Introduction
PYROXD2, also known as YueF, (GenBank accession no.
BC006131), was initially identified as a novel hepatitis B virus
X-interacting protein (HBx) in studies conducted using a yeast
two-hybrid screening system (1,2). As a
putative tumor-suppressor protein, the overexpression of PYROXD2
can cause cell-cycle arrest in the G1 phase, induce cell apoptosis,
enhance the expression of p53 and p21WAF1/Cip1, decrease cyclin D1
and pRb expression and suppress the growth of hepatocellular
carcinoma (HCC) tumors in nude mice in vivo (3). PYROXD2 is highly expressed in the
cytoplasm of normal cells and tissues but is expressed at lower
levels in corresponding cancer cells, including liver, lung and
renal cell carcinoma and bladder cancer cells (1,2). The
biological functions of PYROXD2 and the mechanism which regulates
its expression remain largely unknown.
Myeloid zinc finger 1 (MZF1) is a member of the
SCAN-zinc finger (SCAN-ZF) family of transcription factors, and has
finger-like molecular structures that bind in a sequence-specific
manner into the groove of the DNA (4). MZF1 has been implicated in
tumorigenicity and it is thought to mediate the migration and
invasion of cancer cells by suppressing the activity of certain
gene promoter regions in vivo and in vitro (5–8).
Moreover, higher levels of MZF1 RNA were revealed in a series of
human cancer tissue than in normal tissue (5). MZF1 binds with the proteins found in
promyelocytic leukemia nuclear bodies (9). Promyelocytic leukemia nuclear bodies
strongly influence gene transcription activity and chromosomal
structure through their interaction with other factors and their
formation is dependent on the oligomerization of promyelocytic
leukemia proteins (10,11). The MZF1 protein is a
promoter/enhancer binding-type transcription factor, which
functions both as a trans-activator and a
trans-repressor. This observation revealed that the relative
oncogenic activity of MZF1 is determined by the aggregated effects
produced by the increase and decrease in gene expression (12), phosphorylation modifications,
SUMOylation modifications, and co-activating and co-repressing
molecules (5). The MZF1 protein
must become phosphorylated in order to respond to the stimulating
effects of transforming growth factor-β (TGF-β) (13), which is a growth factor known to be
important for facilitating the migration and invasion of cancer
cells and the development of the epithelial-mesenchymal transition
phenotype (14). SUMOylation of
transcription factors usually requires the participation of
co-repressors and may thus mediate certain suppressive processes
orchestrated by MZF1 during cellular differentiation and oncolytic
processes (15).
In the present study, we examined the expression
levels of both PYROXD2 and MZF1 using RT-PCR and western blot
analysis. We found increased levels of MZF1 mRNA and protein
expression and decreased levels of PYROXD2 mRNA and protein
expression in cancer cell lines and HCC tissues compared to the
expression levels in normal cell lines and liver tissue. We also
sought to identify the cis-elements and transcription
factors which activate PYROXD2 transcription in liver cancer
cells. To accomplish this goal, we performed a deletion analysis of
the PYROXD2 gene promoter region, followed by a mutant
analysis of that region to identify transcription factors that may
regulate PYROXD2 transcription. We then evaluated the
influence of MZF1 on PYROXD2 protein expression. Our results
revealed that MZF1 is a transcription factor crucial in the
regulation of PYROXD2 gene expression. Moreover, an
MZF1 gene binding site (TGGGGA) located in the −320/−312
region was significant for the functioning of the PYROXD2
promoter.
Materials and methods
Ethics statement and human tissue
preparation
The experiments involving humans were approved by
the Ethics Committee, and each study participant provided a signed
written informed consent document. All the tissue samples were
obtained from the Department of Surgery, Zhejiang Provincial
Peoples Hospital, Hangzhou, China.
Twelve samples of live human HCC tissues and 12
samples of corresponding adjacent normal liver tissues were
obtained from 12 HCC patients (2 females and 10 males) and examined
by a pathologist. All 12 HCC samples displayed a distinct cellular
subtype and all of the adjacent tissue samples appeared to be
normal and did not have fibrosis or other non-neoplastic changes.
Four tumors were at stage II and eight were at stage III. All
tissue samples were immediately dissected into several sections
(~100 mg/section), washed with normal saline, frozen in liquid
nitrogen and stored at −80°C.
Cell culture and transfection
Liver carcinoma cell lines HepG2, L02, Sk-hep1 and
293T were obtained from China Center for Type Culture Collection,
Wuhan, China, maintained in our lab and cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) (Gibco, Waltham, MA, USA) at 37°C in a 5%
CO2 atmosphere. Transfections were performed using
Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) according to the
manufacturer's instructions.
Reporter constructs and expression
vectors
Polymerase chain reaction (PCR) was used to amplify
the full length of the PYROXD2 promoter (−1998/−1) present
in the genomic DNA of L02 cells. Subsequently a directional PCR
cloning strategy was employed to clone the amplified promoter
region into pGL3 basic vectors (luciferase reporter plasmids;
Promega, Madison, WI, USA) at locations between the KpnI and
the XholI restriction enzyme sites. The luciferase reporter
plasmids were designated as (−1998/−1)-PYROXD2 promoter vectors.
Based on the selection made for the starting and ending nucleotide
base in the PYROXD2 promoter sequence, the serial
PYROXD2 promoter deletion mutants were designated as
(−1800/−1)-PYROXD2 promoter vector,
(−1600/−1)-PYROXD2 promoter vector,
(−1200/−1)-PYROXD2 promoter vector,
(−1000/−1)-PYROXD2 promoter vector, (−800/−1)-PYROXD2
promoter vector, (−600/−1)-PYROXD2 promoter vector,
(−400/−1)-PYROXD2 promoter vector, (−200/−1)-PYROXD2
promoter vector, (−1998/−200)-PYROXD2 promoter vector,
(−1998/−400)-PYROXD2 promoter vector, (−1998/−600)-PYROXD2
promoter vector, (−1998/−800)-PYROXD2 promoter vector,
(−1998/−1000)-PYROXD2 promoter vector and
(−1998/−1200)-PYROXD2 promoter vector, respectively. Two
siRNA sequences (GATCCGTACACAAGGGGACCATTC
ATTCTTCAAGAGAGAATGAATGGTCCCCTTGTGTATT TTTTACGCGTG and
GATCCGGCAGGTCCAGGTAGT GTAATTCAAGAGATTACACTACCTGGACCTGCTTTTT
TACGCGTA) were cloned into pLVX-U6 between the BamHI and the
EcoRI restriction enzyme sites with the purpose of silencing
the MZF1 protein expression. A control sequence
(GATCCGGCAACCTATGGGTGGGTAATTTTCAAGAG
AAATTACCCACCCATAGGTTGCTTTTTTACGCGTA) was cloned into the same
vector at the same restriction enzyme site. The MZF1 gene
was cloned into N-p3xflag-CMV (then designated as MZF1-Flag) to
force the overexpression of the MZF1 protein. All deletion and
mutant constructs were checked using DNA sequencing methods prior
to being used in any experiments.
Site-directed mutagenesis
Constructs bearing the mutant promoter variants of
PYROXD2 were generated by PCR, with the (−1998/−1)-PYROXD2
promoter vector as a template. Potential transcription factor
binding sites were identified using TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and the
results are listed in Table I.
Site-directed mutagenesis was performed with a KOD-Plus-Mutagenesis
kit (Toyobo, Osaka, Japan) according to the manufacturer's
instructions. The mutant primers used for site-directed mutagenesis
(information provided upon request) were designed and produced by
Generay Biotech Co., Ltd. (Shanghai, China). All mutants were
verified by sequencing.
 | Table I.TFs in each group whose binding site
was mutated in Fig. 3B are
listed. |
Table I.
TFs in each group whose binding site
was mutated in Fig. 3B are
listed.
| Group no. | TFs | Group no. | TFs | Group no. | TFs |
|---|
| 1 | MZF1 | 14 | GATA-2 | 29 | MZF1 |
|
| GATA-1 |
| MZF1 |
| STRE |
| 2 | Dfd |
| STRE | 30 | SRY |
|
| AML-1a |
| deltaE |
| CdxA |
|
| RORalp | 15 | Skn-1 |
| Lyf-1 |
|
| CdxA |
| CRE-BP | 31 | AML-1a |
|
| Sox-5 | 16 | cap | 32 | CRE-BP |
| 3 | HSF |
| C/EBPb |
| GATA-1 |
|
| HSF | 17 | d1 |
| CREB |
|
| HSF |
| CdxA | 33 | GATA-1 |
| 4 | cap | 18 | CF1 |
| CRE-BP |
|
| GATA-X |
| ADR1 |
| GATA-2 |
|
| NIT2 |
| SRY | 34 | NF-E2 |
| 5 | Sp1 | 19 | GATA-1 |
| p300 |
|
| ADR1 |
| NIT2 | 35 | Nkx-2 |
| 6 | ADR1 |
| BR-C Z |
| GATA-2 |
|
| cap |
| SRY | 36 | AP-1 |
| 7 | GATA-3 |
| cap | 37 | MZF1 |
| 8 | MATa1 | 20 | cap | 38 | E2F |
| 9 | ADR1 |
| c-Ets |
| C/EBPb |
|
| HSF |
| AP-1 |
| Ik-2 |
|
| c-Ets- |
| GCM | 39 | deltaE |
|
| HSF | 21 | c-Myb |
| TATA |
| 10 | p300 | 22 | NF-Y |
| MZF1 |
|
| cap |
| CREB | 40 | AML-1a |
| 11 | GATA-2 |
| GATA-2 | 41 | GATA-2 |
|
| AhR/Ar |
| GATA-2 |
| GATA-1 |
|
| Ttk 69 | 23 | Ttk 69 |
| GATA-3 |
| 12 | cap |
| dl |
| CdxA |
|
| E2F |
| Hb |
| c-Ets- |
|
| C/EBPa |
| Dfd |
| MZF1 |
|
| cap | 24 | STRE | 42 | CdxA |
| 13 | E2F |
| Nkx-2 | 43 | MZF1 |
|
| ADR1 | 25 | ADR1 | 44 | USF |
|
| SP1 |
| CdxA | 45 | GATA-1 |
|
| ADR1 | 26 | RORalp | 46 | p300 |
|
| MZF1 |
| MZF1 |
|
|
|
| HSF | 27 | NF-1 |
|
|
| 14 | USF |
| E2F |
|
|
|
| MZF1 | 28 | c-Myb |
|
|
|
| HSF2 |
| BR-C Z |
|
|
|
| GATA-1 |
| Dfd |
|
|
Dual-luciferase reporter gene
assay
Cells were seeded into 96-well plates and cultured
for 12 h, after which they were transfected with luciferase
reporter plasmids which had the selected serial PYROXD2
promoter. Subsequently, each sample was co-transfected with 20 ng
of Renilla luciferase control vector pGL4.70 (Promega) to
monitor the transfection efficiency. The pGL3 vectors were used as
controls. The luciferase activity was assessed at 24 h
post-transfection using a Dual-Luciferase Reporter Assay system
(Promega) according to the manufacturer's instructions. The assay
results were assessed with a Varioskan Flash Spectra Scanning
Multimode Reader (Thermo Fisher 3001; Thermo Fisher Scientific,
Waltham, MA, USA).
DNA binding assay
A DNA binding assay was used to detect the
interactions between the MZF1 and the putative promoter core
binding DNA sequence (AGGGGA, −320/−312). The biotinylated positive
DNA sequence was biotin-TCTCC TCCCCTGTGCATCTACCTTC-3′. The putative
positive DNA duplexes, MZF1 binding duplexes, were created by
annealing biotin-TCTCCTCCCCTGTGCATCTACCTTC-3′ and
5′-GAAGGTAGATGCACAGGGGAGGAGA-3′. The control DNA duplexes were
produced by annealing biotin-TCTCCTCCCCTGTGCATCTACCTTC-3′ and
5′-GAAGGTAGATGCACAGTAGAGGAGA-3′. All oligonucleotides were coupled
to M-280 Streptavidin Dynabeads (Miltenyi Biotec, Bergisch
Gladbach, Germany), according to the manufacturer's instructions. A
Nuclear and Cytoplasmic Protein Extraction kit (Beyotime
Biotechnology, Jiangsu, China) was used to extract total nuclear
proteins from the L02 cells, according to the manufacturer's
instructions. The DNA binding assays were performed as described by
Plotz et al (16) and the
MZF1 protein antibody (Abcam, Cambridge, UK) was used to
immunoprecipitate the protein-DNA complex.
Real-time PCR
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen) and from liver tumor and normal tissue using
the PureLink® RNA Mini kit (Thermo Fischer Scientific)
according to the manufacturer's instructions. The extracted mRNA
was reverse-transcribed using a Transcriptor First Strand cDNA
Synthesis kit (Roche Diagnostics, Pleasanton, CA, USA). First
Universal SYBR-Green Master Reagent (Rox; Roche) and KOD FX enzymes
(Toyobo) were used to perform comparative Ct analyses with a CFX96™
Real-Time system (Bio-Rad, Hercules, CA, USA). The sense and
antisense primers used to detect PYROXD2 mRNA were:
5-AAGTGCTCCATTGGATCAGC-3 and 5-GAGGCATGGGCATAAGGTCA-3,
respectively. The sense and antisense primers used to detect MZF1
mRNA were: 5-GAAACTGAGCCTCCAACTCC-3 and 5-GGGTGGGTACAGACTCCTG-3,
respectively. The sense and antisense primers used to detect actin
mRNA were: 5-ATCAGCAAGCAGGAGTATGACGAGT-3 and
5-ATGCCAATCTCATCTTGTTTTCTGC-3, respectively.
Western blot analysis
The cells were lysed by incubation in a RIPA buffer
(Beyotime Biotechnology) and their total soluble proteins were
isolated by centrifugation. The soluble proteins were then
separated by electrophoresis on a 12% SDS gel and the individual
protein bands were transferred onto nitrocellulose membranes for
analysis using western blotting and standard antibody detection
procedures. The proteins were extracted from tissue samples using a
T-PER® Tissue Protein Extraction kit (Thermo Fisher
Scientific). The primary antibodies used for immunostaining were
anti-MZF1 (rabbit), anti-PYROXD2 (rabbit) and anti-GAPDH (mouse)
(all from Abcam Cambridge, MA, USA). The membranes were incubated
with the primary antibodies, washed with TBST buffer and then
incubated with anti-mouse IgG (H+L) conjugate (Dylight™ 800) or
anti-rabbit IgG (H+L) conjugate (Dylight™ 800) (both from Cell
Signaling Technology, Danvers, MA, USA), depending on which primary
antibody was used during the first incubation. The
LI-COR® Biosciences Odyssey® Infrared Imaging
system (LI-COR Biosciences, Lincoln, NE, USA) was used to detect
antibody binding and quantify the individual protein bands.
Statistical analysis
Each data point represents the mean ± SD obtained
from at least three independent experiments. The Student's t-test
was used to analyze differences between two independent groups; a
two-sided P-value <0.05 was considered to indicate a
statistically significant difference.
Results
PYROXD2 expression in liver tumor
tissue and liver cancer cell lines compared with its expression in
normal liver tissue and normal cell lines
Previous studies had found lower levels of PYROXD2
expression in several carcinoma tissues than in corresponding
normal tissues. This finding revealed that PYROXD2 plays an
important role in tumor suppression (3). In order to further assess the
different expression models of PYROXD2, we collected a sample of
hepatic carcinoma tissue and a sample of normal liver tissue from
each of the 12 liver carcinoma patients who underwent therapeutic
surgery. We then used reverse transcription-polymerase chain
reaction (RT-PCR) to determine the relative levels of PYROXD2 and
GAPDH mRNA expression and western blotting techniques in order to
determine the relative levels of PYROXD2 and GAPDH protein
expression in each tissue sample. We also examined these expression
levels in several liver cell lines.
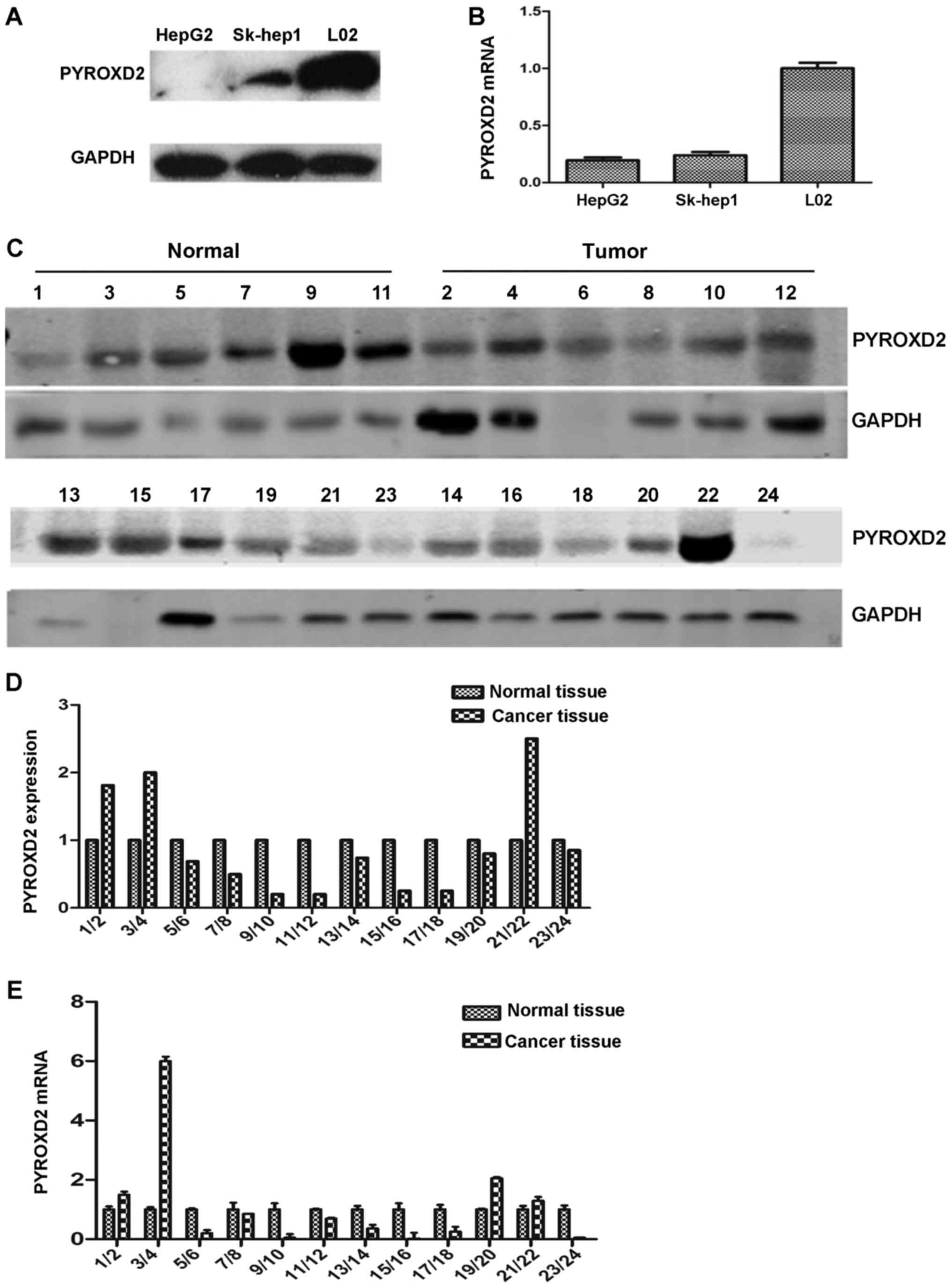
We obtained the same results observed in previous
studies (3). The PYROXD2 protein
was highly expressed in the normal hepatic cell line L02 but it was
barely detectable in the HepG2 and Sk-hep1 hepatoma cell lines
(Fig. 1A). The level of PYROXD2
mRNA expression in the three cell lines, as assessed by RT-PCR,
displayed the same pattern as the level of PYROXD2 protein
expression (Fig. 1B). The level of
PYROXD2 protein expression in normal tissue samples
(Levelnormal) and the corresponding HCC tissue samples
(LevelHCC) as detected by western blotting (Fig. 1C) were calculated based on the ratio
of the gray value for PYROXD2 and the gray value for GAPDH in the
same tissue sample
(gray-valuePYROXD2/gray-valueGAPDH). By
establishing the Levelnormal value as 1, the relative
expression levels of PYROXD2 protein in the corresponding HCC
tissue could be expressed as the ratio:
LevelHCC/Levelnormal. The results of these
calculations are illustrated in Fig.
1D. While 9 of the 12 patients had a level of PYROXD2 protein
expression in their HCC tissue sample that was 10 to 90% lower than
that in their corresponding sample of normal liver tissue, 3 of the
12 HCC patients had a higher level of PYROXD2 expression in their
HCC sample than in their normal tissue sample. Furthermore, similar
to the trend revealed by the PYROXD2 protein expression, the levels
of PYROXD2 mRNA in the samples of normal liver tissue were higher
than the levels in the corresponding samples of HCC tissue
(Fig. 1E). These results
demonstrated that although PYROXD2 was highly expressed in normal
liver tissue and cells, its expression was decreased in HCC tissue
and certain HCC cell types, a finding which revealed that decreased
PYROXD2 expression plays a role in hepatocarcinogenesis.
MZF1 expression in HCC tissue and
various liver cell lines
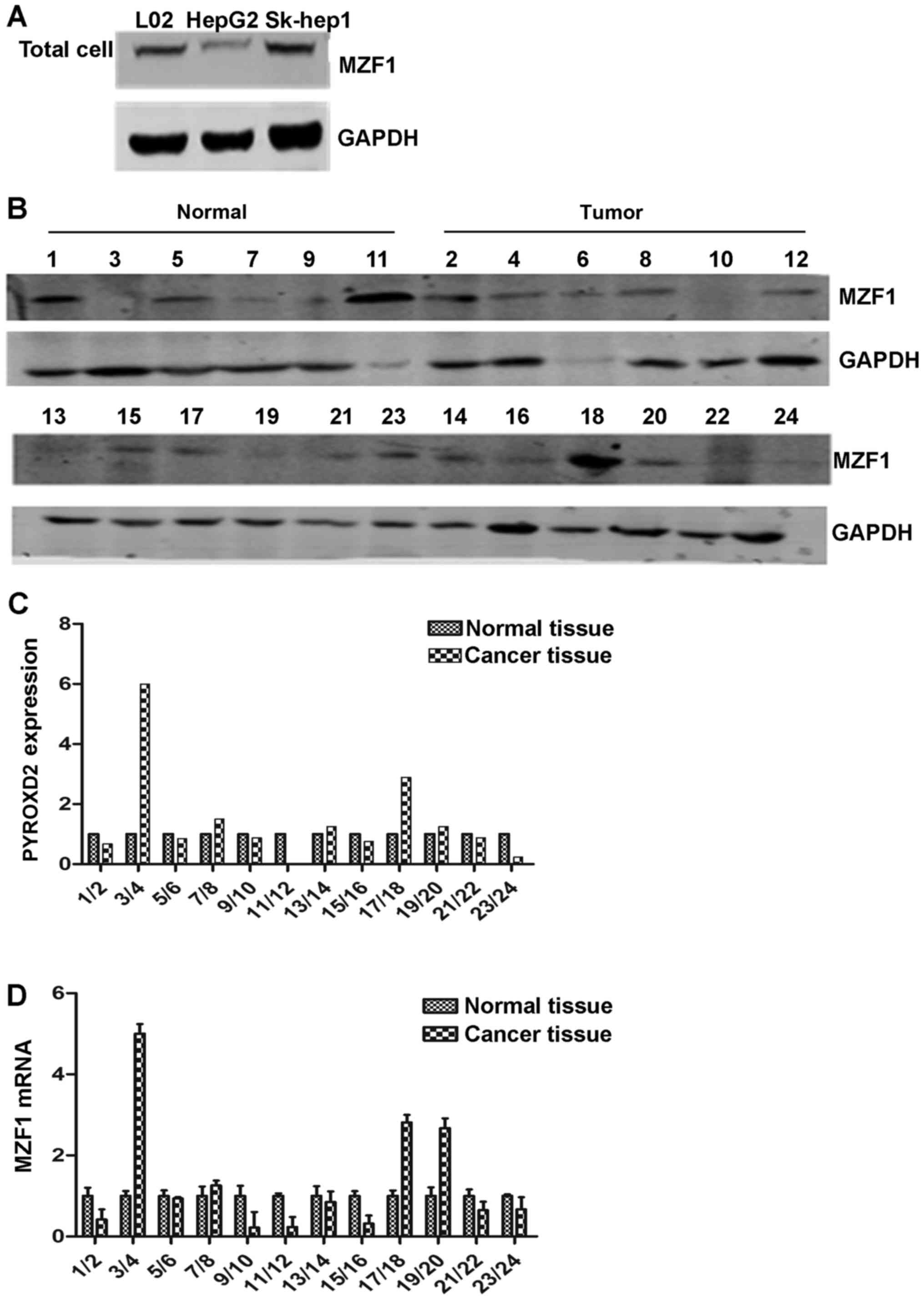
We examined the relative levels of MZF1 protein
expression in L02, HepG2 and Sk-hep1 cells using western blotting
(Fig. 1A). MZF1 which served as a
transcription factor was mainly detected as a component of cell
nuclear proteins and exhibited different expression in diverse
tissues and cell lines (5). The
same result was obtained by immunofluorescence analysis (data not
shown) and MZF1 also displayed different expression levels in
HepG2, L02 and Sk-hep1 cells (Fig.
2B), which indicated that MZF1 was less expressed in the HepG2
cells than in the L02 and Sk-hep1 cells as determined by western
blotting. We also examined the endogenous levels of the MZF1
protein and mRNA expression in the 12 resected HCC tissue samples
and the 12 corresponding normal tissue samples using western
blotting and RT-PCR, respectively. The levels of MZF1 protein and
mRNA expression were calculated using the same methods as those
used to calculate the levels of PYROXD2 protein and mRNA
expression; the results are shown in Fig. 2C-E. The tumors in 6 of the 12
patients had a significantly increased level of MZF1 expression
compared with the level in the corresponding normal liver sample;
however, the tumors in the other 6 patients had 10–90% lower levels
of MZF1 expression than those in the corresponding normal tissue
samples.
Sequence AGGGGA (−320/−312) in the
PYROXD2 promoter is the main element controlling PYROXD2
expression
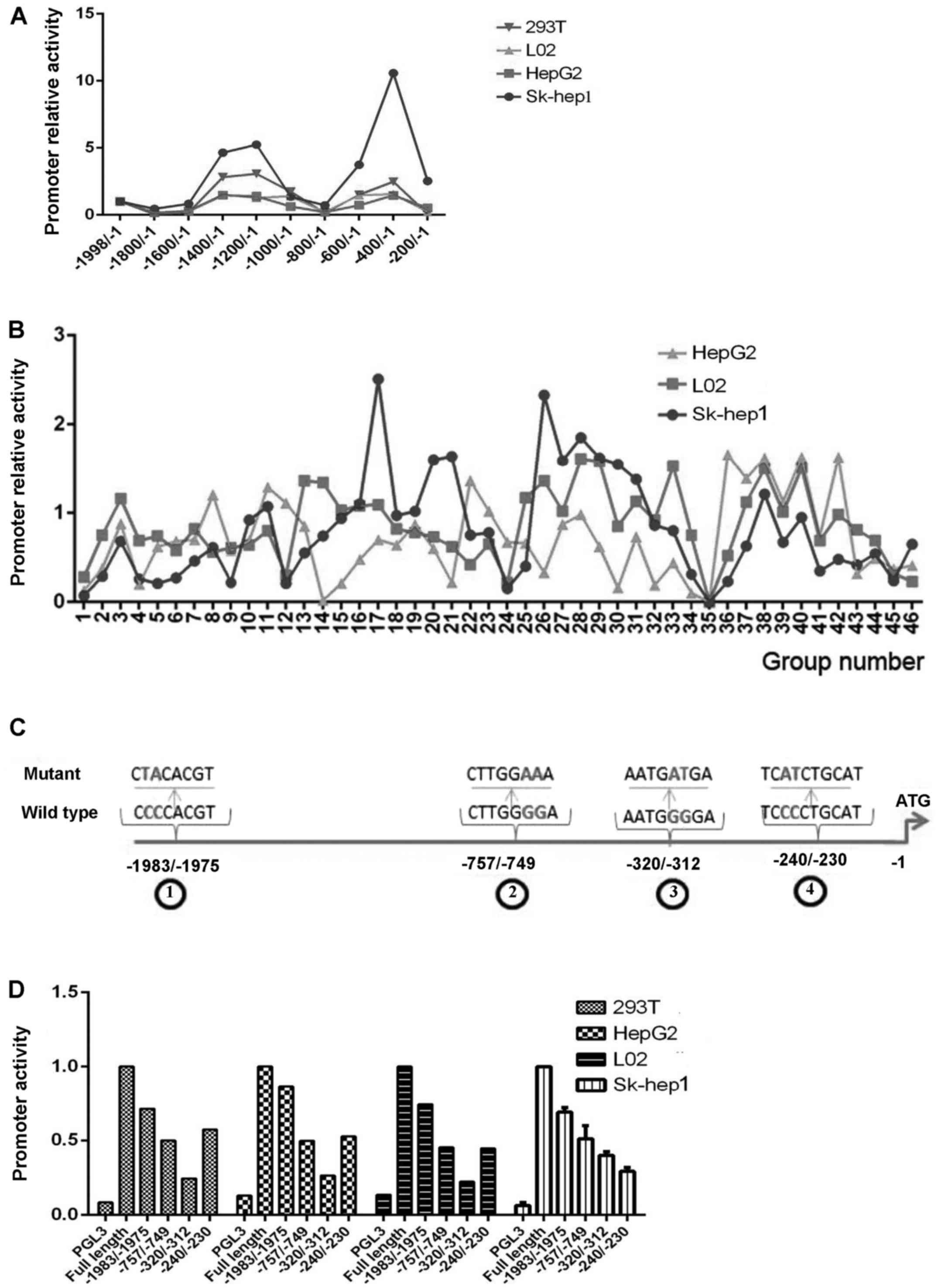
We cloned the 1999 base pair (bp) fragment
(−1998/−1) of the promoter region of the PYROXD2 gene
located upstream of the ATG transcription initiation codon of exon
1, with the purpose of analyzing the promoter and its regulatory
elements. A search for potential regulatory motifs which was
performed using the TFSEARCH identified the putative transcription
factor binding sites and is listed in Table I. Subsequently, the serial deletion
mutants were constructed using strategies and primers (relevant
information provided upon request). A nucleotide deletion in the
promoter regulatory region could potentially affect the binding of
transcription factors and alter the transcription rate of a gene.
Promoter activity was determined with a luciferase assay system and
the results were normalized by h-galactosidase activity. The
absorbance value of a blank control sample was subtracted from each
assay result. After the activity of the (−1998/−1)-PYROXD2 promoter
was defined as 1, the relative activities of other serial mutant
promoters were expressed as the ratio
Imutant/Ifulllength, in which I is the
intensity of an absorbance value, the superscript ‘mutant’
signifies a serial mutant promoter and the superscript ‘full
length’ signifies the (−1998/−1)-PYROXD2 promoter.
To analyze various characteristics of the PYROXD2
promoter and identify the region most commonly activated, we
constructed a series of 200 bp fragment deletions which ranged from
base −1998 to base −1. As revealed in Fig. 3A, deletion of bases −1998/−1801
produced only a slight decrease in promoter activity. The most
remarkable change in promoter activity occurred when the deletions
were produced between the −1600/−1201 and −800/−401 regions, in
which case the promoter exhibited increased activity. In contrast,
the (−1998/−1)-PYROXD2 promoter displayed decreased activity when
the deletions were produced in both the −1200/−801 and −400/−201
regions. It should be emphasized that the PYROXD2 promoter lost all
its activity when the deletions were produced in either the
−400/−201 or −200/−1 regions. Our results indicated that
cis-elements located in the −400/−1 region constitute the
core promoter responsible for basal transcription of the PYROXD2
gene. Next, we used site-directed mutagenesis to generate a series
of mutant reporters based on the (−1998/−1)-PYROXD2 promoter
vector, with the purpose of identifying critical
cis-elements in the promoter region. As revealed in Fig. 3B and Table I, the different mutants produced
different effects on the promoter activity in the three cell lines.
Notably, insertion of a mutation into the −320/−312 region produced
a complete loss of promoter activity in all three cell lines
(Fig. 3B and Table I). These results indicate that the
putative MZF1 binding site (TGGGGA) located in the −320/−312 region
of the PYROXD2 promoter may be crucial for the transcription
activity of the promoter.
A preliminary sequence analysis of the −1998/−1
domain revealed the presence of four cis-elements that may
bind with MZF1. Among them, the −1983/−1975 and −320/−312 regions
were non-overlapping, whereas the −757/−749 and −240/−230 regions
overlapped with other transcription factors. To assess how the four
MZF1 binding cis-elements of the PYROXD2 promoter may
affect PYROXD2 gene transcription, we constructed mutant
promoters in which only two nucleotides were substituted in the
core MZF1 binding sequence. As revealed in Fig. 3C, the substitutions did not affect
the binding ability of other overlapping transcriptional effectors.
When compared with the effects produced by other mutations, a
mutation in the −320/−312 region appeared to produce the largest
decrease in luciferase activity (Fig.
3D).
MZF1 is a key trans-acting factor
controlling PYROXD2 expression
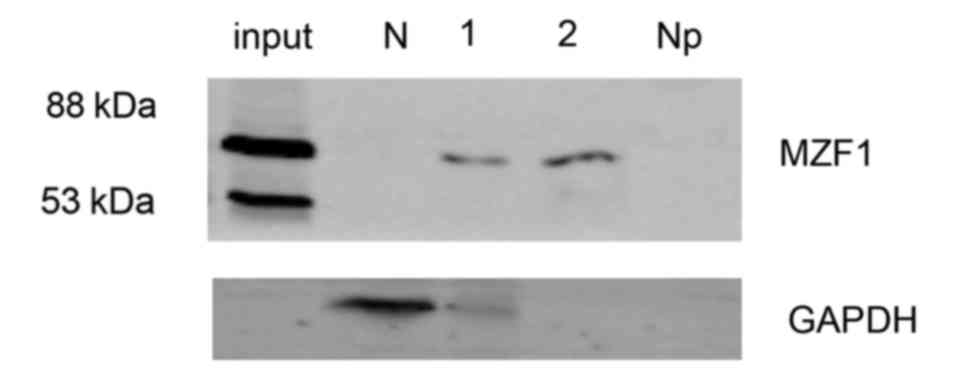
A separate set of experiments was conducted to
elucidate how the −320/−312 region of the PYROXD2 promoter
interacts with MZF1 and to determine whether endogenous MZF1 binds
to the −320/−312 region of the PYROXD2 promoter. To
accomplish these goals, we performed DNA binding assays using
primers that spanned the putative MZF1-binding site of the region
(Fig. 4). The specific sequence
within the −320/−312 region was precipitated from cell lysates by
the addition of the anti-MZF1 antibody but not by the addition of
the control IgG. Our data markedly indicated that MZF1 binds to the
TGGGGA domain in the proximal promoter of PYROXD2.
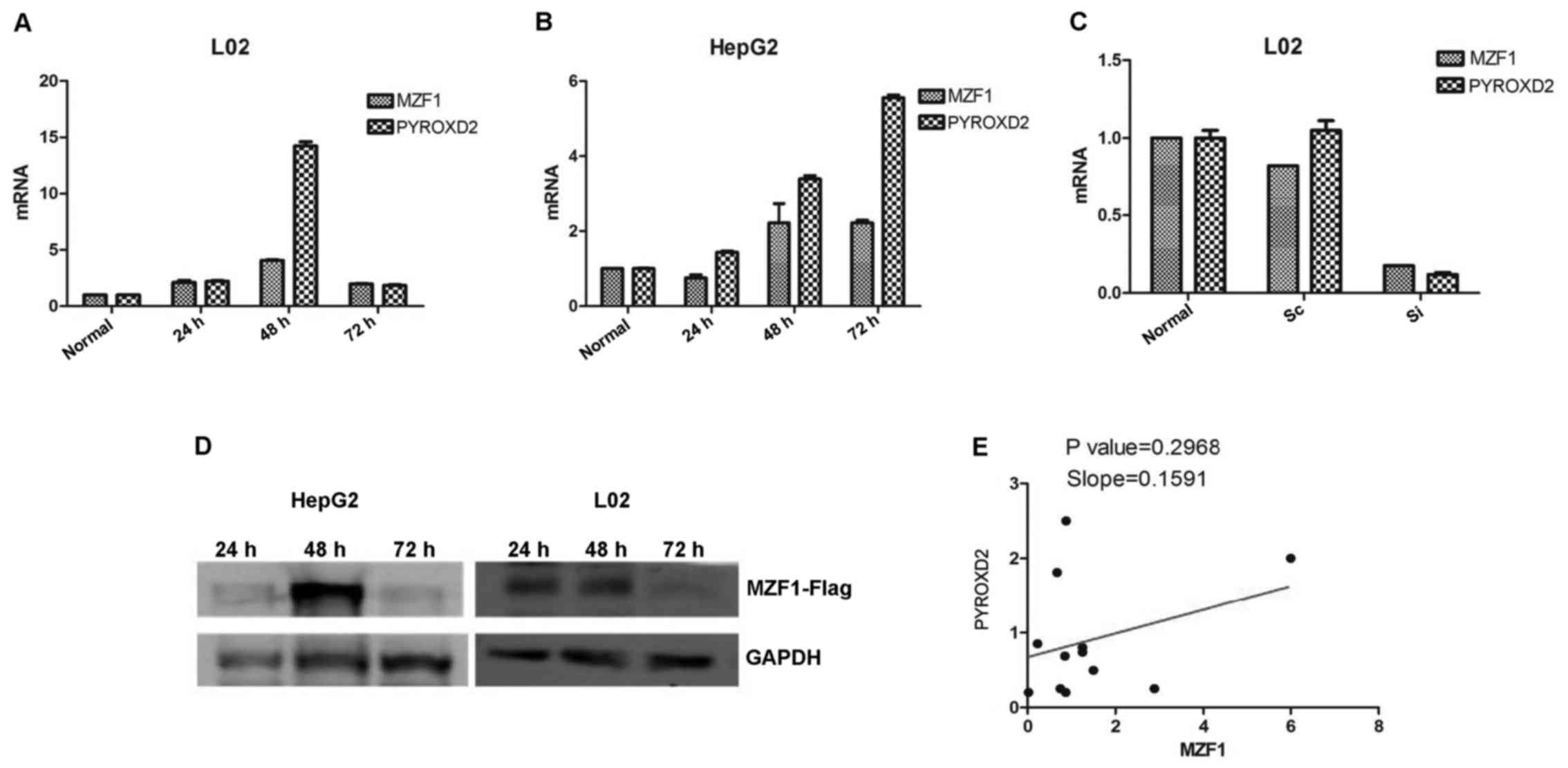
To further ascertain the involvement of MZF1 in
PYROXD2 transcription, we assessed the levels of endogenous
PYROXD2 expression in the HepG2 and L02 cells after transfecting
them with MZF1 expressing plasmids, or by silencing their
endogenous MZF1 expression with siRNA (Fig. 5). Increased endogenous PYROXD2
expression was observed in MZF1 overexpressing the HepG2 and L02
cells, and decreased endogenous PYROXD2 expression was observed in
the L02 cells transfected with the MZF1 siRNA (Fig. 5). These findings indicate that MZF1
functions as a key regulator of the PYROXD2
transcription.
Discussion
The tumor-suppressive activity of PYROXD2 and its
different expression levels in normal tissues and several
corresponding tumor tissues have been reported (1,2). In
the present study, we collected specimens of cancerous liver tissue
and adjacent normal liver tissue from 12 patients, and
quantitatively assessed the endogenous levels of MZF1 and PYROXD2
expression in those tissues by western blotting and RT-PCR. We
found that the PYROXD2 protein was highly expressed in normal liver
tissue and in a normal human liver cell line; however, its
expression was either absent or decreased in a large proportion of
HCC tissue and hepatocarcinoma cell lines, indicating that
decreasing PYROXD2 expression may be involved in
hepatocarcinogenesis.
The molecular mechanism which regulates
PYROXD2 transcription was not elucidated in previous
studies. We sought to examine various characteristics of the
PYROXD2 promoter and identify the most commonly activated
PYROXD2 promoter region in liver cells. To accomplish this
goal, we constructed a series of luciferase reporter plasmids that
contained 5′ and 3′-deletions in the PYROXD2 promoter, and
then used them to perform luciferase-based reporter assays in HepG2
and Sk-hep1 liver cancer cell lines as well as in two normal
control cell lines (L02 and 293T). The most remarkable change in
the promoter activity occurred when the deletions were produced in
both the −400/−199 and −200/−1 regions in which case, there was an
almost complete loss of PYROXD2 promoter activity. Our
site-directed mutagenesis studies conducted with three different
cell lines demonstrated that the putative MZF1 binding site
(TGGGGA) located in the −320/−312 region of the PYROXD2
promoter was largely responsible for the loss of PYROXD2 promoter
activity. The results of the DNA binding assays also indicated the
occurrence of interactions between MZF1 and cis-elements
located in the −317/−313 region of the PYROXD2 promoter.
Ectopic expression of MZF1 induced an increased expression of
PYROXD2; accordingly, silencing of MZF1 inhibited PYROXD2
expression in the same cell lines. These findings suggest that MZF1
is critical for the transcription activity of the PYROXD2
promoter and functions as a trans-activator in regulating
PYROXD2 expression. In addition these findings provided novel
insights into the mechanism underlying the tumorigenic effect of
PYROXD2.
Our results indicated that MZF1 was expressed at
higher levels in samples of human cancer tissue than in samples of
normal tissue, which were consistent with previous studies
(5). Based on the previous cellular
experiment results, we analyzed the activating function of MZF1 on
the PYROXD2 promoter in tissues by comparing the expression level
of MZF1 with the PYROXD2 expression level. Notably MZF1 gene
expression was not significantly correlated with PYROXD2 expression
in samples of resected tumor tissues. The protein expression in
human cells is regulated at several levels such as in transcription
factors and mRNA stability. Promoter transcription activities were
affected by the amount and type of the transcription factor family
members and their function antagonism associations. Additively, the
untranslated regions (UTR) and the AU-rich elements (ARE) render
mRNA unstable in cells and tissues leading to a gradual decrease in
protein production (17). Although
the present study revealed that the MZF1 gene expression was not
significantly correlated with the PYROXD2 expression in samples of
resected tumor tissues, we speculated that the main mechanisms were
relative to the diversity of the transcription factors involved in
the regulation of the PYROXD2 promoter activity and MZF1 is only
one of these factors. It should be pointed out that MZF1 is an
activating factor of the PYROXD2 promoter, and decreased PYROXD2
expression may contribute to cancer progression. Therefore, it is
important to understand the underlying mechanism that regulates the
expression level of PYROXD2 by other transcription factor members
and their interaction relationships.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Zhejiang province (LQ12C03003), the Natural
Science Foundation of China (project nos. 31260621 and 31160240)
and the Hangzhou Normal University supporting project (no.
PE13002004042).
References
|
1
|
Zhang JL, Zhao WG, Wu KL, Wang K, Zhang X,
Gu CF, Li Y, Zhu Y and Wu JG: Human hepatitis B virus X protein
promotes cell proliferation and inhibits cell apoptosis through
interacting with a serine protease Hepsin. Arch Virol. 150:721–741.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang J, Wu K, Zhang J, Si W, Zhu Y and Wu
J: Putative tumor suppressor YueF affects the functions of
hepatitis B virus X protein in hepatoma cell apoptosis and p53
expression. Biotechnol Lett. 30:235–242. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang HW, Peng JP and Zhang J: YueF
overexpression inhibits cell proliferation partly through p21
upregulation in renal cell carcinoma. Int J Mol Sci. 12:2477–2487.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deng Y, Wang J, Wang G, Jin Y, Luo X, Xia
X, Gong J and Hu J: p55PIK transcriptionally activated by MZF1
promotes colorectal cancer cell proliferation. Biomed Res Int.
2013:8681312013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eguchi T, Prince T, Wegiel B and
Calderwood SK: Role and regulation of myeloid zinc finger protein 1
in cancer. J Cell Biochem. 116:2146–2154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsieh YH, Wu TT, Huang CY, Hsieh YS and
Liu JY: Suppression of tumorigenicity of human hepatocellular
carcinoma cells by antisense oligonucleotide MZF-1. Chin J Physiol.
50:9–15. 2007.PubMed/NCBI
|
|
7
|
Asiedu MK, Beauchamp-Perez FD, Ingle JN,
Behrens MD, Radisky DC and Knutson KL: AXL induces
epithelial-to-mesenchymal transition and regulates the function of
breast cancer stem cells. Oncogene. 33:1316–1324. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mudduluru G, Vajkoczy P and Allgayer H:
Myeloid zinc finger 1 induces migration, invasion, and in vivo
metastasis through Axl gene expression in solid cancer. Mol Cancer
Res. 8:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jensen K, Shiels C and Freemont PS: PML
protein isoforms and the RBCC/TRIM motif. Oncogene. 20:7223–7233.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dellaire G and Bazett-Jones DP: PML
nuclear bodies: Dynamic sensors of DNA damage and cellular stress.
BioEssays. 26:963–977. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Damme E, Laukens K, Dang TH and Van
Ostade X: A manually curated network of the PML nuclear body
interactome reveals an important role for PML-NBs in SUMOylation
dynamics. Int J Biol Sci. 6:51–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edelstein LC and Collins T: The SCAN
domain family of zinc finger transcription factors. Gene. 359:1–17.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Driver J, Weber CE, Callaci JJ, Kothari
AN, Zapf MA, Roper PM, Borys D, Franzen CA, Gupta GN, Wai PY, et
al: Alcohol inhibits osteopontin-dependent transforming growth
factor-β1 expression in human mesenchymal stem cells. J Biol Chem.
290:9959–9973. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Massagué J: A very private TGF-beta
receptor embrace. Mol Cell. 29:149–150. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stielow B, Krüger I, Diezko R, Finkernagel
F, Gillemans N, Kong-a-San J, Philipsen S and Suske G: Epigenetic
silencing of spermatocyte-specific and neuronal genes by SUMO
modification of the transcription factor Sp3. PLoS Genet.
6:e10012032010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Plotz G, Raedle J, Brieger A, Trojan J and
Zeuzem S: hMutSalpha forms an ATP-dependent complex with hMutLalpha
and hMutLbeta on DNA. Nucleic Acids Res. 30:711–718. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sahoo A and Im SH: Molecular mechanisms
governing IL-24 gene expression. Immune Netw. 12:1–7. 2012.
View Article : Google Scholar : PubMed/NCBI
|