Introduction
Non-small cell lung cancer (NSCLC), composed of
adenocarcinoma and squamous cell carcinoma, is the preponderant
form of lung cancer and accounts for the majority of lung
cancer-related deaths worldwide (1–4).
Despite advances in the diagnosis and treatment of patients with
NSCLC, the overall 5-year survival rate of NSCLC is still less than
15% due to the vast majority of patients that are diagnosed at an
advanced stage with metastasis or relapse (5,6). Tumor
metastasis is a complex process involving multiple steps, including
changes in tumor cell adhesion, matrix degradation, cell migration
and invasion, angiogenesis and the ability of the immune system to
combat invasion. (7–14). Lack of biomarkers for early
diagnosis and metastasis markers are still some of the challenges
in NSCLC. Therefore, identification of new functional genes and
demonstration of their roles in NSCLC is essential for the
development of specific diagnostic methods and the design of more
effective therapeutic strategies for NSCLC patients (15). Placenta-specific protein 1 (PLAC1)
protein is encoded by placenta-specific gene 1, which is mainly
localized in the cell membrane and involved in the regulation of
trophoblast growth and differentiation. PLAC1 has been found to
play an important role in placental and embryo development
(particularly the brain development) (16–21).
During embryo implantation, the invasion of trophocytes into the
endometrium and the formation of blood vessels are very similar to
the growth, invasion and migration of tumors (22). Recently, increasing evidence
revealed that PLAC1 expression is activated in a variety of human
cancers, including gastric, non-small cell lung, liver and
colorectal cancer, primary colorectal adenocarcinoma, epithelial
ovarian and breast cancer (16,23–30).
In addition, increased expression of PLAC1 was found to be
positively correlated with the degree of tumor invasion, lymph node
metastasis and distant metastasis (31). However, the expression pattern and
functional role of PLAC1 in NSCLC have not yet been well
documented.
In the present study, we performed
immunohistochemistry and qRT-PCR analysis of PLAC1 expression
levels in NSCLC tumor and adjacent non-tumor tissues, and NSCLC
cell lines. We found that PLAC1 expression was significantly
upregulated in NSCLC tissues, and its increased level was
associated with poor prognosis and shorter survival time. Applying
loss of function analysis, we investigated the biological function
of PLAC1 in NSCLC cells. Finally, mechanistic investigation was
performed to determine its regulation of underlying targets and
pathways in NSCLC cells.
Materials and methods
Tissue collection
We obtained 88 NSCLC lung tissues from patients who
underwent surgery at Jiangsu Province Hospital between 2012 and
2014 and were diagnosed with NSCLC based on histopathological
evaluation. Clinicopathological characteristics, including
tumor-node-metastasis (TNM) staging, were recorded. No local or
systemic treatment was conducted in these patients before surgery.
All collected tissue samples were immediately snap-frozen in liquid
nitrogen and stored at −80°C until required. The present study was
approved by the Research Ethics Committee of Nanjing Medical
University, China. Written informed consent was obtained from all
patients.
Immunochemistry
Tumor core samples were transferred to blank wax
block holes, with 3–5 independent sample points and 40–45
points/chip array. Serial sections (5-µm thick) were deparaffinized
and rehydrated with xylene, and a series of grades of alcohol.
After epitope retrieval (it can re-expose the antigen epitope or
amend the denaturing effects of chemical reagents and thermal
applications); and inactivation of endogenous peroxidase, sections
were blocked with 10% normal goat serum for 30 min, and then
sequentially incubated with a rabbit anti-PLAC1/CP1 antibody
(R&D Systems, Minneapolis, MN, USA) and horseradish
peroxidase-conjugated anti-rabbit IgG [Cell Signaling Technology
(CST), Danvers, MA, USA]. After being washed, the slides were
developed using diaminobenzidine tetrahydrochloride and
counterstained with hematoxylin.
Cell lines
Five NSCLC adenocarcinoma cell lines (PC-9, SPC-A1,
H1299, A549 and H1650), 2 NSCLC squamous carcinomas cell lines
(H520 and SK-MES-1) and human bronchial epithelial cells 16HBE were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences (Shanghai, China). A549, H1975,
H1299, H1650 and H520 cells were cultured in RPMI-1640 medium;
16HBE, PC-9 and SPC-A1 cells were cultured in Dulbeccos modified
Eagles medium (DMEM) (Gibco-BRL, Grand Island, NY, USA) medium
supplemented with 10% fetal bovine serum (FBS) 100 U/ml penicillin
and 100 mg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) at
37°C/5% CO2.
RNA extraction and qPCR assays
Total RNA was isolated using TRIzol reagent
(Invitrogen) according to the manufacturer's instructions. Total
RNA (500 ng) was reverse-transcribed in a final volume of 10 µl
using random primers under standard conditions for the PrimeScript
RT Reagent kit (Takara, Dalian, China). We used the SYBR Premix Ex
Taq (TaKaRa) to determine PLAC1 expression levels, following the
manufacturer's instructions. Results were normalized to the
expression of β-actin (Invitrogen). The specific primers used were
as follows: PLAC1 sense, 5′-CTGTCTTAGTCGCCTTCATGC-3′ and antisense,
5′-TGAACCAATCTGTCGAGCACA-3′; β-actin sense, 5′-TCA CCC ACA CTG TGC
CCA TCT ACG A-3′ and antisense, 5′-CAG CGG AAC CGC TCA TTG CCA ATG
G-3′ (Invitrogen). The quantitative PCR (qPCR) assays were
conducted on an ABI 7300 (Applied Biosystems, Foster City, CA,
USA), and data was collected with this instrument. qPCR mainly
included denaturation and annealing, the product length of which
was expected to be between 80–150 bp. Our qPCR results were
analyzed and expressed relative to threshold cycle (CT) values and
then converted to fold changes.
Small interfering RNA duplexes and
cell transfection
The si-PLAC1 (5′-GGGCACGCCAUCUAAGUUUTT-3′) or si-NC
(5′-UUCUCCGAACGUGUCACGUTT-3′) (Invitrogen) were transfected into
PC-9 and H1299 cells. PC-9 and H1299 cells were grown on 6-well
plates to confluency and transfected using Lipofectamine 2000
(Invitrogen) according to the manufacturer's instructions. At 24 h
post-transfection, the cells were harvested for qPCR or 48 h for
western blot analysis.
Cell viability assays
Cell viability was monitored using a cell
proliferation reagent kit I [Cell Counting Kit-8 (CCK-8)] (KeyGen
Biotech, Shanghai, China). The PC-9 and H1299 cells transfected
with si-PLAC1 (3,000 cells/well) were grown in 96-well plates. Cell
viability was assessed every 24 h following the manufacturer's
protocol. All experiments were performed in quadruplicate. For each
treatment group, the wells were assessed in triplicate.
Flow cytometry
H1299 or PC-9 cells transfected with si-PLAC1 were
harvested 48 h after transfection by trypsinization. After double
staining with FITC-Annexin V and propidium iodide (PI) was carried
out using the FITC-Annexin V apoptosis detection kit (BD
Biosciences, San Jose, CA, USA) according to the manufacturer's
recommendations, the cells were analyzed with a flow cytometer
(FACScan) equipped with CellQuest software (both from BD
Biosciences). Cells were discriminated into viable cells (Annexin
V-negative and PI-negative), early apoptotic (Annexin V-positive
and PI-negative), dead and apoptotic cells (Annexin V-positive and
PI-positive), and then the relative ratio of early apoptotic cells
was compared with the control transfectant from each experiment.
Cells for cell cycle analysis were stained with PI using the
Cycletest Plus DNA reagent kit (BD Biosciences) following the
manufacturers protocol and analyzed by FACScan. The percentage of
the cells in the G0/G1, S and G2/M phases were counted and
compared.
Cell migration and invasion
assays
For the migration assays, at 48 h post-transfection,
5×104 cells in serum-free media were placed into the
upper chamber of an insert (8-µm pore size; Millipore, Billerica,
MA, USA). For the invasion assays, 1×105 cells in
serum-free medium were placed into the upper chamber of an insert
coated with Matrigel (Sigma-Aldrich, St. Louis, MO, USA). Medium
containing 10% FBS (Gibco-BRL) was added to the lower chamber.
After incubation for 24 h, the cells remaining on the upper
membrane were removed with cotton wool. Cells that had migrated or
invaded through the membrane were stained with methanol and 0.1%
crystal violet, imaged and counted using an IX71 inverted
microscope (Olympus, Tokyo, Japan). Experiments were independently
repeated 3 times.
Western blot assay and antibodies
Cells protein lysates (Millipore) were separated by
10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), transferred to 0.22 µm NC membranes (Sigma, St. Louis,
MO, USA), and incubated with specific antibodies. Enhanced
chemiluminescence (ECL) chromogenic substrate (Beyotime, Shanghai,
China) was used to visualize the bands and the intensity of the
bands were quantified by densitometry (Quantity One Software;
Bio-Rad, Hercules, CA, USA). β-actin antibody (1:5,000) was used as
a control, and anti-PLAC1 (1:500) (ab117528), anti-vimentin
(1:2,000) (AF2105), anti-AKT (1:5,000) (MAB2055), anti-pAKT (S473)
(1:2,000) (AF887) and anti-E-cadherin (1:10,000) (AF648) were
purchased from R&D Systems. Donkey anti-goat IgG-HRP (1:5,000)
(sc-2020) was purchased from Santa Cruz Biotechnology (Santa Cruz,
CA, USA). Goat anti-mouse IgG-HRP (1:5,000) (#7076s) and goat
anti-rabbit IgG-HRP (1:5,000) (7074s) were purchased from CST.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 5.01 software (GraphPad Software, Inc., La Jolla,
CA, USA). The significance of differences between groups was
estimated using the Students t-test or χ2 test.
Progression-free survival (PFS) rates were calculated using the
Kaplan-Meier method with the log-rank test applied for comparison.
Variables with P<0.05 in univariate analysis were used in
subsequent multivariate analysis on the basis of Cox regression
analyses. Two-sided P-values were calculated, and a probability
level of 0.05 was chosen for statistical significance.
Results
PLAC1 expression is upregulated in
NSCLC and is correlated with poor prognosis
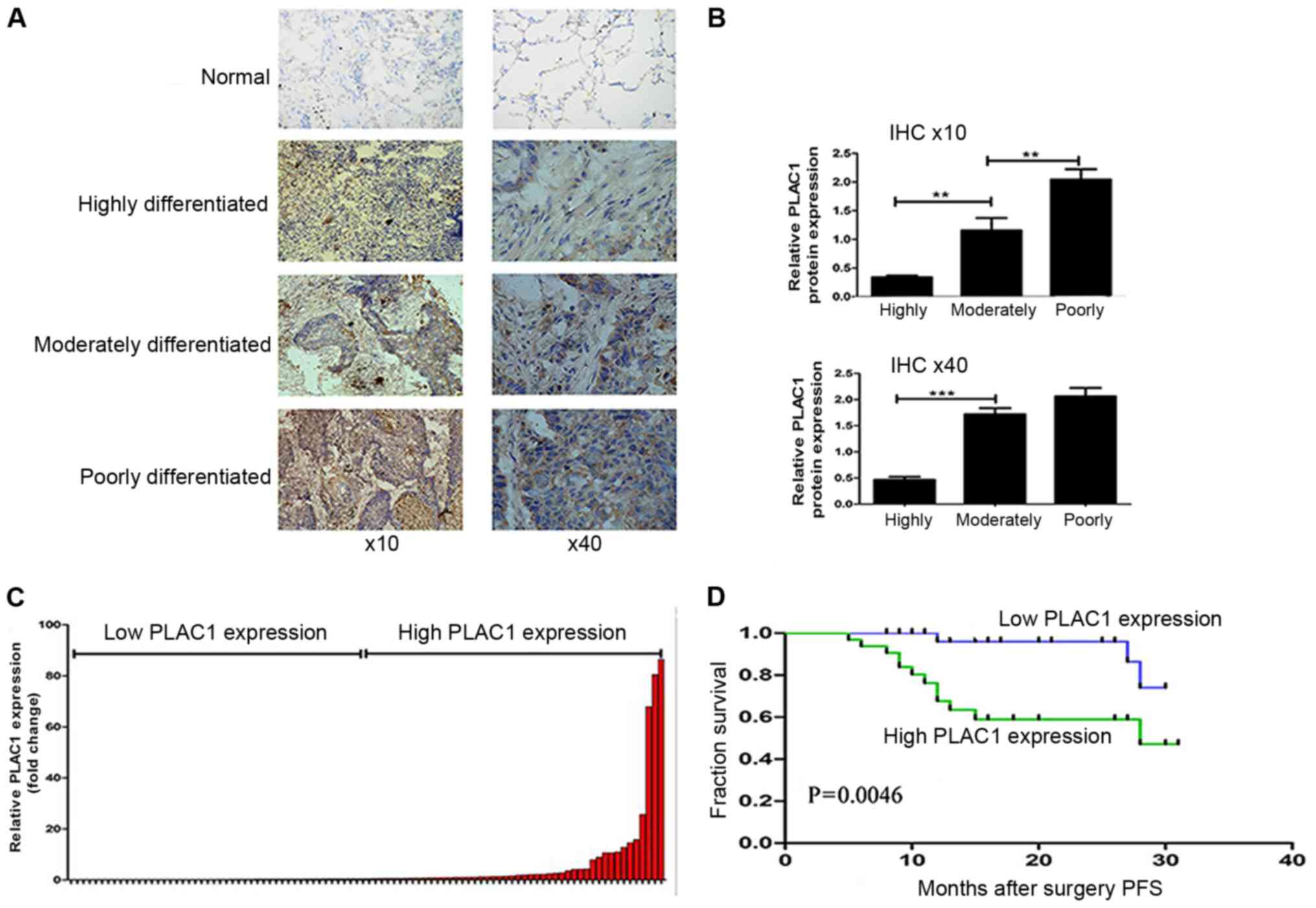
The expression and localization of PLAC1 protein was
determined by IHC staining using anti-PLAC1 antibody. As shown in
Fig. 1A, PLAC1-positive signaling
was confined to the neoplastic cell population, but not in adjacent
stromal and non-neoplastic epithelial cells. Moreover, the PLAC1
protein level was higher in the moderately- and
poorly-differentiated tumor tissues than that in the
highly-differentiated tumor tissues (Fig. 1B). In terms of clinical stage, the
expression of PLAC1 in patients with advanced-stage tissue was
higher than that in patients with early-stage tissue (Table I). Furthermore, the level of PLAC1
mRNA expression was determined in 88 NSCLC tissue samples by
qRT-PCR. However, according to the histological classification,
there was no significant difference in the expression of PLAC1 at
the RNA level. To investigate the relationship between the
expression level of PLAC1 and the clinical features of patients we
classified these patients into 2 groups: the high-PLAC1 expression
(n=44, fold change ≥ mean ratio); and the low-PLAC1 expression
groups (n=44, fold change ≤ mean ratio) (Fig. 1C). The association analysis revealed
that higher PLAC1 expression was related with TNM stage (P=0.0335),
and lymph node metastasis (P=0.0165) (Table I).
 | Table I.Correlation between PLAC1 expression
and clinicopathological characteristics of NSCLC patients. |
Table I.
Correlation between PLAC1 expression
and clinicopathological characteristics of NSCLC patients.
|
| PLAC1 |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low, no. of cases
(44) | High, no. of cases
(44) | χ2 test
(P-value) |
|---|
| Age, years |
|
| 0.3812 |
|
≤65 | 25 | 29 |
|
|
>65 | 19 | 15 |
|
| Sex |
|
| 0.1697 |
|
Male | 27 | 33 |
|
|
Female | 17 | 11 |
|
| Histologic
subtype |
|
| 0.8131 |
|
SCC | 12 | 13 |
|
|
Adenocarcinoma | 32 | 31 |
|
| TNM stage |
|
| 0.0335a |
|
Ia+Ib | 25 | 17 |
|
|
IIa+IIb | 12 | 9 |
|
|
III+IV | 7 | 18 |
|
| Tumor size
(cm) |
|
| 0.0314a |
| ≤5 | 30 | 20 |
|
|
>5 | 14 | 24 |
|
| Lymph node
metastasis |
|
| 0.0165a |
|
Negative | 32 | 20 |
|
|
Positive | 12 | 24 |
|
| Smoking
history |
|
| 0.6669 |
|
Smokers | 18 | 20 |
|
| Non
smokers | 26 | 24 |
|
Furthermore, Kaplan-Meier survival analysis was
conducted to investigate the correlation between PLAC1 expression
and NSCLC patient prognosis. With respect to PFS, the median
survival time for the low PLAC1 group was obviously far longer than
that for the high PLAC1 group (Fig.
1D). The Kaplan-Meier survival curve revealed that 2 years of
overall survival for low PLAC1 expression was ~95% while for the
high PLAC1 expression it was 55%. These findings support the
hypothesis that PLAC1 overexpression plays an important role in
NSCLC development and progression.
Modulation of PLAC1 expression in
NSCLC cells
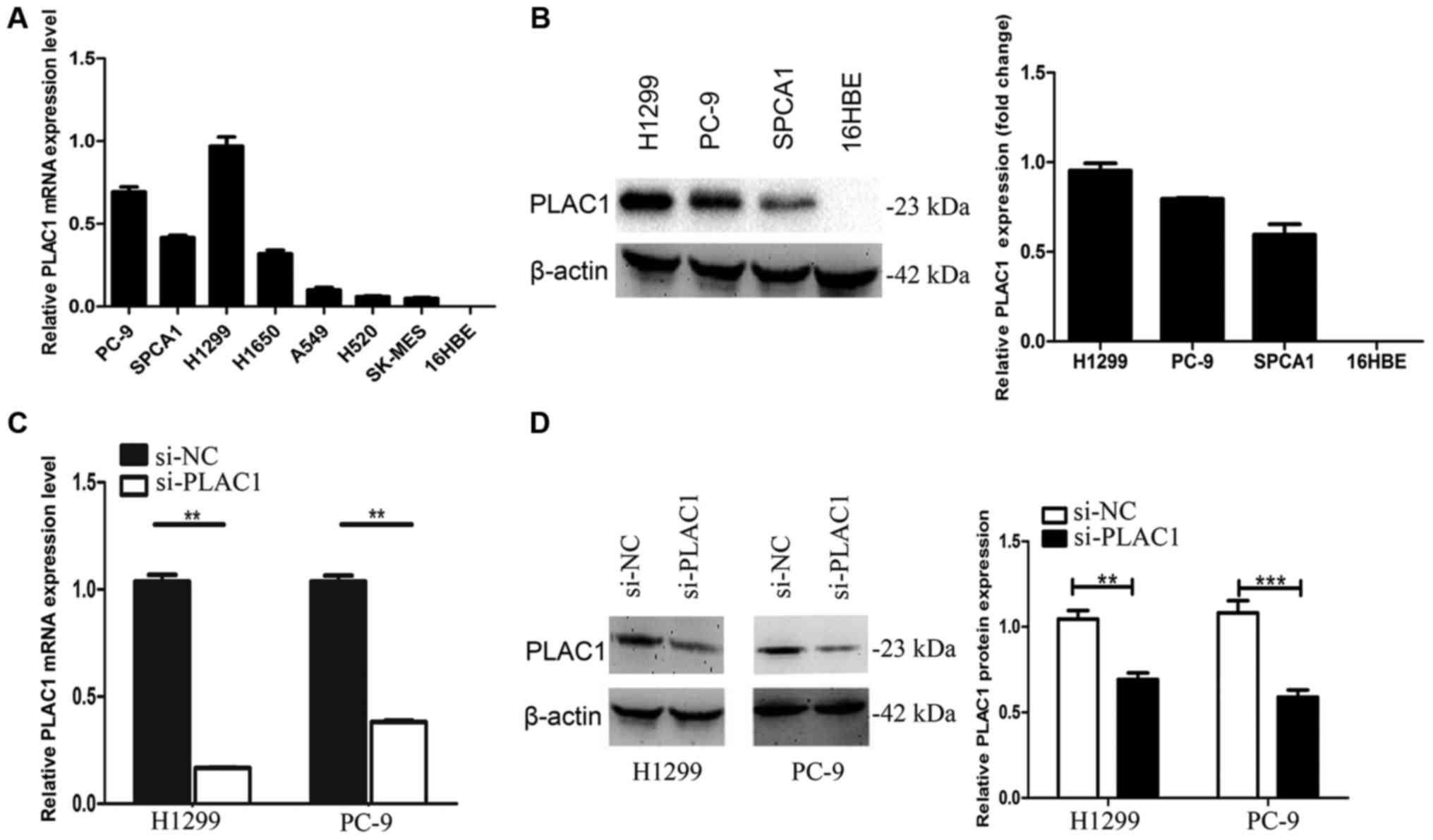
We next performed qRT-PCR analysis to examine the
expression of PLAC1 in 7 human NSCLC cell lines, including both
adenocarcinoma and squamous carcinoma subtypes. The results
revealed that PLAC1 expression was higher in the NSCLC cells than
that in the normal cell line 16HBE. As H1299 and PC-9 had higher
PLAC1 expression level, we chose these 2 cells lines for further
investigation (Fig. 2A). Next, we
detected the protein level of PLAC1 in H1299, PC-9 and SPC-A1
cells, and the results were consistent with the qRT-PCR results
(Fig. 2B). To investigate the
functional effects of PLAC1 in NSCLC cells, we knocked down its
expression through transfection with PLAC1-specific siRNA duplexes,
and a scrambled siRNA was used as a negative control. Then, qRT-PCR
analysis determined that PLAC1 mRNA expression was decreased by 85%
in H1299 cells and 75% in PC-9 cells compared with the controls
(Fig. 2C). Consistent with this
observation, the PLAC1 protein was significantly decreased in both
cell lines transfected with PLAC1 siRNA (Fig. 2D).
Knockdown of PLAC1 impairs NSCLC cell
proliferation and induces apoptosis
To assess the role of PLAC1 in NSCLC, we
investigated the effect of targeted knockdown of PLAC1 on cell
proliferation. CCK-8 assays revealed that cell growth was inhibited
in H1299 and PC-9 cells transiently transfected with si-PLAC1
compared with the controls (Fig.
3A). Colony formation assay results revealed that clonogenic
survival was inhibited following downregulation of PLAC1 in H1299
and PC-9 cells (Fig. 3B).
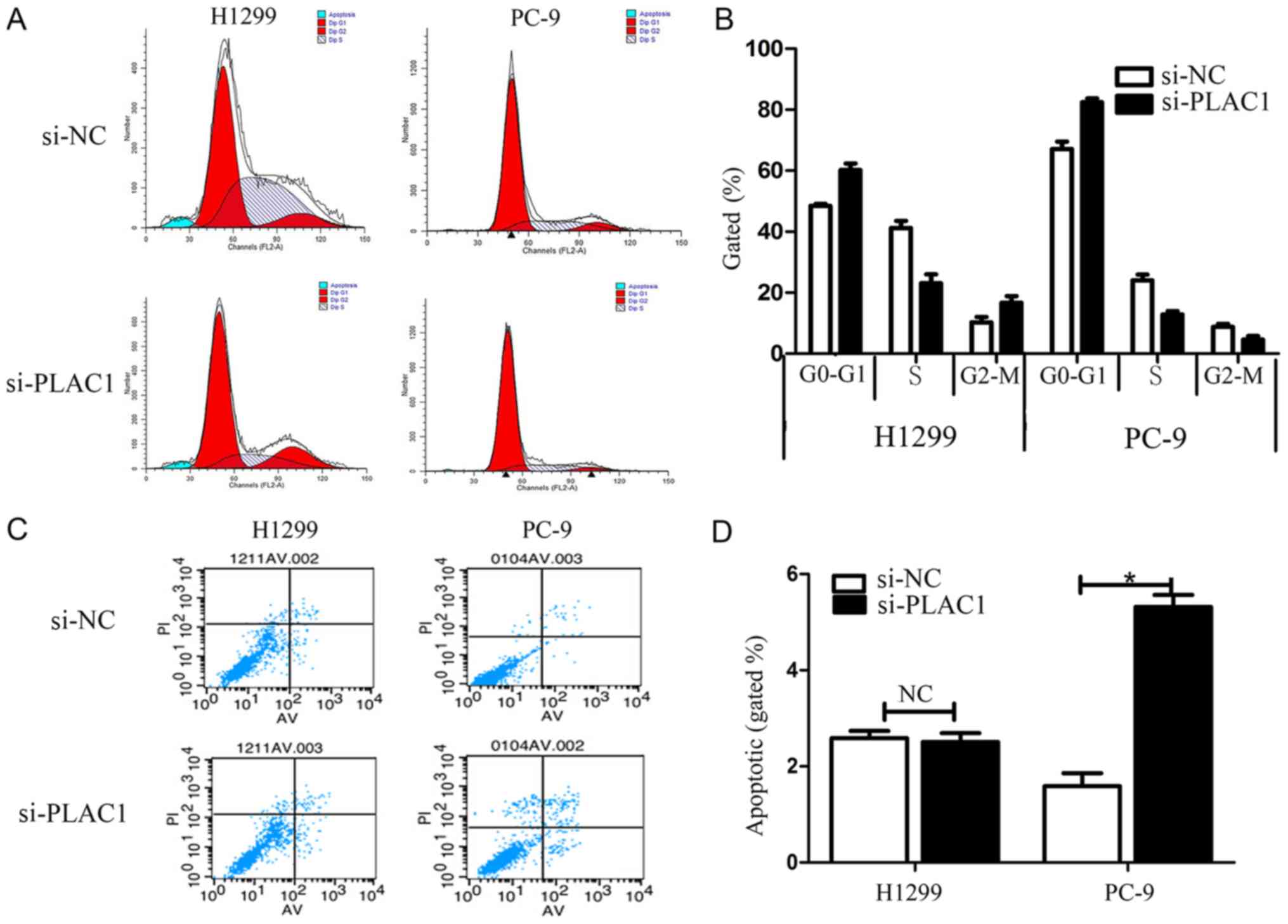
To further examine the effect of PLAC1 knockdown on
the cell cycle in NSCLC cells, cell cycle progression was analyzed
by flow cytometry. The results revealed that H1299 and PC-9 cells
transfected with si-PLAC1 had a marked cell cycle arrest at the
G1/G0 phase and a decreased G2/S phase (Fig. 4A and B). To determine whether NSCLC
cell proliferation was influenced by cell apoptosis, we performed
flow cytometry. The results revealed that NSCLC cells transfected
with PLAC1 siRNA exhibited a significant effect on apoptosis in
PC-9 cells in comparison with that in the control cells, but not in
the H1299 cells (Fig. 4C and D).
Furthermore, in the H1299 cells a G2/M arrest was noted in the
si-PLAC1 group, but the opposite phenomenon was observed in PC-9
cells due to the difference in apoptosis between the 2 types of
cells. Specifically, since the apoptosis in the si-PLAC1 group of
PC-9 cells was significantly increased than that in the si-NC
group, thus the total number of active cells in the cell cycle in
the si-PLAC1 group was relatively less than that in the si-NC
group, leading to the number of PC-9 cells in the G2/M phase to be
relatively less in the si-PLAC1 group.
Decreased PLAC1 expression inhibits
NSCLC cell migration and invasion
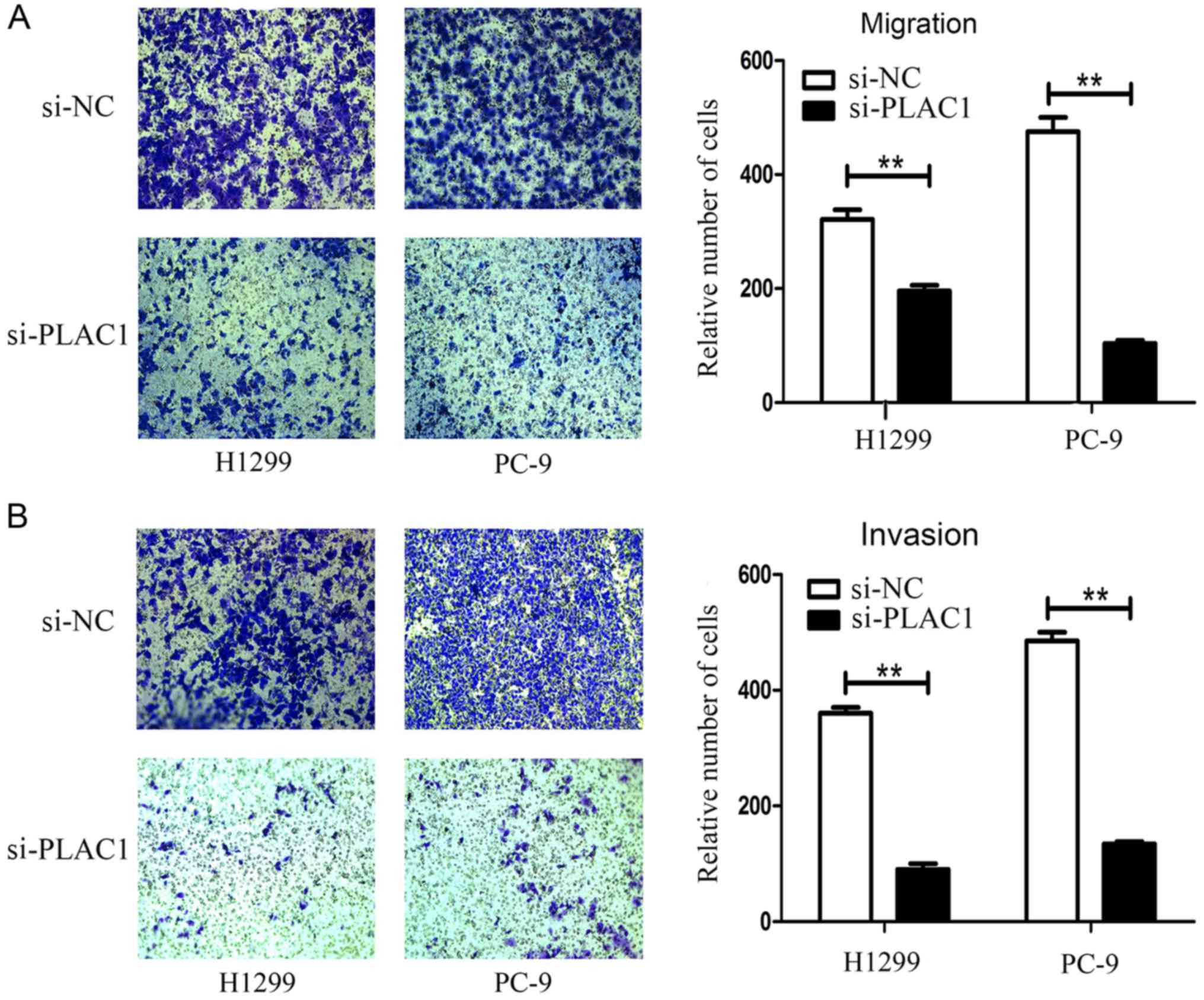
Cancer cell migration and invasion is a significant
aspect of tumor progression. To investigate the effect of PLAC1 on
NSCLC cell migration and invasion, we performed Transwell assays.
After knockdown of PLAC1 in H1299 and PC-9 cells, we observed a
marked reduction of the migratory capacity of these cells compared
with the control cells (Fig. 5A).
Moreover, invasive activity of H1299 and PC-9 cells was also
inhibited by PLAC1 siRNA treatment (Fig. 5B). The results indicated that PLAC1
plays an important role in cancer cell migration, invasion and
tumor progression.
PLAC1 is involved in the regulation of
the AKT pathway and EMT in NSCLC cells
To explore the molecular mechanisms by which PLAC1
contributes to the phenotypes of NSCLC cells, we investigated
potential targets involved in cancer cell proliferation, invasion
and metastasis. The AKT pathway is an important regulator of cancer
cell motility and migration (32–34).
Thus this prompted us to analyze whether PLAC1 had an effect on the
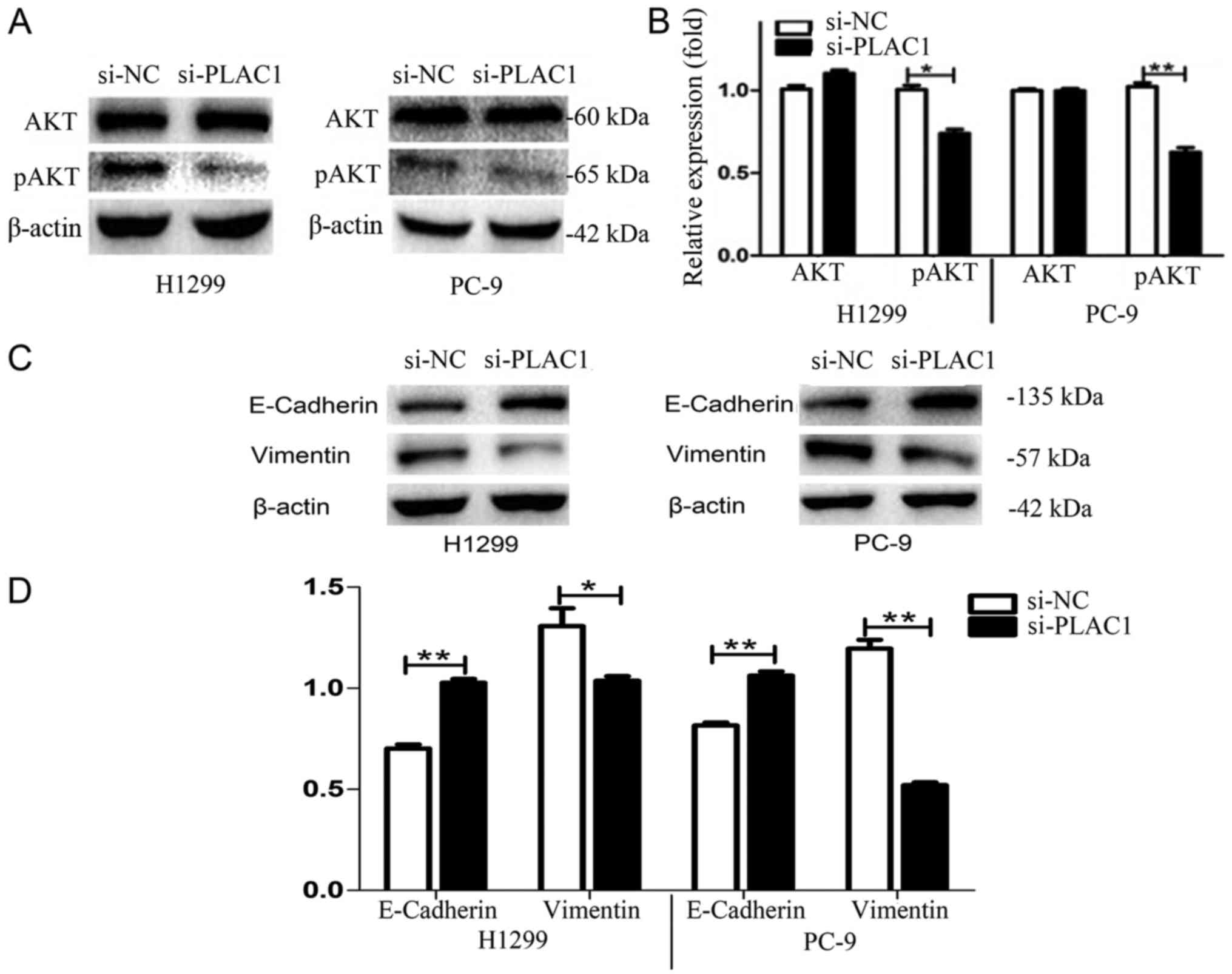
regulation of AKT kinase in H1299 and PC-9 cells. Western blot
analysis revealed that silencing of PLAC1 led to a marked reduction
of phosphorylated AKT levels in H1299 and PC-9 cells, suggesting
that AKT kinase activation was involved in the execution of
downstream effects of PLAC1 (Fig. 6A
and B). In addition, previous studies indicated that
epithelial-mesenchymal transition (EMT) was closely related to
tumor migration and invasion. EMT is an evolutionarily conserved
developmental process, which is induced during cancer progression
and contributes to metastatic colonization. EMT endows metastatic
properties in cancer cells by enhancing mobility, invasion and
resistance to apoptotic stimuli (35–38).
To assess the effects of PLAC1 on EMT-related protein expression
such as E-cadherin and vimentin, siRNA specifically targeting PLAC1
was transfected into H1299 and PC-9 cells. As shown in Fig. 6C and D, the expression of E-cadherin
was increased in PLAC1-knockdown H1299 and PC-9 cells.
Additionally, the expression of vimentin was decreased in
PLAC1-knockdown cells. These data indicated that PLAC1 may
influence the proliferation, invasive ability of NSCLC cells partly
by altering the AKT pathway and EMT process.
Discussion
Effective control of cell proliferation and invasion
is critical to the prevention of oncogenesis and successful cancer
therapy (7). Therefore,
identification of NSCLC-associated genes and investigation of their
clinical significance and functions may provide a missing piece of
the well-known oncogenic and tumor-suppressor network puzzle
(15). In the present study, we
evaluated RNA and protein expression patterns of PLAC1 in NSCLC
cell lines and primary tumor samples from NSCLC patients. We found
that the expression of PLAC1 was upregulated in NSCLC tissues and
cells compared to normal tissues and cells. Moreover, an increased
PLAC1 expression level was related to cancer cell differentiation
and short survival time of patients. These findings indicated that
PLAC1 played a direct role in the modulation of NSCLC progression
and may be considered as a novel prognostic marker for NSCLC.
PLAC1 is a new tumor-associated gene, which was
originally described as an X-linked gene (39,40).
PLAC1 expression is restricted primarily to cells derived from the
trophoblast lineage, and is expressed during embryonic development
(18). Several studies have
revealed that PLAC1 expression in normal placenta and cancer cells
is driven by specific interactions involving a combination of
transcription factors. Recently, a number of studies have
demonstrated the role of PLAC1 in placental development, pregnancy
and host immune response against malignant tumors (16,23).
Notably, PLAC1 upregulation contributed to a range of biological
functions and provided a cellular growth advantage, resulting in
progressive and uncontrolled tumor growth.
To further investigate the biological role of PLAC1
in NSCLC, we explored the effects of PLAC1 on various aspects of
NSCLC cell phenotype. RNAi-mediated suppression of PLAC1 in H1299
and PC-9 cells led to a significant inhibition of proliferation,
migration and invasion, and cell growth arrest. To further document
the molecular mechanism by which PLAC1 contributes to the NSCLC
cell function, we investigated the potential target proteins
involved in cell proliferation and invasion. In the present study,
loss of PLAC1 in NSCLC cells led to a significant increase in the
epithelial-mesenchymal transition-related protein levels of
E-cadherin, and a decrease in vimentin. In addition, PLAC1
downregulation also reduced pAKT protein levels. Our findings
indicated that PLAC1 contributed to NSCLC cell proliferation,
migration and invasion possibly and partly via regulation of
epithelial-mesenchymal transition and the AKT pathway.
In summary, the present study revealed that PLAC1
expression was highly expressed in NSCLC tissues, suggesting that
PLAC1 may be a prognostic factor and higher risk for NSCLC
patients. PLAC1 was involved in regulation of the proliferation,
migration, and invasion abilities of NSCLC cells partly through
regulation of epithelial-mesenchymal transition and the AKT
pathway. These findings elucidated NSCLC pathogenesis and
progression, and facilitate the development of tumor-associated
gene-directed diagnostics and therapeutics against this deadly
disease.
Acknowledgements
The present study was supported by the National
Natural Scientific Foundation of China (no. 81372397).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: WHO Panel: The 2015 World Health Organization
Classification of Lung Tumors: Impact of Genetic, Clinical and
Radiologic Advances Since the 2004 Classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jung KW, Park S, Kong HJ, Won YJ, Lee JY,
Seo HG and Lee JS: Cancer statistics in Korea: Incidence,
mortality, survival, and prevalence in 2009. Cancer Res Treat.
44:1–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chaari A, Ben Nasr S, Labidi S, Afrit M
and Boussen H: Metastatic non-small cell lung cancer: A Tunisian
retrospective study about 100 cases. Tunis Med. 93:294–296.
2015.(In French). PubMed/NCBI
|
|
7
|
Geiger TR and Peeper DS: Metastasis
mechanisms. Biochim Biophys Acta. 1796:293–308. 2009.PubMed/NCBI
|
|
8
|
Chan DA and Giaccia AJ: Hypoxia, gene
expression, and metastasis. Cancer Metastasis Rev. 26:333–339.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Headley MB, Bins A, Nip A, Roberts EW,
Looney MR, Gerard A and Krummel MF: Visualization of immediate
immune responses to pioneer metastatic cells in the lung. Nature.
531:513–517. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rankin EB and Giaccia AJ: Hypoxic control
of metastasis. Science. 352:175–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Turajlic S and Swanton C: Metastasis as an
evolutionary process. Science. 352:169–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheung KJ and Ewald AJ: A collective route
to metastasis: Seeding by tumor cell clusters. Science.
352:167–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kiberstis PA: Metastasis: An evolving
story. Science. 352:162–163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tüting T and de Visser KE: CANCER. How
neutrophils promote metastasis. Science. 352:145–146. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Swanton C and Govindan R: Clinical
implications of genomic discoveries in lung cancer. N Engl J Med.
374:1864–1873. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Baddoo MC and Yin Q: The placental
specific gene, PLAC1, is induced by the Epstein-Barr virus and is
expressed in human tumor cells. Virol J. 11:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Silva WA Jr, Gnjatic S, Ritter E, Chua R,
Cohen T, Hsu M, Jungbluth AA, Altorki NK, Chen YT, Old LJ, et al:
PLAC1, a trophoblast-specific cell surface protein, is expressed in
a range of human tumors and elicits spontaneous antibody responses.
Cancer Immun. 7:182007.PubMed/NCBI
|
|
18
|
Fant M, Barerra-Saldana H, Dubinsky W,
Poindexter B and Bick R: The PLAC1 protein localizes to membranous
compartments in the apical region of the syncytiotrophoblast. Mol
Reprod Dev. 74:922–929. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Massabbal E, Parveen S, Weisoly DL, Nelson
DM, Smith SD and Fant M: PLAC1 expression increases during
trophoblast differentiation: Evidence for regulatory interactions
with the fibroblast growth factor-7 (FGF-7) axis. Mol Reprod Dev.
71:299–304. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rawn SM and Cross JC: The evolution,
regulation, and function of placenta-specific genes. Annu Rev Cell
Dev Biol. 24:159–181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fant M, Weisoly DL, Cocchia M, Huber R,
Khan S, Lunt T and Schlessinger D: PLAC1, a trophoblast-specific
gene, is expressed throughout pregnancy in the human placenta and
modulated by keratinocyte growth factor. Mol Reprod Dev.
63:430–436. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang WL, Yang Q, Zhang H, Lin HY, Zhou Z,
Lu X, Zhu C, Xue LQ and Wang H: Role of placenta-specific protein 1
in trophoblast invasion and migration. Reproduction. 148:343–352.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dong XY, Peng JR, Ye YJ, Chen HS, Zhang
LJ, Pang XW, Li Y, Zhang Y, Wang S, Fant ME, et al: Plac1 is a
tumor-specific antigen capable of eliciting spontaneous antibody
responses in human cancer patients. Int J Cancer. 122:2038–2043.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu F, Shen D, Kang X, Zhang C and Song Q:
New tumour antigen PLAC1/CP1, a potentially useful prognostic
marker and immunotherapy target for gastric adenocarcinoma. J Clin
Pathol. 68:913–916. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F, Zhang H, Shen D, Wang S, Ye Y, Chen
H, Pang X, Song Q and He P: Identification of two new
HLA-A*0201-restricted cytotoxic T lymphocyte epitopes from
colorectal carcinoma-associated antigen PLAC1/CP1. J Gastroenterol.
49:419–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu FF, Shen DH, Wang S, Ye YJ and Song
QJ: Expression of PLAC1/CP1 genes in primary colorectal carcinoma
and its clinical significance. Zhonghua Bing Li Xue Za Zhi.
39:810–813. 2010.(In Chinese). PubMed/NCBI
|
|
27
|
Ghods R, Ghahremani MH, Madjd Z, Asgari M,
Abolhasani M, Tavasoli S, Mahmoudi AR, Darzi M, Pasalar P,
Jeddi-Tehrani M, et al: High placenta-specific 1/low
prostate-specific antigen expression pattern in high-grade prostate
adenocarcinoma. Cancer Immunol Immunother. 63:1319–1327. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu W, Zhai M, Wu Z, Qi Y, Wu Y, Dai C,
Sun M, Li L and Gao Y: Identification of a novel HLA-A2-restricted
cytotoxic T lymphocyte epitope from cancer-testis antigen PLAC1 in
breast cancer. Amino Acids. 42:2257–2265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tchabo NE, Mhawech-Fauceglia P, Caballero
OL, Villella J, Beck AF, Miliotto AJ, Liao J, Andrews C, Lele S,
Old LJ, et al: Expression and serum immunoreactivity of
developmentally restricted differentiation antigens in epithelial
ovarian cancer. Cancer Immun. 9:62009.PubMed/NCBI
|
|
30
|
Koslowski M, Türeci O, Biesterfeld S,
Seitz G, Huber C and Sahin U: Selective activation of
trophoblast-specific PLAC1 in breast cancer by
CCAAT/enhancer-binding protein beta (C/EBPbeta) isoform 2. J Biol
Chem. 284:28607–28615. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koslowski M, Sahin U, Mitnacht-Kraus R,
Seitz G, Huber C and Türeci O: A placenta-specific gene ectopically
activated in many human cancers is essentially involved in
malignant cell processes. Cancer Res. 67:9528–9534. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cantley LC: The phosphoinositide 3-kinase
pathway. Science. 296:1655–1657. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo J, Manning BD and Cantley LC:
Targeting the PI3K-Akt pathway in human cancer: Rationale and
promise. Cancer Cell. 4:257–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yeh PS, Wang W, Chang YA, Lin CJ, Wang JJ
and Chen RM: Honokiol induces autophagy of neuroblastoma cells
through activating the PI3K/Akt/mTOR and endoplasmic reticular
stress/ERK1/2 signaling pathways and suppressing cell migration.
Cancer Lett. 370:66–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen X, Guan X, Zhang H, Xie X, Wang H,
Long J, Cai T, Li S, Liu Z and Zhang Y: DAL-1 attenuates
epithelial-to mesenchymal transition in lung cancer. J Exp Clin
Cancer Res. 34:32015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mittal V: Epithelial mesenchymal
transition in aggressive lung cancers. Adv Exp Med Biol. 890:37–56.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pattabiraman DR, Bierie B, Kober KI, Thiru
P, Krall JA, Zill C, Reinhardt F, Tam WL and Weinberg RA:
Activation of PKA leads to mesenchymal-to-epithelial transition and
loss of tumor-initiating ability. Science. 351:aad36802016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Seton-Rogers S: Epithelial-mesenchymal
transition: Untangling EMT's functions. Nat Rev Cancer. 16:12016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cocchia M, Huber R, Pantano S, Chen EY, Ma
P, Forabosco A, Ko MS and Schlessinger D: PLAC1, an Xq26 gene with
placenta-specific expression. Genomics. 68:305–312. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fant M, Farina A, Nagaraja R and
Schlessinger D: PLAC1 (Placenta-specific 1): A novel, X-linked gene
with roles in reproductive and cancer biology. Prenat Diagn.
30:497–502. 2010.PubMed/NCBI
|