Introduction
Acute lung injury (ALI) is a complicated pathogenic
process with high mortality due to sepsis, pulmonary infection and
shock (1). Lipopolysaccharide
(LPS), a component found in the cell membrane of gram-negative
bacteria, induces the production of numerous inflammatory cytokines
and chemokines and has been shown as a major cause of ALI (2). Intraperitoneal administration of LPS
has been widely used to model the pathogenesis of ALI and has
yielded reproducible results.
Short palate, lung and nasal epithelium clone 1
(SPLUNC1) is a protein mainly secreted by large airway epithelia
and submucosal glands (3,4). Accordingly, we can find this protein
in the human oral cavity (5), nasal
secretions (6), tracheal aspirates
(7) as well as in rat lung
(8), suggesting that SPLUNC1 is a
highly abundant secretory product of the mammalian respiratory
system. Recent studies by our group demonstrated that SPLUNC1
inhibits the overall process of Epstein Barr virus replication and
is associated with nasopharyngeal carcinoma prognosis (9–12).
Although SPLUNC1-deficient mice do not develop spontaneous lung
disease without a bacterial insult, they do exhibit more severe
impaired intrapulmonary responses than wild-type controls when
infected by respiratory pathogens such as Mycoplasma
pneumoniae (13),
Pseudomonas aeruginosa (14)
and Klebsiella pneumoniae (15). In addition, loss of SPLUNC1
expression predisposes mice to develop otitis media (16). Although SPLUNC1 protein has been
predicted to exert host defense and has been proposed as a valuable
marker in non-small cell lung cancer (NSCLC) (17), the related immune cells regulated by
SPLUNC1 remain elusive.
ALI triggers innate immune responses and is
associated with the induction and promotion of immune-suppressive
mechanisms, such as the accumulation of myeloid-derived suppressor
cells (MDSCs). MDSCs have been described as a heterogenic
population of immature myeloid cells that consist of myeloid
progenitors and precursors of macrophages, granulocytes and
dendritic cells (DCs) and that, mainly by multiple mechanisms,
suppress T cell and NK cell functions (18,19).
In healthy individuals, immature myeloid cells (IMCs) can be
generated in bone marrow and differentiate into mature
granulocytes, macrophages or DCs. In pathological conditions such
as cancer, sepsis, various infectious diseases, trauma, certain
autoimmune disorders or in bone marrow transplantation, a partial
block in the differentiation of IMCs into mature myeloid cells
results in an expansion of this population. In mice, MDSCs are
identified as cells that co-express the myeloid lineage
differentiation antigen Gr-1 and CD11b (20), which make up only a small proportion
(2–4%) of spleen cells and are absent from the lymph nodes
(18). Recently, several
researchers have demonstrated that the accumulation of MDSCs plays
a vital role in LPS-induced lung injury (21). However, the molecules that are
involved in the generation and expansion of MDSCs during acute
inflammation are not completely understood.
In the present study, we sought to investigate the
effects of SPLUNC1 on lung injury using SPLUNC1-knockout
(SPLUNC1−/−) mice, and to clarify the function of
SPLUNC1- mediated recruitment of CD11b+Gr-1+
MDSCs in the spleen of LPS-induced ALI mice.
Materials and methods
Animals
To obtain SPLUNC1-knockout mice that allowed
conditional and global disruption of the SPLUNC1 gene, we used the
Cre/loxP and flp/FRT recombination systems to target
exons 3 of SPLUNC1. A single loxp site was inserted before exon 3
and before exon 4, and an frt-flanked PGK neo cassette was inserted
between exons 3 and 4 to serve as a positive selectable marker. The
targeting construct was electroporated into embryonic stem (ES)
cells and the germline SPLUNC1loxp-neo chimeras were
obtained by homologous recombination. Then, the FRT-flanked PGK neo
cassette was deleted by crossing female SPLUNC1loxp-neo
mice with transgenic Flp male mice to obtain the homozygous mice
for the floxed SPLUNC1 allele (SPLUNC1flox/flox). To
generate SPLUNC1−/− mice, the
SPLUNC1flox/flox mice were mated with EIIα-Cre mice
(purchased from the Jackson Laboratory, Bar Harbor, ME, USA).
To produce animals for experiments, heterozygous
mice were crossed to generate wild-type littermate controls. All
mice used in the present study were male, 8–12 weeks old, and
housed under specific pathogen-free conditions. The present study
was performed in strict accordance with the recommendations of the
Guide for the Care and Use of Laboratory Animals (National
Institutes of Health). Experimental protocols were approved by the
Animal Ethics Committee of Central South University.
Mouse model of LPS-induced ALI
Mice were randomly divided into four groups: WT
control, KO control, WT-LPS, and KO-LPS. The LPS group was
intraperitoneally injected with a single dose of 5 mg/kg body
weight of LPS (Sigma, Carlsbad, CA, USA) which was dissolved in
saline at a concentration of 1 mg/ml. Meanwhile, the mice in the
control group were intraperitoneally injected with saline alone.
The mice were euthanized at 24, 48, 72 and 96 h after LPS or saline
administration.
Western blot analysis
The proteins were extracted from mouse lung tissues
and then separated by 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to a nitrocellulose
membrane. The membrane was blocked using 5% non-fat milk in
Tris-buffered saline containing 0.1% Tween-20 and then incubated
with primary antibodies: SPLUNC1 (R&D Systems, Minneapolis, MN,
USA) and GAPDH (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) at 4°C overnight. After washing three times with TBST, the
membranes were incubated with HRP-conjugated secondary antibody.
Bands were detected with an electrochemiluminescence detection
system.
Flow cytometric analysis
Single cell suspensions were prepared by
homogenization through a sterile stainless steel mesh from the
spleens. Erythrocytes were depleted using RBC lysing buffer
(Sigma), and splenocytes were washed with phosphate-buffered saline
(PBS). Then, the cells were stained with anti-CD11b and anti-Gr-1
antibodies (eBioscience, San Diego, CA, USA) according to the
manufacturer's protocol. Fluorescence was measured using a MoFlo™
XDP High-Performance Cell Sorter and data were analyzed by Summit
5.2 software (both from Beckman Coulter, Inc., Brea, CA, USA).
RNA extraction and real-time PCR
analysis
Reverse transcription reaction was performed using a
Fermentas Revert Aid First Strand cDNA Synthesis kit (Fermentas,
Burlington, ON, Canada), according to the manufacturer's
instructions. qRT-PCR was performed using an iQ5 Multicolor
Detection system (Bio-Rad, Hercules, CA, USA). The sequences of the
primers were as follows: β-actin forward, 5′-TTTCCAGCCTTCCTTCTT-3′
and reverse, 5′-GGTCTTTACGGATGTCAACG-3′; IL-6 forward,
5′-CTGATGCTGGTGACAACCAC-3′ and reverse, 5′-CAGAATTGCCATTGCACAAC-3′;
IL-8 forward, 5′-CGTCCCTGTGACACTCAAGA-3′ and reverse,
5′-GGAGCATCAGGATCCAAACA-3′; CCL-2 forward,
5′-ATGCAGTTAACGCCCCACTC-3′ and reverse, 5′-ACCCATTCCTTCTTGGGGTC-3′;
CCL-3 forward, 5′-GCAACCAAGTCTTCTCAGCG-3′ and reverse,
5′-TTGGACCCAGGTCTCTTTGG-3′; CCL-5 forward,
5′-ACCATATGGCTCGGACACCA-3′ and reverse, 5′-GCGGTTCCTTCGAGTGACA-3′;
CXCL1 forward, 5′-TGGCTGGGATTCACCTCAAG-3′ and reverse,
5′-CCGTTACTTGGGGACACCTT-3′; IL-1β forward,
5′-TGCCACCTTTTGACAGTGATG-3′ and reverse,
5′-AAGGTCCACGGGAAAGACAC-3′. The expression of mRNA was assessed by
evaluated threshold cycle (CT) values. The CT values were
normalized with the expression levels of β-actin and the relative
amount of mRNA specific to each of the target genes was calculated
using the 2−ΔΔCt method (22–25).
β-actin was used as an internal control.
Histology
Lungs of the differentially treated mice were fixed
with 4% paraformaldehyde, and embedded in paraffin. Subsequently,
the lung tissues were sliced into 4-µm sections, and then stained
with hematoxylin and eosin to observe the inflammatory response and
pathological changes using an Olympus BX51 microscope (Olympus,
Tokyo, Japan). The degree of histologic lung injury was assessed
based on the following criteria: neutrophil infiltration, alveolar
congestion, interstitial edema, necrosis and atelectasis. The
severity of injury was evaluated via a numerical rating system that
ranges from 0 to 4 (score): no injury, 0; injury to 25% of the
field, 1; injury to 50% of the field, 2; injury to 75% of the
field, 3; and diffuse injury, 4. The mean score was then analyzed
for comparison between groups.
Immunohistochemistry (IHC) and
evaluation of staining
IHC was carried out using the peroxidase
anti-peroxidase technique following a microwave antigen retrieval
procedure. Anti-CD11b (MAB1124; 1:300), anti-Gr-1 (MAB1037; 1:300)
(both from R&D Systems) was overlaid on the tissue sections and
incubated overnight at 4°C. Secondary antibody incubation (Santa
Cruz Biotechnology, Inc.) was performed at room temperature for 30
min. Color reaction was developed using 3,3′-diaminobenzidine
tetrachloride (DAB) chromogen solution. All slides were
counterstained with hematoxylin. Positive control slides were
included in every experiment in addition to the internal positive
controls. The specificity of the antibody was determined with
matched IgG isotype antibody as a negative control.
Immunostaining was evaluated by two investigators in
a blinded manner in an effort to provide a consensus on staining
patterns by light microscopy (Olympus). CD11b or Gr-1 staining was
assessed according to the methods described by Hara and Okayasu
(26) with minor modifications.
Each case was rated according to a score that added a scale of
intensity of staining to the area of staining. At least 10
high-power fields were randomly chosen, and >1,000 cells were
counted for each section. The intensity of staining was graded on
the following scale: 0, no staining; 1+, mild staining; 2+,
moderate staining; 3+, intense staining. The area of staining was
evaluated as follows: 0, no staining of cells in any microscopic
fields; 1+, <30% of tissue stained positive; 2+, between 30 and
60% stained positive; 3+, >60% stained positive. The minimum
score when summed (extension + intensity) was, therefore, 0, and
the maximum, 6. A combined staining score (extension + intensity)
of ≤2 was considered to be a negative staining (low staining); a
score between 3 and 4 was considered to be a moderate staining;
whereas a score between 5 and 6 was considered to be a strong
staining. An optimal cut-off level was identified as follows: a
staining index score of 0–2 was used to define tumors with negative
expression and 3–7 indicated positive expression of these two
proteins. Agreement between the two evaluators was 95%, and all
scoring discrepancies were resolved by discussion between the two
evaluators.
Statistical analysis
The data obtained in the present study are expressed
as the mean values ± standard deviation (SD) from at least three
independent experiments using triplicate samples for the individual
treatments. Statistical analyses were performed using two-way ANOVA
or Fisher's exact test (Prism 6; GraphPad Software, San Diego, CA,
USA). A P-value of <0.05 was considered to indicate a
statistically significant difference.
Results
Generation of SPLUNC1-knockout
mice
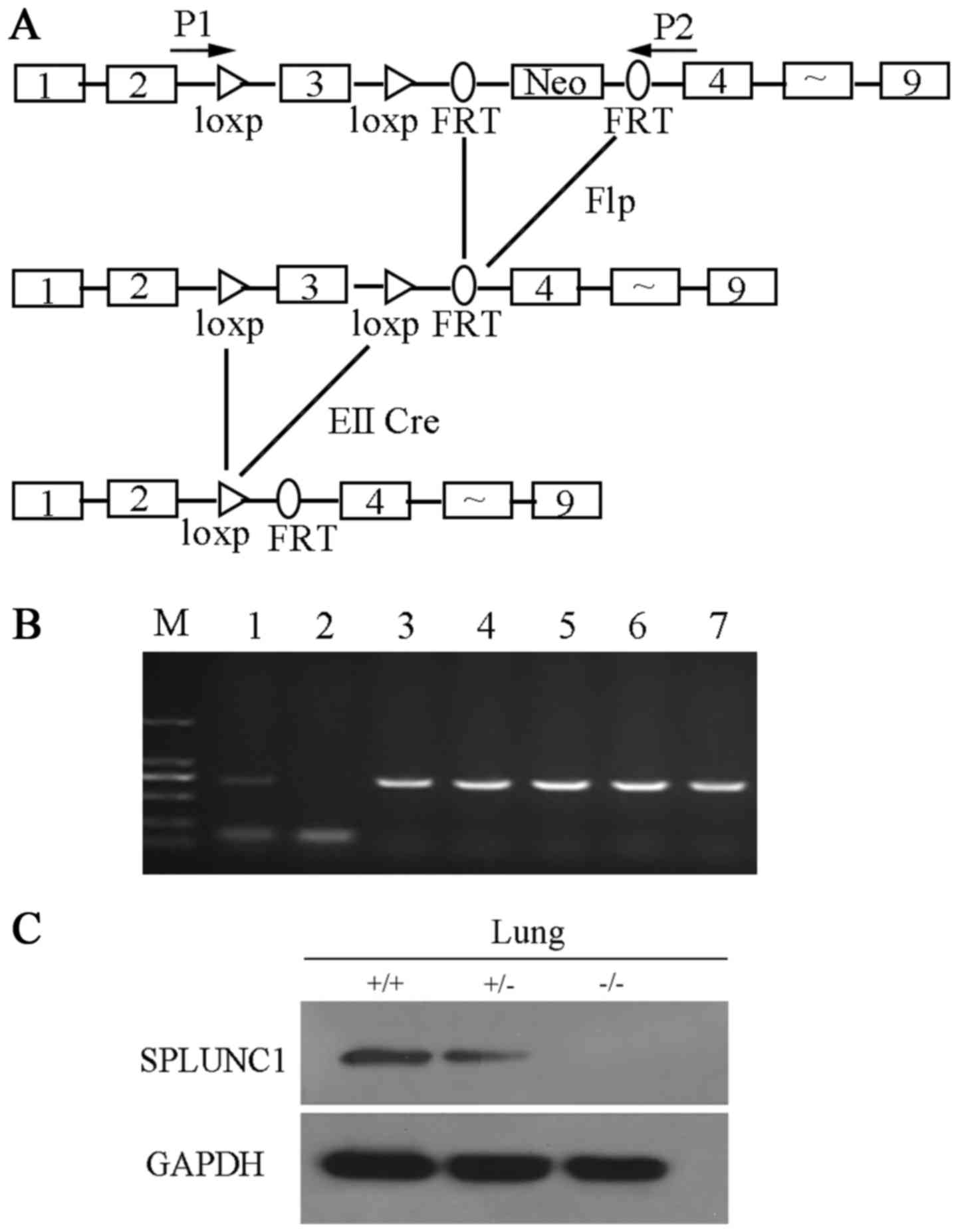
To identify the role of SPLUNC1 in vivo, we
generated SPLUNC1-knockout mice. In this procedure, Cre/loxp
and Flp/FRT recombination systems were used to target exon 3
of the mouse SPLUNC1 gene (Fig.
1A). The genomic DNA was extracted from the mouse tail and used
as a template for PCR analysis with primer P1 and P2 to identify
genotypes (Fig. 1B). SPLUNC1
protein expression in wild-type and SPLUNC1−/− mice was
shown by western blot analysis (Fig.
1C). A specific SPLUNC1 band was detected in the
SPLUNC1+/+ and SPLUNC1+/− lung, while no
SPLUNC1 protein was detected in the lung of the
SPLUNC1−/− mice. Then, we observed that knockout of
SPLUNC1 did not cause embryonic death, and there was no obvious
phenotype difference between the wild-type and
SPLUNC1−/− mice. Similar results were reported by Gally
et al (13) that
SPLUNC1−/− mice were viable, fertile, and largely
indistinguishable from WT littermates in general appearance, body
weight, locomotion and overt behavior.
Knockout of SPLUNC1 affects the
distribution of CD11b+Gr-1+ MDSCs in the
spleen of LPS-induced ALI mice using flow cytometric analysis
MDSCs commonly express the markers CD11b and Gr-1 in
mice; such immature myeloid cells are present in the bone marrow of
healthy mice. However, they differentiate into mature myeloid cells
and accumulate in the spleen under acute inflammation conditions.
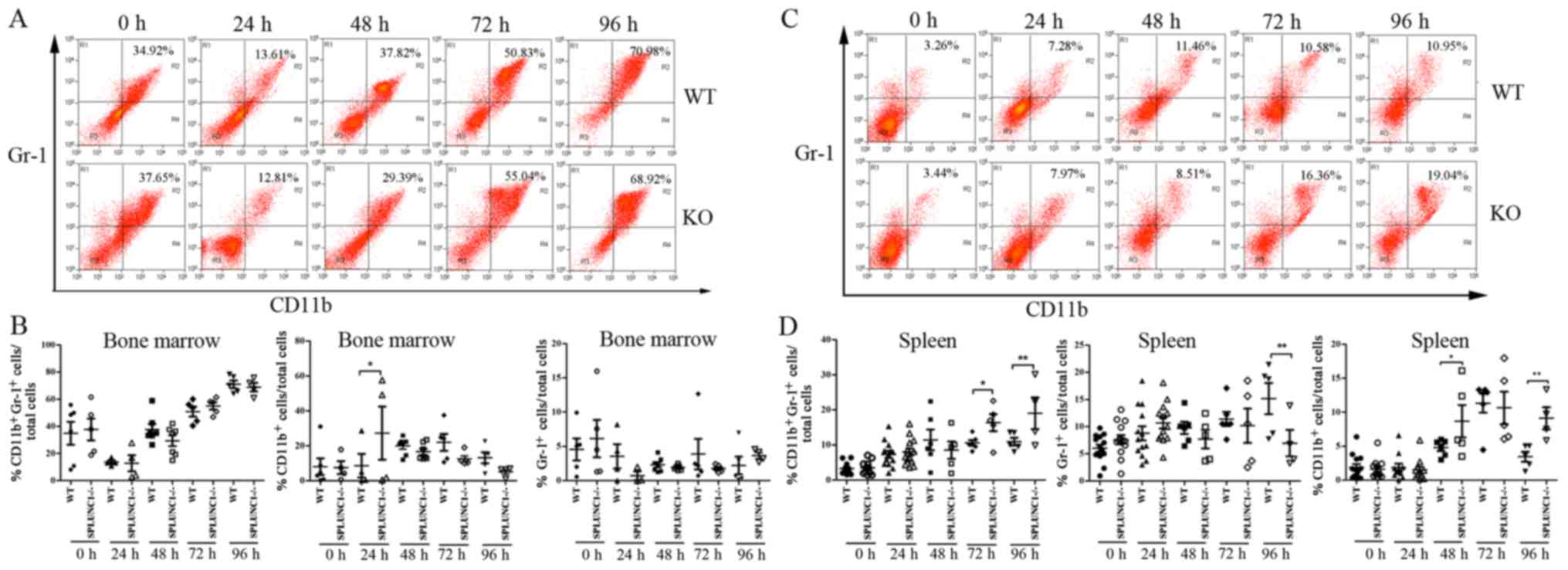
In the present study, we first measured the percentage of
CD11b+Gr-1+ MDSCs in the bone marrow of the
mice at different time points after LPS administration. The number
of CD11b+Gr-1+ MDSCs in the WT group was
reduced from 34.92±8.48 to 13.61±1.33% and in the KO group, from
37.65±8.06 to 12.81±5.92% (P<0.05; Fig. 2A) after LPS injection for 24 h, and
then it rapidly recovered to the base level after injection for 48
h. Meanwhile, the number of these cells gradually increased at 72
and 96 h post LPS injection; it reached the highest level both in
the WT and KO groups (70.98±2.84% in WT vs. 68.92±3.24% in KO mice
at 96 h). Although the percentage of CD11b+ cells in KO
mice was higher than that in WT mice at 24 h after LPS induction
(P<0.05; Fig. 2B), no
significant differences were observed between WT and KO groups at
the different time-points of 0, 24, 48, 72 and 96 h after LPS
injection (P>0.05; Fig. 2B).
These results indicated that knockout of SPLUNC1 had no influence
on the production of MDSCs in the bone marrow of the mice with
ALI.
To investigate whether SPLUNC1 affects the
recruitment of MDSCs from the bone marrow to the spleen in acute
inflammatory response, we next analyzed the proportion of splenetic
CD11b+Gr-1+ MDSCs in the WT and KO groups.
There was significant accumulation of
CD11b+Gr-1+ MDSCs after LPS injection,
increasing from 3.26±0.41% of all splenocytes at baseline to a peak
of 11.46±2.97% by 48 h in the WT group and from 3.44±0.46% of all
splenocytes at baseline to a peak of 19.04±4.53% by 96 h in the KO
group (Fig. 2C). The percentage of
CD11b+ cells in the KO mice was higher than that in the
WT mice at the time-points of 48 h (4.79±0.49% in WT mice vs.
8.67±2.38% in KO mice) and 96 h (3.44±0.74% in WT mice vs.
9.10±1.62% in KO mice) after LPS injection (P<0.05; Fig. 2D). Although the number of
Gr-1+ cells in the WT mice was higher than that in the
KO mice 96 h (15.18±2.87% in WT mice vs. 6.97±2.41% in KO mice)
post LPS injection, the percentage of
CD11b+Gr-1+ MDSCs in the KO mice was higher
than that in the WT mice at the time point of 72 h (10.58±0.79% in
WT mice vs. 16.36±2.42% in KO mice) and 96 h (10.95±1.11% in WT
mice vs. 19.04±4.53% in KO mice) after LPS injection (P<0.05;
Fig. 2D). These data revealed that
the presence of SPLUNC1 suppresses the recruitment of
CD11b+Gr-1+ MDSCs to the spleen in
LPS-induced ALI.
Expression of CD11b/Gr-1 in cells in
the mouse spleen as detected by IHC
Granulocyte differentiation antigen 1 (Gr-1) is
highly expressed in neutrophils and Gr-1+ cells play a
vital role in host defense, functioning as immunoregulators in the
immune system. CD11b+ cells, specifically express M-CSF
receptor (CD115) or IL-4R (CD124), having immunosuppressive
activity. CD11b+ cells are distributed throughout the
body and act as sentinels for the immune system. The frequency of
CD11b+ myeloid cells varies in different organs, ranging
from ~48% of brain myeloid (microglia) cells, 30% of bone marrow
and 6% of total nucleated cells in the gut and spleen. In mice,
MDSCs are identified as cells that co-express the myeloid lineage
differentiation antigens Gr-1 and CD11b, which have been shown to
accumulate quickly in mouse spleen and possess immunosuppressive
effect in response to LPS administration. In the present study, we
aimed to investigate whether SPLUNC1 affects the recruitment of
MDSCs from the bone marrow to the spleen in acute inflammation;
thus, it was important to distinguish between
CD11b+Gr-1+ cells from CD11b+ and
Gr-1+ cells.
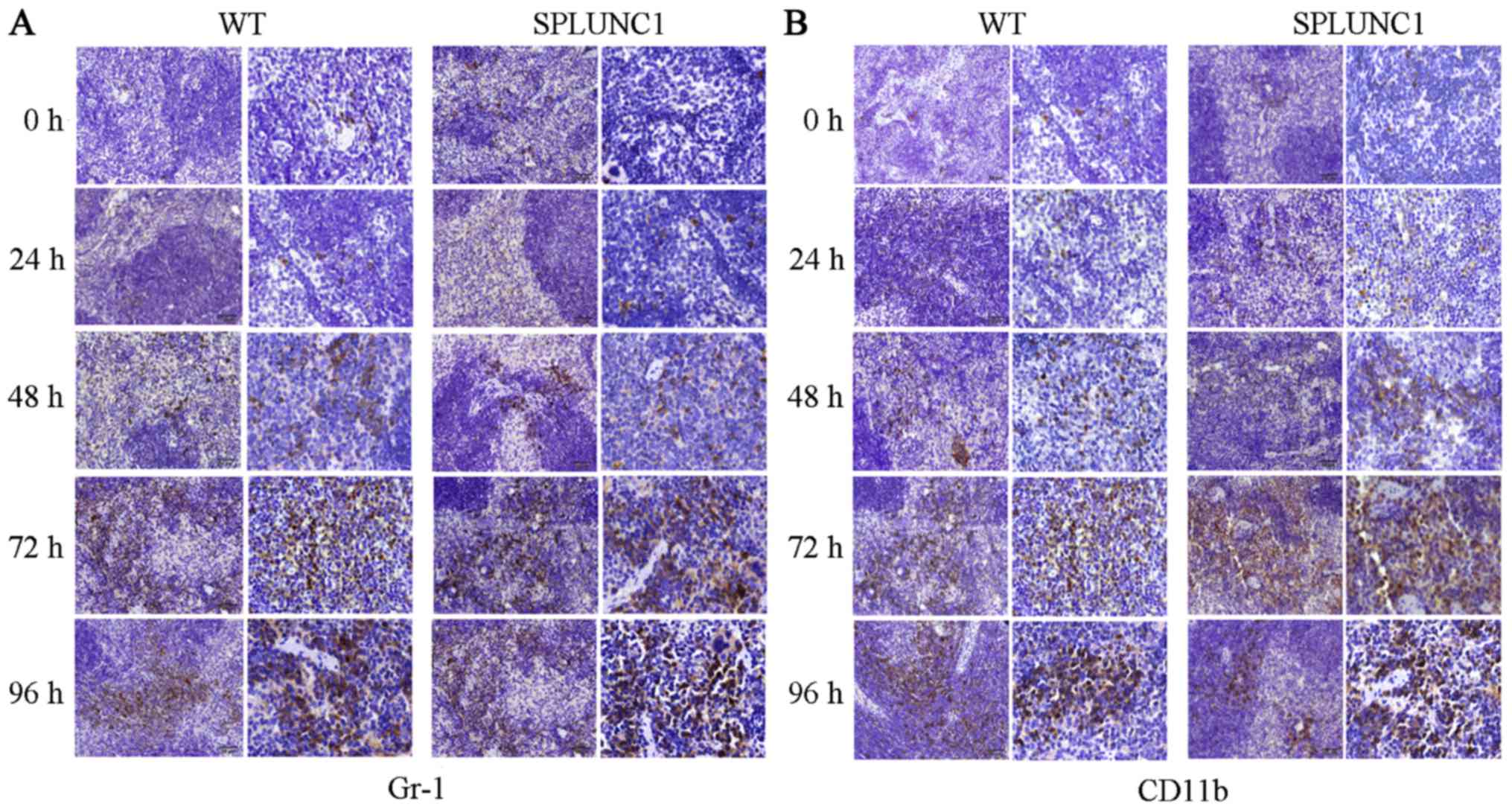
In the present study, we examined the protein
expression of Gr-1 and CD11b in the mouse spleen in response to LPS
injection. In the WT and KO groups, few Gr-1+ cells were
observed in the splenic tissues at 0 and 24 h after LPS injection,
but a marked increase in Gr-1+ cells was noted by 48 h,
particularly 72 and 96 h after LPS injection (Fig. 3A). Similarly, CD11b exhibited low
expression at 0 and 24 h in cells of splenic tissues after LPS
injection, while 48 h post injection, the number of cells
expressing CD11b gradually increased. However, statistically
significant differences between the WT and KO groups were not
detected by IHC at the different time points after LPS injection
(Fig. 3B).
Knockout of SPLUNC1 enhances
LPS-induced lung injury
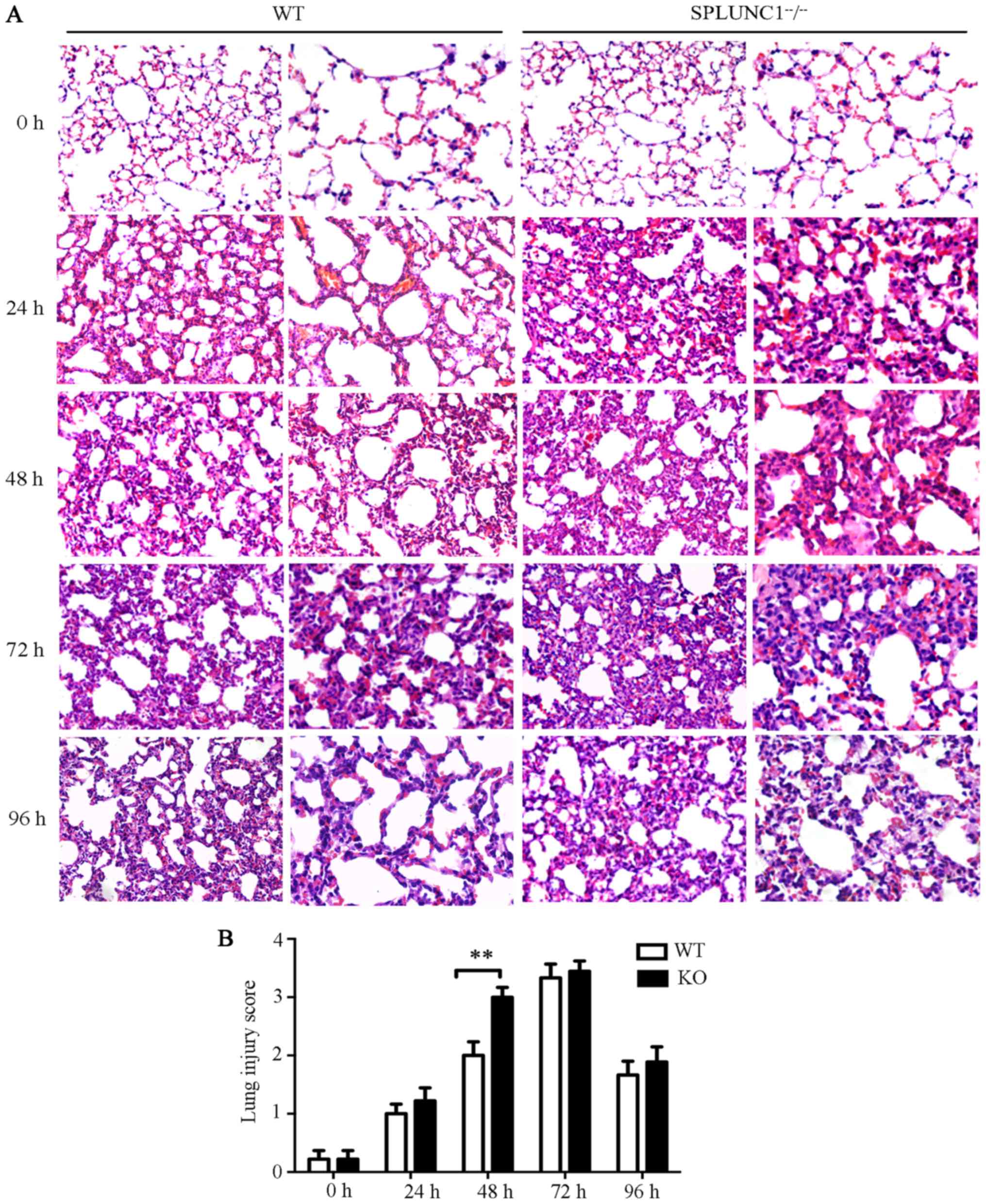
To investigate the role of SPLUNC1 in acute lung
inflammation, WT and SPLUNC1−/− mice were treated with
LPS as described in Materials and methods. The severity of lung
injury was evaluated by histopathological analysis using
hematoxylin and eosin (H&E) staining. In the WT and KO groups,
the lungs of the mice injected with LPS showed neutrophil
infiltration, thickened alveolar walls and lung congestion, while
no such pathological changes were found in the control group (LPS-0
h). Meanwhile, when treated with LPS, the SPLUNC1−/−
group appeared to have more severe pulmonary damage and infiltrated
inflammatory cells compared with the WT mice after LPS treatment
for 48 h (Fig. 4).
Knockout of SPLUNC1 promotes lung
inflammation in response to LPS induction
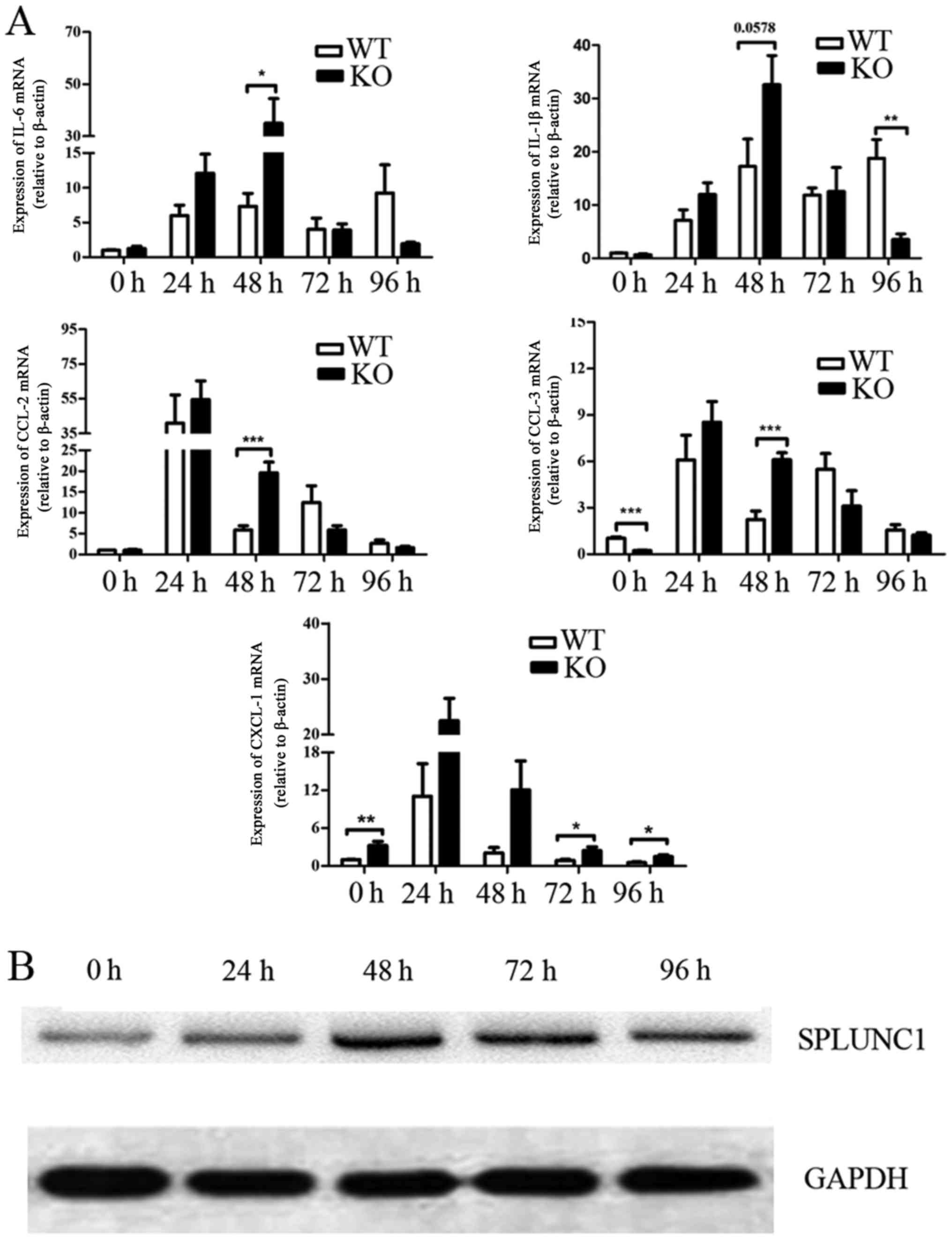
Furthermore, we compared the differential expression
of these cytokines in the WT and SPLUNC1−/− groups at
different time points after LPS treatment. The mRNA expression
levels of IL-6, CCL-2 and CCL-3 in the SPLUNC1−/− group
were higher than these levels in the WT group 48 h post LPS
injection. The level of CXCL-1 mRNA expression in the
SPLUNC1−/− group was higher than the level in the WT
group after LPS injection at 72 and 96 h (P<0.05; Fig. 5A). The SPLUNC1 expression levels
were increased in the lung tissues of the WT mice after LPS
injection (Fig. 5B). Taken
together, these data demonstrated that SPLUNC1 deficiency
upregulated ALI-related cytokines, which may promote early
inflammation leading to more severe lung injury after LPS
injection.
Discussion
The majority of recent studies on SPLUNC1 have
attempted to explore its role in inflammation or infection, while
the immune cells associated with SPLUNC1 regulation remain largely
unknown. In the present study, SPLUNC1 protein was undetectable in
SPLUNC1−/− mouse lung tissues, which confirmed gene
knockout. Our daily observation found that there was no obvious
phenotype difference between the wild-type and
SPLUNC1−/− mice under normal physiological conditions,
which was in accordance with a study reported by Gally et al
(13).
SPLUNC1 plays a significant role in the host immune
system with immunomodulatory (27),
anti-biofilm (28) and/or
chemotactic properties (29). Our
in vitro studies have demonstrated that SPLUNC1 protein can
bind to bacterial lipopolysaccharide (LPS), inhibit the growth of
P. aeruginosa (10),
regulate cell progression and apoptosis (12) and weaken the inflammatory response
induced by EBV infection (11). To
further explore its immune function in acute inflammation, we
established an acute lung injury (ALI) mouse model by LPS
intraperitoneal injection to WT and SPLUNC1-knockout mice,
respectively. The symptoms of LPS-induced ALI mice closely resemble
the observed pathology in humans (30); therefore, intraperitoneal injection
of LPS has been widely used to study the pathogenesis of ALI.
ALI is a severe inflammatory disease that can lead
to clinical syndromes including hypoxemia, pulmonary inflammation,
alveolar-capillary barrier damage and multiple organ injury
(2,31,32).
LPS can be recognized by macrophages via their surface CD14/TLR4
receptor complexes. Activated macrophages and other immune cells
then express multiple cytokines, which initiate, amplify and
perpetuate the inflammatory response in ALI (33,34).
As expected, the present study found that when treated with LPS,
the SPLUNC1−/− group appeared to have more severe
pulmonary damage compared with the WT group after LPS treatment.
The characteristic of ALI is the release of chemokines, such as
CCL-2, CCL-3 and CXCL-1, which recruit macrophage and neutrophil
migration from the intravascular space across the endothelium and
epithelium into the airspaces (35). In the present study, we found that
the mRNA expression levels of IL-6, CCL-2, CCL-3 and CXCL-1 in the
SPLUNC1−/− group were higher than these levels in the WT
group at different time points after LPS injection and the period
of increase of these cytokine in the KO group was longer than that
in the WT group. Moreover, immunohistochemical results indicated
that the expression of SPLUNC1 in the WT group also increased after
LPS injection. These results demonstrated that SPLUNC1 deficiency
upregulated ALI-related cytokines, which may promote early
inflammation leading to more severe lung injury after LPS
injection.
There are two types of innate immune molecular
proteins that play a key role in the defense of gram-negative
bacteria: lipopolysaccharide-binding protein (LBP) and
bactericidal/permeability increasing protein (BPI). The two
structures are similar, and all can be combined with LPS. LBP
presents LPS to the surface of immune cells and stimulates them to
produce an immune response. However, as an antagonist of LPS, BPI
can block the combination of LPS and LBP (36,37).
Our previous study found that SPLUNC1 has a similar folding
structure to BPI and LBP, which can bind to bacterial LPS (10). Therefore, we concluded that the
homology of the BPI domain of the SPLUNC1 molecule may be one of
the important molecular basis of its protective function in
LPS-induced lung injury in mice.
Various studies have suggested that
Gr-1+/CD11b+ cells can play a vital role in
the immune system and Gr-1+CD11b+ MDSCs have
been shown to accumulate quickly in the mouse spleen and possess
immunosuppressive effect in response to LPS administration
(38–41). In the present study, we found that
the expression of those cells in mouse spleen was increased 48 h
after LPS injection; the percentage of
Gr-1+CD11b+ MDSCs in the spleen showed an
increasing production trend post LPS injection both in the WT and
KO groups. In the mouse bone marrow, the number of
CD11b+Gr-1+ MDSCs declined 24 h post LPS
injection, and then it recovered rapidly. Although there were no
differences in mouse bone marrow between WT and KO groups after LPS
injection, the percentage of splenic
CD11b+Gr-1+ MDSCs in the KO mice was
significant higher than that in the WT mice at the time-points of
72 and 96 h post LPS injection. Many factors can induce MDSC
activation and expansion such as macrophage colony-stimulating
factor (M-CSF), IL-6 (42),
vascular endothelial growth factor (VEGF) (43), prostaglandins (44) and granulocyte/macrophage
colony-stimulating factor (GM-CSF) (45). It has been reported that the
molecules which are produced in lung injury can be linked to the
bone marrow and show significant associations with the expansion
and activation of MDSCs (46,47).
These results indicate that SPLUNC1 can influence the recruitment
of MDSCs by upregulating the secretion of cytokines in impaired
lung tissue.
In summary, based on SPLUNC1-knockout mice we
established the LPS-induced ALI mouse model to study the
anti-inflammatory effect of SPLUNC1. Our results found that SPLUNC1
deficiency upregulated levels of ALI-related cytokines leading to
more severe lung injury after LPS injection. Moreover, we first
discovered that the presence of SPLUNC1 can suppress the
recruitment of CD11b+Gr-1+ MDSCs to the
spleen in LPS-induced ALI. Nevertheless, the mechanism of the
CD11b+Gr-1+ MDSC recruitment needs to be
further studied.
Acknowledgements
The present study was supported by the National
Natural Sciences Foundation of China (nos. 81672685, 81402270,
81272975, 81672993 and 81402307), the Key Project of Hunan
Provincial Natural Science Foundation (no. 12JJ2044), the Key
Planned Science and Technology Project of Hunan Province (nos.
2012FJ2014 and 2011FJ3153); the 111 Project 111-2-12), the Natural
Science Foundation of Hunan Province (2016JC2035), the Planned
Project of Development and Reform Commission of Hunan Province
(2012-1493-1), the Planned Project of Department of health of Hunan
Province (B2011-030, B2012-029), the Open-End Fund for the Valuable
and Precision Instruments of Central South University Doctoral
innovation project of Hunan Province (CX2015B057).
Glossary
Abbreviations
Abbreviations:
|
SPLUNC1
|
short palate, lung and nasal
epithelium clone 1
|
|
MDSCs
|
myeloid-derived suppressor cells
|
|
LPS
|
lipopoly-saccharide
|
|
ALI
|
acute lung injury
|
|
IMCs
|
immature myeloid cells
|
|
DCs
|
dendritic cells
|
|
IHC
|
immunohistochemistry
|
|
GADPH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
WT
|
wild-type
|
|
NSCLC
|
non-small cell lung cancer
|
|
IL-6
|
interleukin-6
|
|
CCL-2
|
chemokine (C-C motif) ligand-2
|
|
CCL-3
|
chemokine (C-C motif) ligand-3
|
|
CXCL-1
|
chemokine (C-X-C motif) ligand-1
|
References
|
1
|
Rubenfeld GD, Caldwell E, Peabody E,
Weaver J, Martin DP, Neff M, Stern EJ and Hudson LD: Incidence and
outcomes of acute lung injury. N Engl J Med. 353:1685–1693. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abraham E: Neutrophils and acute lung
injury. Crit Care Med. 31 Suppl 4:S195–S199. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bingle CD and Bingle L: Characterisation
of the human plunc gene, a gene product with an upper airways and
nasopharyngeal restricted expression pattern. Biochim Biophys Acta.
1493:363–367. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bingle CD and Craven CJ: Comparative
analysis of the PLUNC (palate, lung and nasal epithelium clone)
protein families. Biochem Soc Trans. 31:806–809. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang B, Nie X, Xiao B, Xiang J, Shen S,
Gong J, Zhou M, Zhu S, Zhou J, Qian J, et al: Identification of
tissue-specific genes in nasopharyngeal epithelial tissue and
differentially expressed genes in nasopharyngeal carcinoma by
suppression subtractive hybridization and cDNA microarray. Genes
Chromosomes Cancer. 38:80–90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim CH, Kim K, Jik Kim H, Kim J Kook, Lee
JG and Yoon JH: Expression and regulation of PLUNC in human nasal
epithelium. Acta Otolaryngol. 126:1073–1078. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Campos MA, Abreu AR, Nlend MC, Cobas MA,
Conner GE and Whitney PL: Purification and characterization of
PLUNC from human tracheobronchial secretions. Am J Respir Cell Mol
Biol. 30:184–192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sung YK, Moon C, Yoo JY, Moon C, Pearse D,
Pevsner J and Ronnett GV: Plunc, a member of the secretory gland
protein family, is up-regulated in nasal respiratory epithelium
after olfactory bulbectomy. J Biol Chem. 277:12762–12769. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Zeng Z, Wei F, Chen P, Schmitt
DC, Fan S, Guo X, Liang F, Shi L, Liu Z, et al: SPLUNC1 is
associated with nasopharyngeal carcinoma prognosis and plays an
important role in all-trans-retinoic acid-induced growth inhibition
and differentiation in nasopharyngeal cancer cells. FEBS J.
281:4815–4829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou HD, Li XL, Li GY, Zhou M, Liu HY,
Yang YX, Deng T, Ma J and Sheng SR: Effect of SPLUNC1 protein on
the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell
Biochem. 309:191–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ou C, Sun Z, Zhang H, Xiong W, Ma J, Zhou
M, Lu J, Zeng Z, Bo X, Chen P, et al: SPLUNC1 reduces the
inflammatory response of nasopharyngeal carcinoma cells infected
with the EB virus by inhibiting the TLR9/NF-κB pathway. Oncol Rep.
33:2779–2788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen P, Guo X, Zhou H, Zhang W, Zeng Z,
Liao Q, Li X, Xiang B, Yang J, Ma J, et al: SPLUNC1 regulates cell
progression and apoptosis through the miR-141-PTEN/p27 pathway, but
is hindered by LMP1. PLoS One. 8:e569292013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gally F, Di YP, Smith SK, Minor MN, Liu Y,
Bratton DL, Frasch SC, Michels NM, Case SR and Chu HW: SPLUNC1
promotes lung innate defense against Mycoplasma pneumoniae
infection in mice. Am J Pathol. 178:2159–2167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lukinskiene L, Liu Y, Reynolds SD, Steele
C, Stripp BR, Leikauf GD, Kolls JK and Di YP: Antimicrobial
activity of PLUNC protects against Pseudomonas aeruginosa
infection. J Immunol. 187:382–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Bartlett JA, Di ME, Bomberger JM,
Chan YR, Gakhar L, Mallampalli RK, McCray PB Jr and Di YP:
SPLUNC1/BPIFA1 contributes to pulmonary host defense against
Klebsiella pneumoniae respiratory infection. Am J Pathol.
182:1519–1531. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartlett JA, Meyerholz DK, Wohlford-Lenane
CL, Naumann PW, Salzman NH and McCray PB Jr: Increased
susceptibility to otitis media in a Splunc1-deficient mouse model.
Dis Model Mech. 8:501–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mitas M, Hoover L, Silvestri G, Reed C,
Green M, Turrisi AT, Sherman C, Mikhitarian K, Cole DJ, Block MI,
et al: Lunx is a superior molecular marker for detection of
non-small cell lung cancer in peripheral blood [corrected]. J Mol
Diagn. 5:237–242. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gabrilovich DI and Nagaraj S:
Myeloid-derived suppressor cells as regulators of the immune
system. Nat Rev Immunol. 9:162–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rabinovich GA, Gabrilovich D and Sotomayor
EM: Immunosuppressive strategies that are mediated by tumor cells.
Annu Rev Immunol. 25:267–296. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kusmartsev S, Nefedova Y, Yoder D and
Gabrilovich DI: Antigen-specific inhibition of CD8+ T cell response
by immature myeloid cells in cancer is mediated by reactive oxygen
species. J Immunol. 172:989–999. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu C, Jiang L, Xu X, Zhu F, Zhang S, Wu X,
Liu Z, Yang X and Li S: STAT4 knockout protects LPS-induced lung
injury by increasing of MDSC and promoting of macrophage
differentiation. Respir Physiol Neurobiol. 223:16–22. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou Y, Wang W, Zheng D, Peng S, Xiong W,
Ma J, Zeng Z, Wu M, Zhou M, Xiang J, et al: Risk of nasopharyngeal
carcinoma associated with polymorphic lactotransferrin haplotypes.
Med Oncol. 29:1456–1462. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao S, Zhou Y, Yi W, Luo G, Jiang B, Tian
Q, Li Y and Xue M: Fra-1 is downregulated in cervical cancer
tissues and promotes cervical cancer cell apoptosis by p53
signaling pathway in vitro. Int J Oncol. 46:1677–1684. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng D, Liao S, Zhu G, Luo G, Xiao S, He
J, Pei Z, Li G and Zhou Y: CD38 is a putative functional marker for
side population cells in human nasopharyngeal carcinoma cell lines.
Mol Carcinog. 55:300–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hara A and Okayasu I: Cyclooxygenase-2 and
inducible nitric oxide synthase expression in human astrocytic
gliomas: Correlation with angiogenesis and prognostic significance.
Acta Neuropathol. 108:43–48. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thaikoottathil JV, Martin RJ, Di PY, Minor
M, Case S, Zhang B, Zhang G, Huang H and Chu HW: SPLUNC1 deficiency
enhances airway eosinophilic inflammation in mice. Am J Respir Cell
Mol Biol. 47:253–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gakhar L, Bartlett JA, Penterman J,
Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S and McCray PB Jr:
PLUNC is a novel airway surfactant protein with anti-biofilm
activity. PLoS One. 5:e90982010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sayeed S, Nistico L, St Croix C and Di YP:
Multifunctional role of human SPLUNC1 in Pseudomonas aeruginosa
infection. Infect Immun. 81:285–291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen H, Bai C and Wang X: The value of the
lipopolysaccharide-induced acute lung injury model in respiratory
medicine. Expert Rev Respir Med. 4:773–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matuschak GM and Lechner AJ: Acute lung
injury and the acute respiratory distress syndrome: Pathophysiology
and treatment. Mo Med. 107:252–258. 2010.PubMed/NCBI
|
|
32
|
Gaudry S, Ricard JD and Dreyfuss D: Acute
respiratory distress syndrome. Rev Prat. 62:1197–1203. 2012.(In
French). PubMed/NCBI
|
|
33
|
Mei SH, McCarter SD, Deng Y, Parker CH,
Liles WC and Stewart DJ: Prevention of LPS-induced acute lung
injury in mice by mesenchymal stem cells overexpressing
angiopoietin 1. PLoS Med. 4:e2692007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Sun CY, Zhang YB, Guo HZ, Feng
XX, Peng SZ, Yuan J, Zheng RB, Chen WP, Su ZR, et al: Kegan Liyan
oral liquid ameliorates lipopolysaccharide-induced acute lung
injury through inhibition of TLR4-mediated NF-κB signaling pathway
and MMP-9 expression. J Ethnopharmacol. 186:91–102. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bhatia M, Zemans RL and Jeyaseelan S: Role
of chemokines in the pathogenesis of acute lung injury. Am J Respir
Cell Mol Biol. 46:566–572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JW, Gerwick L and Park CI: Molecular
identification and expression analysis of two distinct BPI/LBPs
(bactericidal permeability-increasing protein/LPS-binding protein)
from rock bream, Oplegnathus fasciatus. Fish Shellfish Immunol.
33:75–84. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Krasity BC, Troll JV, Weiss JP and
McFall-Ngai MJ: LBP/BPI proteins and their relatives: Conservation
over evolution and roles in mutualism. Biochem Soc Trans.
39:1039–1044. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vaknin I, Blinder L, Wang L, Gazit R,
Shapira E, Genina O, Pines M, Pikarsky E and Baniyash M: A common
pathway mediated through Toll-like receptors leads to T- and
natural killer-cell immunosuppression. Blood. 111:1437–1447. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Delano MJ, Scumpia PO, Weinstein JS, Coco
D, Nagaraj S, Kelly-Scumpia KM, O'Malley KA, Wynn JL, Antonenko S,
Al-Quran SZ, et al: MyD88-dependent expansion of an immature
GR-1+CD11b+ population induces T cell suppression and Th2
polarization in sepsis. J Exp Med. 204:1463–1474. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Wilde V, van Rompaey N, Hill M, Lebrun
JF, Lemaître P, Lhommé F, Kubjak C, Vokaer B, Oldenhove G,
Charbonnier LM, et al: Endotoxin-induced myeloid-derived suppressor
cells inhibit alloimmune responses via heme oxygenase-1. Am J
Transplant. 9:2034–2047. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McCubbrey AL, Barthel L, Mould KJ, Mohning
MP, Redente EF and Janssen WJ: Selective and inducible targeting of
CD11b+ mononuclear phagocytes in the murine lung with hCD68-rtTA
transgenic systems. Am J Physiol Lung Cell Mol Physiol.
311:L87–L100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bunt SK, Yang L, Sinha P, Clements VK,
Leips J and Ostrand-Rosenberg S: Reduced inflammation in the tumor
microenvironment delays the accumulation of myeloid-derived
suppressor cells and limits tumor progression. Cancer Res.
67:10019–10026. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gabrilovich D, Ishida T, Oyama T, Ran S,
Kravtsov V, Nadaf S and Carbone DP: Vascular endothelial growth
factor inhibits the development of dendritic cells and dramatically
affects the differentiation of multiple hematopoietic lineages in
vivo. Blood. 92:4150–4166. 1998.PubMed/NCBI
|
|
44
|
Serafini P, Carbley R, Noonan KA, Tan G,
Bronte V and Borrello I: High-dose granulocyte-macrophage
colony-stimulating factor-producing vaccines impair the immune
response through the recruitment of myeloid suppressor cells.
Cancer Res. 64:6337–6343. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bronte V, Chappell DB, Apolloni E,
Cabrelle A, Wang M, Hwu P and Restifo NP: Unopposed production of
granulocyte-macrophage colony-stimulating factor by tumors inhibits
CD8+ T cell responses by dysregulating antigen-presenting cell
maturation. J Immunol. 162:5728–5737. 1999.PubMed/NCBI
|
|
46
|
Islam MN, Das SR, Emin MT, Wei M, Sun L,
Westphalen K, Rowlands DJ, Quadri SK, Bhattacharya S and
Bhattacharya J: Mitochondrial transfer from bone-marrow-derived
stromal cells to pulmonary alveoli protects against acute lung
injury. Nat Med. 18:759–765. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kotton DN, Ma BY, Cardoso WV, Sanderson
EA, Summer RS, Williams MC and Fine A: Bone marrow-derived cells as
progenitors of lung alveolar epithelium. Development.
128:5181–5188. 2001.PubMed/NCBI
|