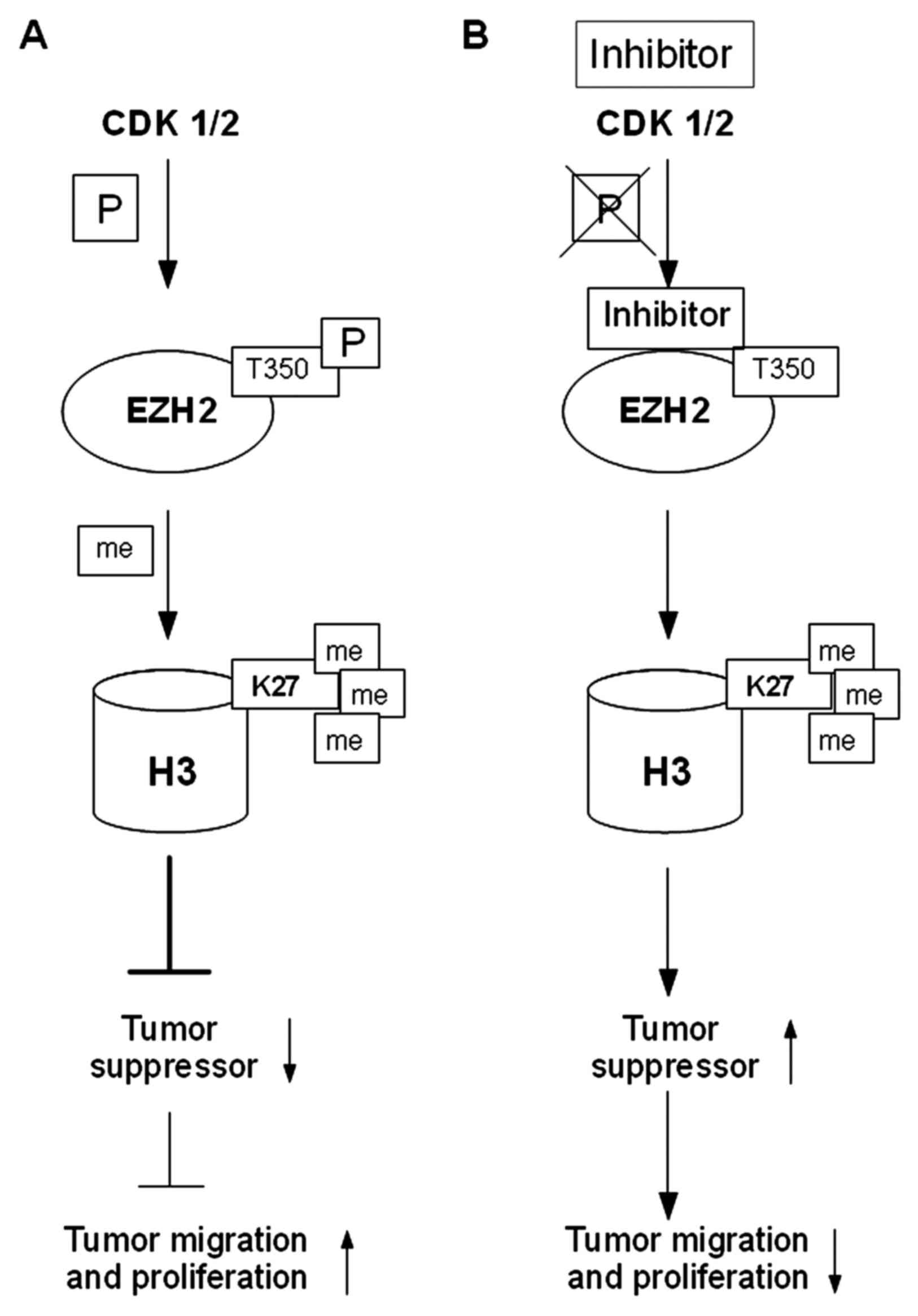
|
1
|
Damaschke NA, Yang B, Bhusari S, Svaren JP
and Jarrard DF: Epigenetic susceptibility factors for prostate
cancer with aging. Prostate. 73:1721–1730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Willard SS and Koochekpour S: Regulators
of gene expression as biomarkers for prostate cancer. Am J Cancer
Res. 2:620–657. 2012.PubMed/NCBI
|
|
3
|
Chin SP, Dickinson JL and Holloway AF:
Epigenetic regulation of prostate cancer. Clin Epigenetics.
2:151–169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Day TK and Bianco-Miotto T: Common gene
pathways and families altered by DNA methylation in breast and
prostate cancers. Endocr Relat Cancer. 20:R215–R232. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Majumdar S, Buckles E, Estrada J and
Koochekpour S: Aberrant DNA methylation and prostate cancer. Curr
Genomics. 12:486–505. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Subramaniam D, Thombre R, Dhar A and Anant
S: DNA methyltransferases: A novel target for prevention and
therapy. Front Oncol. 4:802014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buck-Koehntop BA and Defossez PA: On how
mammalian transcription factors recognize methylated DNA.
Epigenetics. 8:131–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Albany C, Alva AS, Aparicio AM, Singal R,
Yellapragada S, Sonpavde G and Hahn NM: Epigenetics in prostate
cancer. Prostate Cancer. 2011:5803182011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang M and Park JY: DNA methylation in
promoter region as biomarkers in prostate cancer. Methods Mol Biol.
863:67–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashour N, Angulo JC, Andrés G, Alelú R,
González-Corpas A, Toledo MV, Rodríguez-Barbero JM, López JI,
Sánchez-Chapado M and Ropero S: A DNA hypermethylation profile
reveals new potential biomarkers for prostate cancer diagnosis and
prognosis. Prostate. 74:1171–1182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gonzalgo ML, Pavlovich CP, Lee SM and
Nelson WG: Prostate cancer detection by GSTP1 methylation analysis
of postbiopsy urine specimens. Clin Cancer Res. 9:2673–2677.
2003.PubMed/NCBI
|
|
12
|
Rouprêt M, Hupertan V, Yates DR, Catto JW,
Rehman I, Meuth M, Ricci S, Lacave R, Cancel-Tassin G, de la Taille
A, et al: Molecular detection of localized prostate cancer using
quantitative methylation-specific PCR on urinary cells obtained
following prostate massage. Clin Cancer Res. 13:1720–1725. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dumache R, Puiu M, Motoc M, Vernic C and
Dumitrascu V: Prostate cancer molecular detection in plasma samples
by glutathione S-transferase P1 (GSTP1) methylation analysis. Clin
Lab. 60:847–852. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bastian PJ, Palapattu GS, Lin X,
Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW
and Nelson WG: Preoperative serum DNA GSTP1 CpG island
hypermethylation and the risk of early prostate-specific antigen
recurrence following radical prostatectomy. Clin Cancer Res.
11:4037–4043. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reibenwein J, Pils D, Horak P, Tomicek B,
Goldner G, Worel N, Elandt K and Krainer M: Promoter
hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate
cancer patients and its clinical relevance. Prostate. 67:427–432.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mahon KL, Qu W, Devaney J, Paul C,
Castillo L, Wykes RJ, Chatfield MD, Boyer MJ, Stockler MR, Marx G,
et al: PRIMe consortium: Methylated Glutathione S-transferase 1
(mGSTP1) is a potential plasma free DNA epigenetic marker of
prognosis and response to chemotherapy in castrate-resistant
prostate cancer. Br J Cancer. 111:1802–1809. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang GH, Lee S, Lee HJ and Hwang KS:
Aberrant CpG island hypermethylation of multiple genes in prostate
cancer and prostatic intraepithelial neoplasia. J Pathol.
202:233–240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sidhu S, Deep JS, Sobti RC, Sharma VL and
Thakur H: Methylation pattern of MGMT gene in relation to age,
smoking, drinking and dietary habits as epigenetic biomarker in
prostate cancer patients. GEBJ. 8:1–11. 2010.
|
|
19
|
Tang D, Kryvenko ON, Mitrache N, Do KC,
Jankowski M, Chitale DA, Trudeau S, Rundle A, Belinsky SA and
Rybicki BA: Methylation of the RARB gene increases prostate cancer
risk in black Americans. J Urol. 190:317–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keil KP, Abler LL, Mehta V, Altmann HM,
Laporta J, Plisch EH, Suresh M, Hernandez LL and Vezina CM: DNA
methylation of E-cadherin is a priming mechanism for prostate
development. Dev Biol. 387:142–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kito H, Suzuki H, Ichikawa T, Sekita N,
Kamiya N, Akakura K, Igarashi T, Nakayama T, Watanabe M, Harigaya
K, et al: Hypermethylation of the CD44 gene is associated with
progression and metastasis of human prostate cancer. Prostate.
49:110–115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Singal R, Ferdinand L, Reis IM and
Schlesselman JJ: Methylation of multiple genes in prostate cancer
and the relationship with clinicopathological features of disease.
Oncol Rep. 12:631–637. 2004.PubMed/NCBI
|
|
23
|
Woodson K, Hayes R, Wideroff L, Villaruz L
and Tangrea J: Hypermethylation of GSTP1, CD44, and E-cadherin
genes in prostate cancer among US Blacks and Whites. Prostate.
55:199–205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Padar A, Sathyanarayana UG, Suzuki M,
Maruyama R, Hsieh JT, Frenkel EP, Minna JD and Gazdar AF:
Inactivation of cyclin D2 gene in prostate cancers by aberrant
promoter methylation. Clin Cancer Res. 9:4730–4734. 2003.PubMed/NCBI
|
|
25
|
Henrique R, Costa VL, Cerveira N, Carvalho
AL, Hoque MO, Ribeiro FR, Oliveira J, Teixeira MR, Sidransky D and
Jerónimo C: Hypermethylation of Cyclin D2 is associated with loss
of mRNA expression and tumor development in prostate cancer. J Mol
Med. 84:911–918. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Das PM, Ramachandran K, Vanwert J,
Ferdinand L, Gopisetty G, Reis IM and Singal R: Methylation
mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer.
5:282006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Delgado-Cruzata L, Hruby GW, Gonzalez K,
McKiernan J, Benson MC, Santella RM and Shen J: DNA methylation
changes correlate with Gleason score and tumor stage in prostate
cancer. DNA Cell Biol. 31:187–192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Kron KJ, Pethe VV, Demetrashvili N,
Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L,
van der Kwast TH, et al: Association of tissue promoter methylation
levels of APC, TGFβ2, HOXD3 and RASSF1A with prostate cancer
progression. Int J Cancer. 129:2454–2462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jerónimo C, Henrique R, Hoque MO, Ribeiro
FR, Oliveira J, Fonseca D, Teixeira MR, Lopes C and Sidransky D:
Quantitative RARbeta2 hypermethylation: A promising prostate cancer
marker. Clin Cancer Res. 10:4010–4014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Youssef EM, Chen XQ, Higuchi E, Kondo Y,
Garcia-Manero G, Lotan R and Issa JP: Hypermethylation and
silencing of the putative tumor suppressor Tazarotene-induced gene
1 in human cancers. Cancer Res. 64:2411–2417. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang J, Liu L and Pfeifer GP: Methylation
of the retinoid response gene TIG1 in prostate cancer correlates
with methylation of the retinoic acid receptor beta gene. Oncogene.
23:2241–2249. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choudhury JH and Ghosh SK: Promoter
hypermethylation profiling identifies subtypes of head and neck
cancer with distinct viral, environmental, genetic and survival
characteristics. PLoS One. 10:e01298082015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin JC, Wu YC, Wu CC, Shih PY, Wang WY and
Chien YC: DNA methylation markers and serum α-fetoprotein level are
prognostic factors in hepatocellular carcinoma. Ann Hepatol.
14:494–504. 2015.PubMed/NCBI
|
|
34
|
Zhang CY, Zhao YX, Xia RH, Han J, Wang BS,
Tian Z, Wang LZ, Hu YH and Li J: RASSF1A promoter hypermethylation
is a strong biomarker of poor survival in patients with salivary
adenoid cystic carcinoma in a Chinese population. PLoS One.
9:e1101592014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maruyama R, Toyooka S, Toyooka KO, Virmani
AK, Zöchbauer-Müller S, Farinas AJ, Minna JD, McConnell J, Frenkel
EP and Gazdar AF: Aberrant promoter methylation profile of prostate
cancers and its relationship to clinicopathological features. Clin
Cancer Res. 8:514–519. 2002.PubMed/NCBI
|
|
36
|
Ge YZ, Xu LW, Jia RP, Xu Z, Feng YM, Wu R,
Yu P, Zhao Y, Gui ZL, Tan SJ, et al: The association between
RASSF1A promoter methylation and prostate cancer: Evidence from 19
published studies. Tumour Biol. 35:3881–3890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pellacani D, Kestoras D, Droop AP, Frame
FM, Berry PA, Lawrence MG, Stower MJ, Simms MS, Mann VM, Collins
AT, et al: DNA hypermethylation in prostate cancer is a consequence
of aberrant epithelial differentiation and hyperproliferation. Cell
Death Differ. 21:761–773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Florl AR, Steinhoff C, Müller M, Seifert
HH, Hader C, Engers R, Ackermann R and Schulz WA: Coordinate
hypermethylation at specific genes in prostate carcinoma precedes
LINE-1 hypomethylation. Br J Cancer. 91:985–994. 2004.PubMed/NCBI
|
|
39
|
Schulz WA, Elo JP, Florl AR, Pennanen S,
Santourlidis S, Engers R, Buchardt M, Seifert HH and Visakorpi T:
Genomewide DNA hypomethylation is associated with alterations on
chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer.
35:58–65. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gurel B, Iwata T, Koh CM, Jenkins RB, Lan
F, Van Dang C, Hicks JL, Morgan J, Cornish TC, Sutcliffe S, et al:
Nuclear MYC protein overexpression is an early alteration in human
prostate carcinogenesis. Mod Pathol. 21:1156–1167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
LeBeau AM, Sevillano N, Markham K, Winter
MB, Murphy ST, Hostetter DR, West J, Lowman H, Craik CS and
VanBrocklin HF: Imaging active urokinase plasminogen activator in
prostate cancer. Cancer Res. 75:1225–1235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y and Cozzi PJ: Targeting uPA/uPAR in
prostate cancer. Cancer Treat Rev. 33:521–527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ogishima T, Shiina H, Breault JE,
Tabatabai L, Bassett WW, Enokida H, Li LC, Kawakami T, Urakami S,
Ribeiro-Filho LA, et al: Increased heparanase expression is caused
by promoter hypomethylation and up-regulation of transcriptional
factor early growth response-1 in human prostate cancer. Clin
Cancer Res. 11:1028–1036. 2005.PubMed/NCBI
|
|
44
|
Tokizane T, Shiina H, Igawa M, Enokida H,
Urakami S, Kawakami T, Ogishima T, Okino ST, Li LC, Tanaka Y, et
al: Cytochrome P450 1B1 is overexpressed and regulated by
hypomethylation in prostate cancer. Clin Cancer Res. 11:5793–5801.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Q, Williamson M, Bott S,
Brookman-Amissah N, Freeman A, Nariculam J, Hubank MJ, Ahmed A and
Masters JR: Hypomethylation of WNT5A, CRIP1 and S100P in prostate
cancer. Oncogene. 26:6560–6565. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shojima K, Sato A, Hanaki H, Tsujimoto I,
Nakamura M, Hattori K, Sato Y, Dohi K, Hirata M, Yamamoto H, et al:
Wnt5a promotes cancer cell invasion and proliferation by
receptor-mediated endocytosis-dependent and -independent
mechanisms, respectively. Sci Rep. 5:80422015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yamamoto H, Oue N, Sato A, Hasegawa Y,
Yamamoto H, Matsubara A, Yasui W and Kikuchi A: Wnt5a signaling is
involved in the aggressiveness of prostate cancer and expression of
metalloproteinase. Oncogene. 29:2036–2046. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Basu GD, Azorsa DO, Kiefer JA, Rojas AM,
Tuzmen S, Barrett MT, Trent JM, Kallioniemi O and Mousses S:
Functional evidence implicating S100P in prostate cancer
progression. Int J Cancer. 123:330–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lambropoulou M, Deftereou TE, Kynigopoulos
S, Patsias A, Anagnostopoulos C, Alexiadis G, Kotini A, Tsaroucha
A, Nikolaidou C, Kiziridou A, et al: Co-expression of galectin-3
and CRIP-1 in endometrial cancer: Prognostic value and patient
survival. Med Oncol. 33:82016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ludyga N, Englert S, Pflieger K, Rauser S,
Braselmann H, Walch A, Auer G, Höfler H and Aubele M: The impact of
cysteine-rich intestinal protein 1 (CRIP1) in human breast cancer.
Mol Cancer. 12:282013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chervona Y and Costa M: Histone
modifications and cancer: Biomarkers of prognosis? Am J Cancer Res.
2:589–597. 2012.PubMed/NCBI
|
|
52
|
Kurdistani SK: Histone modifications in
cancer biology and prognosis. Prog Drug Res. 67:91–106.
2011.PubMed/NCBI
|
|
53
|
Chen S and Sang N: Histone deacetylase
inhibitors: The epigenetic therapeutics that repress
hypoxia-inducible factors. J Biomed Biotechnol. 2011:1979462011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Crea F, Clermont PL, Mai A and Helgason
CD: Histone modifications, stem cells and prostate cancer. Curr
Pharm Des. 20:1687–1697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cohen I, Poręba E, Kamieniarz K and
Schneider R: Histone modifiers in cancer: Friends or foes? Genes
Cancer. 2:631–647. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sawicka A and Seiser C: Histone H3
phosphorylation - a versatile chromatin modification for different
occasions. Biochimie. 94:2193–2201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nanni S, Priolo C, Grasselli A, D'Eletto
M, Merola R, Moretti F, Gallucci M, De Carli P, Sentinelli S,
Cianciulli AM, et al: Epithelial-restricted gene profile of primary
cultures from human prostate tumors: A molecular approach to
predict clinical behavior of prostate cancer. Mol Cancer Res.
4:79–92. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sedelnikova OA and Bonner WM: GammaH2AX in
cancer cells: A potential biomarker for cancer diagnostics,
prediction and recurrence. Cell Cycle. 5:2909–2913. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shaheen FS, Znojek P, Fisher A, Webster M,
Plummer R, Gaughan L, Smith GC, Leung HY, Curtin NJ and Robson CN:
Targeting the DNA double strand break repair machinery in prostate
cancer. PLoS One. 6:e203112011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Baptista T, Graça I, Sousa EJ, Oliveira
AI, Costa NR, Costa-Pinheiro P, Amado F, Henrique R and Jerónimo C:
Regulation of histone H2A.Z expression is mediated by sirtuin 1 in
prostate cancer. Oncotarget. 4:1673–1685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Monteiro FL, Baptista T, Amado F, Vitorino
R, Jerónimo C and Helguero LA: Expression and functionality of
histone H2A variants in cancer. Oncotarget. 5:3428–3443. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nowak M, Svensson MA, Carlsson J, Vogel W,
Kebschull M, Wernert N, Kristiansen G, Andrén O, Braun M and Perner
S: Prognostic significance of phospho-histone H3 in prostate
carcinoma. World J Urol. 32:703–707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Henrique R, Oliveira AI, Costa VL,
Baptista T, Martins AT, Morais A, Oliveira J and Jerónimo C:
Epigenetic regulation of MDR1 gene through post-translational
histone modifications in prostate cancer. BMC Genomics. 14:8982013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang YA and Yu J: EZH2, an epigenetic
driver of prostate cancer. Protein Cell. 4:331–341. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Schwartz YB and Pirrotta V: A new world of
Polycombs: Unexpected partnerships and emerging functions. Nat Rev
Genet. 14:853–864. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Piunti A and Shilatifard A: Epigenetic
balance of gene expression by Polycomb and COMPASS families.
Science. 352:aad97802016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang QT: Epigenetic regulation of cardiac
development and function by polycomb group and trithorax group
proteins. Dev Dyn. 241:1021–1033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Moison C, Senamaud-Beaufort C, Fourrière
L, Champion C, Ceccaldi A, Lacomme S, Daunay A, Tost J, Arimondo PB
and Guieysse-Peugeot AL: DNA methylation associated with polycomb
repression in retinoic acid receptor β silencing. FASEB J.
27:1468–1478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Simon JA and Lange CA: Roles of the EZH2
histone methyltransferase in cancer epigenetics. Mutat Res.
647:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zeng X, Chen S and Huang H:
Phosphorylation of EZH2 by CDK1 and CDK2: A possible regulatory
mechanism of transmission of the H3K27me3 epigenetic mark through
cell divisions. Cell Cycle. 10:579–583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Clermont PL, Crea F and Helgason CD:
Trithorax Genes in Prostate CancerAdvances in Prostate Cancer.
Hamilton G: InTech; Croatia: pp. 541–564. 2013
|
|
72
|
Daniunaite K, Jarmalaite S, Kalinauskaite
N, Petroska D, Laurinavicius A, Lazutka JR and Jankevicius F:
Prognostic value of RASSF1 promoter methylation in prostate cancer.
J Urol. 192:1849–1855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gurioli G, Salvi S, Martignano F, Foca F,
Gunelli R, Costantini M, Cicchetti G, De Giorgi U, Sbarba PD,
Calistri D, et al: Methylation pattern analysis in prostate cancer
tissue: Identification of biomarkers using an MS-MLPA approach. J
Transl Med. 14:2492016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ikromov O, Alkamal I, Magheli A, Ratert N,
Sendeski M, Miller K, Krause H and Kempkensteffen C: Functional
epigenetic analysis of prostate carcinoma: A role for seryl-tRNA
synthetase? J Biomark 2014. 3621642014.
|
|
75
|
Litovkin K, Van Eynde A, Joniau S, Lerut
E, Laenen A, Gevaert T, Gevaert O, Spahn M, Kneitz B, Gramme P, et
al: DNA methylation-guided prediction of clinical failure in
high-risk prostate cancer. PLoS One. 10:e01306512015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Moritz R, Ellinger J, Nuhn P, Haese A,
Müller SC, Graefen M, Schlomm T and Bastian PJ: DNA
hypermethylation as a predictor of PSA recurrence in patients with
low- and intermediate-grade prostate cancer. Anticancer Res.
33:5249–5254. 2013.PubMed/NCBI
|
|
77
|
Serenaite I, Daniunaite K, Jankevicius F,
Laurinavicius A, Petroska D, Lazutka JR and Jarmalaite S:
Heterogeneity of DNA methylation in multifocal prostate cancer.
Virchows Arch. 466:53–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tan HL, Haffner MC, Esopi DM, Vaghasia AM,
Giannico GA, Ross HM, Ghosh S, Hicks JL, Zheng Q, Sangoi AR, et al:
Prostate adenocarcinomas aberrantly expressing p63 are molecularly
distinct from usual-type prostatic adenocarcinomas. Mod Pathol.
28:446–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tsvetkova A, Todorova A, Todorov T,
Georgiev G, Drandarska I and Mitev V: Molecular and
clinicopathological aspects of prostate cancer in Bulgarian
probands. Pathol Oncol Res. 21:969–976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yoon HY, Kim SK, Kim YW, Kang HW, Lee SC,
Ryu KH, Shon HS, Kim WJ and Kim YJ: Combined hypermethylation of
APC and GSTP1 as a molecular marker for prostate cancer:
Quantitative pyrosequencing analysis. J Biomol Screen. 17:987–992.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yoon HY, Kim YW, Kang HW, Kim WT, Yun SJ,
Lee SC, Kim WJ and Kim YJ: DNA methylation of GSTP1 in human
prostate tissues: Pyrosequencing analysis. Korean J Urol.
53:200–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang W, Jiao H, Zhang X, Zhao R, Wang F,
He W, Zong H, Fan Q and Wang L: Correlation between the expression
of DNMT1, and GSTP1 and APC, and the methylation status of GSTP1
and APC in association with their clinical significance in prostate
cancer. Mol Med Rep. 12:141–146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Bastian PJ, Ellinger J, Heukamp LC, Kahl
P, Müller SC and von Rücker A: Prognostic value of CpG island
hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in
patients undergoing radical prostatectomy. Eur Urol. 51:665–674;
discussion 674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Müller A and Florek M:
5-Azacytidine/Azacitidine. Recent Results Cancer Res. 184:159–170.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yoon HY, Kim YW, Kang HW, Kim WT, Yun SJ,
Lee SC, Kim WJ and Kim YJ: Pyrosequencing analysis of APC
methylation level in human prostate tissues: A molecular marker for
prostate cancer. Korean J Urol. 54:194–198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yaqinuddin A, Qureshi SA, Pervez S, Bashir
MU, Nazir R and Abbas F: Frequent DNA hypermethylation at the
RASSF1A and APC gene loci in prostate cancer patients of Pakistani
Origin. ISRN Urol. 2013:6272492013.PubMed/NCBI
|
|
87
|
Olkhov-Mitsel E, Zdravic D, Kron K, van
der Kwast T, Fleshner N and Bapat B: Novel multiplex MethyLight
protocol for detection of DNA methylation in patient tissues and
bodily fluids. Sci Rep. 4:44322014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Pakneshan P, Szyf M and Rabbani SA:
Hypomethylation of urokinase (uPA) promoter in breast and prostate
cancer: Prognostic and therapeutic implications. Curr Cancer Drug
Targets. 5:471–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hagelgans A, Menschikowski M, Fuessel S,
Nacke B, Arneth BM, Wirth MP and Siegert G: Deregulated expression
of urokinase and its inhibitor type 1 in prostate cancer cells:
Role of epigenetic mechanisms. Exp Mol Pathol. 94:458–465. 2013.
View Article : Google Scholar : PubMed/NCBI
|