Introduction
Osteosarcoma is one of the most common primary
malignant bone tumors in children, young adults and adolescents
(1,2). Despite newly developed multi-agent
chemotherapy and gradually improving surgical techniques, the
prognosis for patients with metastatic osteosarcoma is still poor
(3). It is important to investigate
the complex molecular mechanisms and identify novel biomarkers for
the treatment, diagnosis and prognosis of osteosarcoma. The copines
are a widely distributed class of calcium-dependent
phospholipid-binding proteins that are evolutionally conserved from
Arabidopsis to Homo sapiens (4,5).
Copine 1 (CPNE1), a soluble calcium-dependent membrane-binding
protein, is ubiquitously expressed in various tissues and organs.
CPNE1 is located on chromosome 20q11.21 region in humans and
has several alternative splicing forms coding for the same
537-amino acid protein (6). CPNE1
has a pair of C2 domains (C2A and C2B) at the N-terminus and a Von
Willebrand factor A (VWA) domain (A domain) at the C-terminus
(7). It was previously reported
that the C2 domains of CPNE1 may function in cell signaling and/or
membrane trafficking pathways, and the A domain facilitates the
binding of CPNE1 with various intracellular proteins (8,9).
Lentivirus-based vectors with small hairpin RNA (shRNA) have been
used as a successful tool for silencing target gene expression,
particularly in cancer cells, with high specificity, stability and
efficiency in vitro and in vivo (10,11).
However, no valid evidence concerning the biological function of
CPNE1 in osteosarcoma exists to date. In the present study, we
successfully silenced CPNE1 expression in Saos-2 and HOS
cells using RNA interference (RNAi) technology and investigated the
biological role of CPNE1 in osteosarcoma.
Materials and methods
Main reagents
Rabbit anti-human CPNE1 polyclonal antibody (cat.
no. AB155675) was purchased from Abcam (Cambridge, UK); mouse
anti-human antibodies to Ras (cat. no. YT2960), MEK-1/2 (cat. no.
YT2715), cyclin A1 (cat. no. YT1168) and IRAK2 (cat. no. YT2392)
were purchased from ImmunoWay Biotechnology Company (Plano, TX,
USA); mouse anti-human antibodies to WNT1 (cat. no. YM0649),
β-catenin (cat. no. YM3065) and cIAP2 (cat. no. Ym1343) were also
purchased from ImmunoWay Biotechnology Company; Lipofectamine™
2000, TRIzol reagent and Opti-MEM were purchased from Invitrogen
Corporation (Carlsbad, CA, USA); AgeI, EcoRI and
SYBR-Green Master Mix kits were purchased from New England Biolabs
(NEB; Beijing, China); Taq DNA polymerase was purchased from
Takara Biotechnology Co., Ltd. (Dalian, China). Cisplatin (DDP) was
purchased from Qilu Pharmaceutical (Hainan) Co., Ltd. (Haikou,
China). Adriamycin (ADR) was purchased from the Zhejiang Haizheng
Pharmaceutical Co., Ltd. (Taizhou, China).
Cell culture and tissue
collection
The human osteosarcoma cell lines (Saos-2 and HOS)
were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China) and were identified by short tandem
repeat (STR) method in 2015. Dulbecco's modified Eagle's medium
(DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine serum
(FBS) was used to culture the cells. All cells were maintained at
37°C in a humidified atmosphere with 5% CO2.
Twenty-five osteosarcoma and 8 cartilage tumor
tissues samples were obtained from patients who underwent surgery
at the Affiliated Yixing Hospital of Jiangsu University between
January 2005 and December 2015. All cases had been clinically and
pathologically confirmed. Immunohistochemistry was performed by
using 25 samples of osteosarcoma (13 males and 12 females; mean
age, 37 years) and 8 samples of cartilage tumor (4 males and 4
females; mean age, 28 years). The experimental protocols for the
present study were approved by the Hospital's Protection of Human
Subjects Committee.
Immunohistochemistry
Paraffin-embedded histological specimens were cut
into 4-µm thick sections. Then, sections were routinely dewaxed and
rehydrated in xylol, and graded alcohol. Endogenous peroxidase
activity was blocked with 3% hydrogen peroxide in
phosphate-buffered solution (PBS) for 15 min and non-specific
binding was blocked with 2% bovine serum for 20 min. The slides
were incubated with 1:100 diluted primary antibody against human
CPNE1 for 18 h at 4°C in 2% bovine serum albumin (BSA) in PBS. The
horseradish peroxidase-conjugated goat anti-rabbit IgG secondary
antibody was added and incubated for 1 h at 37°C. The immune
reaction was developed with
3,3′-diaminobenzidine-tetrahydrochloride-dihydrate (DAB). Slides
were washed with distilled water, counterstained with hematoxylin,
dehydrated and mounted. All sections were observed and analyzed
under a light microscope.
Lentiviral plasmid construction,
lentivirus production and cell infection
The human CPNE1 (GenBank accession no.
NM_003915)-specific small interfering RNA (siRNA) sequence, which
was designed using online software from Invitrogen, was
5′-CACACAACTGGTCTCATACTT-3′. The non-silencing (NS) sequence,
5′-TTCTCCGAACGTGTCACGT-3′, was used as a scrambled control
(12). The following
oligonucleotides were synthesized, annealed and ligated into the
pGCSIL-GFP plasmid vector between the AgeI and EcoRI
sites. Then, these plasmids were amplified in DH5α-competent
Escherichia coli cells and purified using the Qiagen
plasmid. Recombinant lentiviruses were produced in 293T cells by
co-transfection of the recombinant pGCSIL-GFP vector, along with
packaging plasmids, pHelper1.0 and pHelper2.0, using Lipofectamine™
2000. For lentivirus transduction, the Saos-2 and HOS cells were
subcultured at a density of 5×104 cells/well into 6-well
culture plates. After growing to 30% confluence, the recombinant
lentiviruses were transfected into cells at a multiplicity of
infection (MOI) of 20. Each cell line was divided into the
following groups: the scr-siRNA (cells infected with Lv-si-CTRL)
and the CPNE1-siRNA group (cells infected with Lv-si-CPNE1). At 48
h post infection, the infection efficiencies were determined using
a fluorescence microscope (Leica Microsystems, Wetzlar, Germany).
CPNE1-knockdown efficiency was evaluated by quantitative
reverse transcription-polymerase chain reaction (RT-qPCR) and
western blot analysis.
RNA extraction and RT-PCR
Total RNA was extracted from the cells after 5 days
of infection using TRIzol reagent (Invitrogen) in accordance with
the manufacturer's instructions, and then cDNA was synthesized from
the total RNA. Reverse transcription was performed using M-MLV
reverse transcriptase (Promega, Madison, WI, USA). The expression
level of CPNE1 was detected by qPCR using an SYBR-Green
Master Mixture (Takara Biotechnology Co., Ltd.). qPCR was performed
on a Bio-Rad Connect Real-Time PCR system. GAPDH was used as
an internal control. Relative gene expression levels were
calculated using 2−ΔΔCt analysis. Two sets of primers
were used for PCR: GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′; CPNE1 forward,
5′-ACCCACTCTGCGTCCTT-3′ and reverse, 5′-TGGCGTCTTGTTGTCTATG-3′.
Cellomics ArrayScan assay
Briefly, Saos-2 cells infected with the
lentiviral-mediated CPNE1-siRNA or scr-siRNA were seeded into
96-well plates at a density of 2×103 cells/well and
cultured at 37°C in a humidified atmosphere with 5% CO2.
The infected Saos-2 cells with green fluorescence were imaged and
counted on the Cellomics ArrayScan high-content screening (HCS)
reader once a day for 5 days. Each experiment was conducted at
least 3 times, independently. The data were collected and analyzed
to create a 5-day growth curve of the infected cells.
Methylthiazolyldiphenyl-tetrazolium
bromide (MTT) assay
After lentivirus infection, the Saos-2 and HOS cells
were plated in a 96-well plate at a density of 1×104
cells/well. At indicated time points, MTT was added to each well at
a final concentration of 5 mg/ml and incubated with the cells at
37°C for an additional 4 h. After removing the supernatants,
dimethyl sulfoxide (DMSO) was added to each well to terminate the
reaction. Absorbance was read at a wavelength of 490 nm using an
ELISA reader (Bio-Rad Systems, Hercules, CA, USA) and data were
analyzed. All experiments were performed in triplicate.
Colony formation assay
Lentivirus-transduced osteosarcoma cells were plated
in 6-well plates at a density of 400 cells/well. The cell culture
medium was changed every other day. After incubation at 37°C for 14
days, the colonies were fixed with 4% paraformaldehyde, stained
with Giemsa staining (Sigma-Aldrich, St. Louis, MO, USA) for 20
min, and rinsed with distilled water. The colonies were counted and
analyzed.
Transwell migration and invasion
assays
Transwell chambers (8.0-µm pore size; Costar,
Cambridge, NY, USA) with Matrigel (BD Biosciences, San Jose, CA,
USA) bedding were placed in 24-well plates. Briefly,
1×105 cells in serum-free DMEM were seeded into the
upper compartment of the chamber. The lower compartment of the
chamber was filled with 600 µl DMEM containing 15% FBS as a
chemoattractant. After incubation at 37°C for 48 h, the non-invaded
cells on the upper surface of the chamber membrane were scraped off
with a cotton swab, and the successfully translocated cells were
then fixed with paraformaldehyde and stained with crystal violet.
The invaded cells were quantified, and images were acquired under
magnification of ×200 (5 randomly selected fields). Cell migration
assay was also performed in the Transwell chambers following the
method for the invasion assay with minor modifications: the upper
compartment of the chamber was not pre-coated with Matrigel (BD
Biosciences), and 5×104 cells suspended in serum-free
DMEM were added to the upper chamber.
Flow cytometric analysis of cell cycle
istribution
The cell cycle distribution was assessed using flow
cytometry using propidium iodide (PI) staining. Briefly,
lentivirus-infected Saos-2 cells were harvested, re-suspended in
ice-cold PBS and fixed with 70% (v/v) cold alcohol at 4°C. After
washing thrice in PBS, Saos-2 cells were resuspended in RNase A
(Sigma)-PBS solution (100 µg/ml PI and 10 µg/ml RNase A) and
incubated in the dark at room temperature for 30 min. The
suspension was then filtered through a nylon mesh, and the DNA
content of stained nuclei was analyzed using a flow cytometer
(FACSCalibur; BD Biosciences) in accordance with the manufacturer's
guidelines.
Western blot analysis
Five days after infection, lentivirus-transduced
Saos-2 cells were collected and lysed in RIPA buffer (100 mM
Tris-HCl, 150 mM NaCl, 1% sodium deoxycholate, 1% Tween-20 and 0.1%
SDS) containing protease inhibitor mixture. After centrifugation
(5,000 rpm, 10 min), the supernatant was collected. The protein
concentration was determined by bicinchoninic acid (BCA) method.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide
gel electrophoresis (SDS-PAGE), and then transferred onto
polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA).
Free binding sites on the membranes were blocked with 5% BSA at
room temperature for 2 h, and subsequently incubated with different
antibodies, including anti-CPNE1 (1:1,000), anti-Ras (1:1,000),
anti-MEK-1/2 (1:1,000), anti-cyclin A1 (1:1,000), and anti-IRAK2
(1:1,000), anti-WNT1 (1:1,000), anti-β-catenin (1:1,000) and
anti-cIAP2 (1:1,000). The signals were detected by enhanced ECL
Plus Western Blotting Detection system (Amersham Biosciences, Inc.,
Piscataway, NJ, USA).
Chemosensitivity assay
In vitro drug sensitivity was evaluated by
MTT assay. After 72 h of transfection with CPNE1 siRNA or control
siRNA, the Saos-2 cells in 96-well plates were treated with various
concentrations of DDP (0.009765625, 0.01953125, 0.0390625,
0.078125, 0.15625, 0.3125, 0.625, 1.25, 2.5, 5 and 10 µg/ml) and
ADR (0.01953125, 0.0390625, 0.078125, 0.15625, 0.3125, 0.625, 1.25,
2.5, 5, 10 and 20 µg/ml). Then, 20 µl MTT (5 mg/ml) was added to
each well and incubated for 4 h at 37°C. Subsequently, DMSO (150
µl) was added to solubilize the formazan crystals. Absorbance was
measured using a microplate reader at 490 nm. The inhibition ratio
(%) and IC50 (drug concentration causing a 50%
inhibition of cell growth) were determined.
Statistical analysis
The results obtained are expressed as mean ±
standard deviations (SD) of at least 3 independent experiments. The
statistical significance of differences between groups was
determined by Student's t-test and one-way ANOVA using GraphPad
Prism 5.0 software (GraphPad Software, Inc., San Diego, CA, USA).
P<0.05 was considered statistically significant.
Results
CPNE1 is overexpressed in osteosarcoma
samples
CPNE1 expression was examined in 25 samples from
patients with osteosarcoma. Of the 25 patients, 21 (84.00%) were
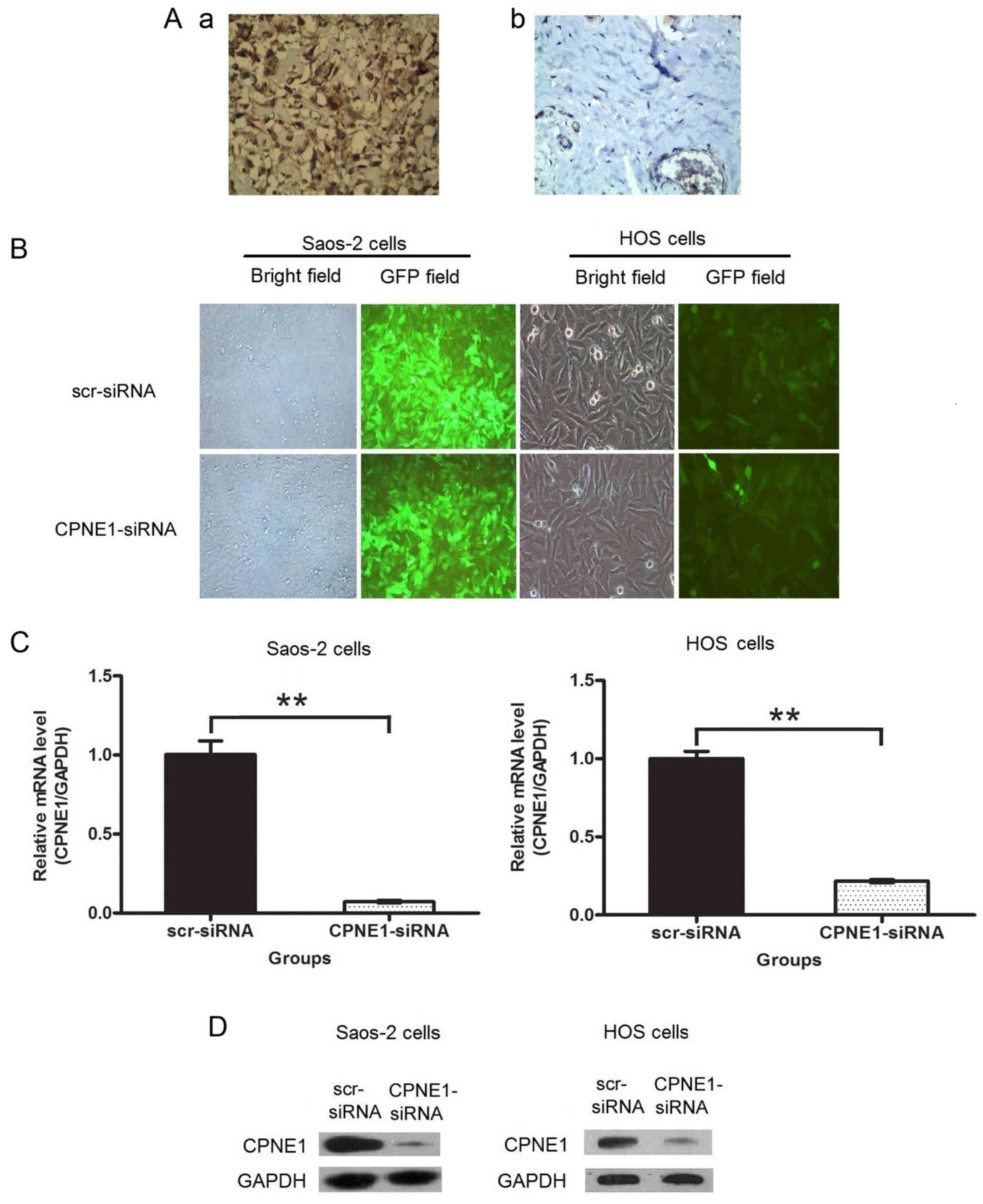
noted to be positive for CPNE1 expression. As shown in Fig. 1A, CPNE1 protein was observed to be
localized in the nuclei and cytoplasm of the osteosarcoma tissues
(a), while it was rarely detected in the cartilage tumor tissues
(b). The CPNE1 expression in the osteosarcoma tissues was
significantly higher than that in the cartilage tumor tissues.
These results demonstrated a correlation between CPNE1
overexpression and osteosarcoma occurrence. The lentiviral vector
infection efficiency was investigated using fluorescence
microscope. The results of transfection demonstrated that >90%
of the treated osteosarcoma cells exhibited green fluorescence
indicative of infection (Fig. 1B).
CPNE1 expression at the mRNA and protein levels were measured by
qPCR and western blot assays, respectively. The results indicate
that CPNE1 expression at the mRNA (Fig.
1C) and protein (Fig. 1D)
levels were significantly decreased in the Saos-2 and HOS cells
compared to that in the scr-siRNA groups. Thus, these results
confirmed that lentiviral-mediated RNAi efficiently downregulated
or blocked CPNE1 expression.
Growth inhibition of human
osteosarcoma cells by CPNE1 depletion
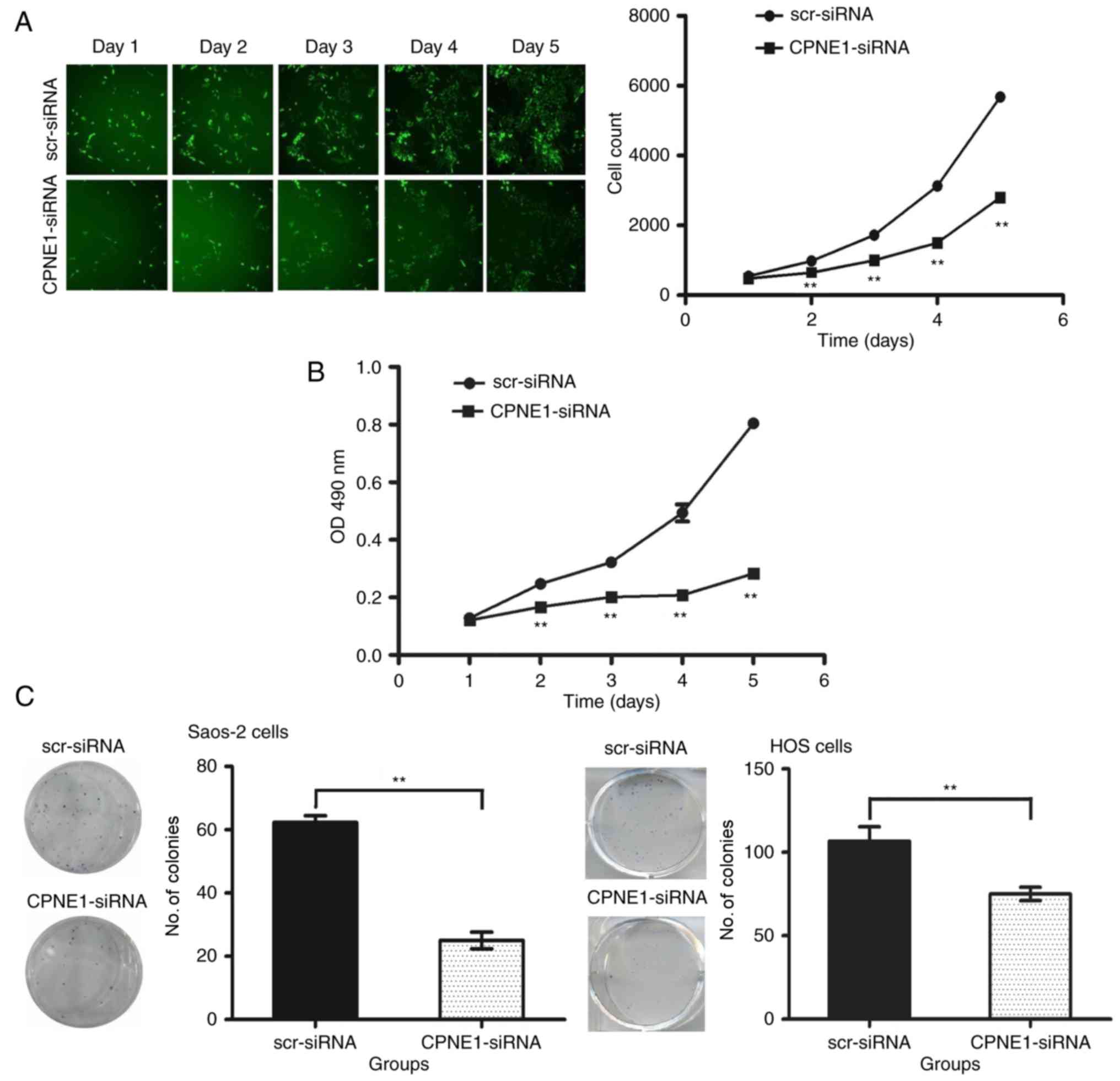
The effects of CPNE1-siRNA on the viability of
Saos-2 and HOS cells were studied in vitro. Cellomics
analysis showed that CPNE1 knockdown significantly inhibited
the growth of Saos-2 cells as compared to that observed after
scr-siRNA treatment, and the difference was more pronounced in a
time-dependent manner (P<0.01) (Fig.
2A). Although the lentiviruses were transfected into the Saos-2
cells and expressed the GFP and CPNE1 siRNA, the siRNA cannot
completely knock down CPNE1 in cells or suppress total CPNE1
expression at the high level. Therefore a small portion of the
cells kept proliferating. MTT assay was performed to investigate
the effect of CPNE1 silencing on osteosarcoma cell growth.
As shown in Fig. 2B, HOS cells
showed a significant (P<0.01) reduction in viability 2 days
after infection. The results suggest that CPNE1 silencing inhibits
the proliferation of osteosarcoma cells.
The results of the colony formation assay showed
that the number of colonies in the CPNE1-siRNA group (25.00±2.65)
was significantly less than that in the scr-siRNA group
(62.33±2.08) in the Saos-2 cells (P<0.01), and the number of
colonies in the CPNE1-siRNA group (75.00±4.00) was significantly
less than that in the scr-siRNA group (106.70±8.62) in the HOS
cells (P<0.01) (Fig. 2C). These
results showed that the reduction in CPNE1 expression decreased the
ability of osteosarcoma cells to form colonies.
siRNA-mediated CPNE1 knockdown
inhibits the invasion and migration of osteosarcoma cells
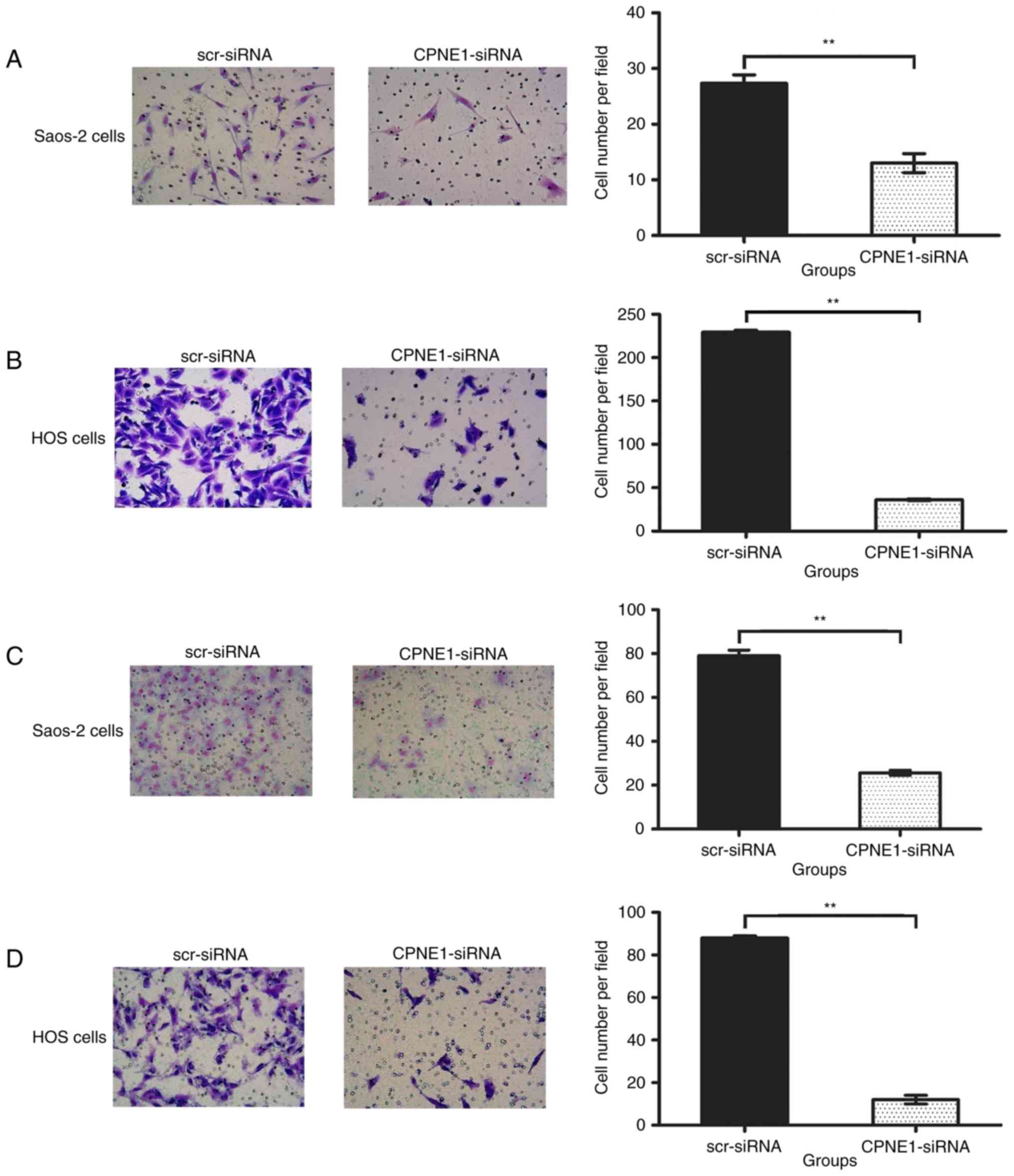
As shown in Fig. 3A and
B, in the Transwell invasion assay, the number of invading
Saos-2 cells was 13.00±1.73 in the CPNE1-siRNA group, which was
significantly less than that in the scr-siRNA group (27.33±1.53;
P<0.01), and the number of invading HOS cells was 36.00±1.00 in
the CPNE1-siRNA group, which was significantly less than that in
the scr-siRNA group (229.30±2.31; P<0.01). As shown in Fig. 3C and D, in the Transwell migration
assay, the number of migrating Saos-2 cells was 25.67±1.16 in the
CPNE1-siRNA group, which was significantly less than that in the
scr-siRNA group (79.00±2.65; P<0.01). The number of migrating
HOS cells was 12.00±2.00 in the CPNE1-siRNA group, which was
significantly less than that in the scr-siRNA group (88.00±1.00;
P<0.01). The result showed that CPNE1 silencing
suppressed invasion and metastasis of osteosarcoma cells.
CPNE1 silencing leads to alterations
in the cell cycle of Saos-2 cells
To elucidate the mechanisms underlying RNAi-mediated
growth inhibition, flow cytometric analysis of the DNA content was
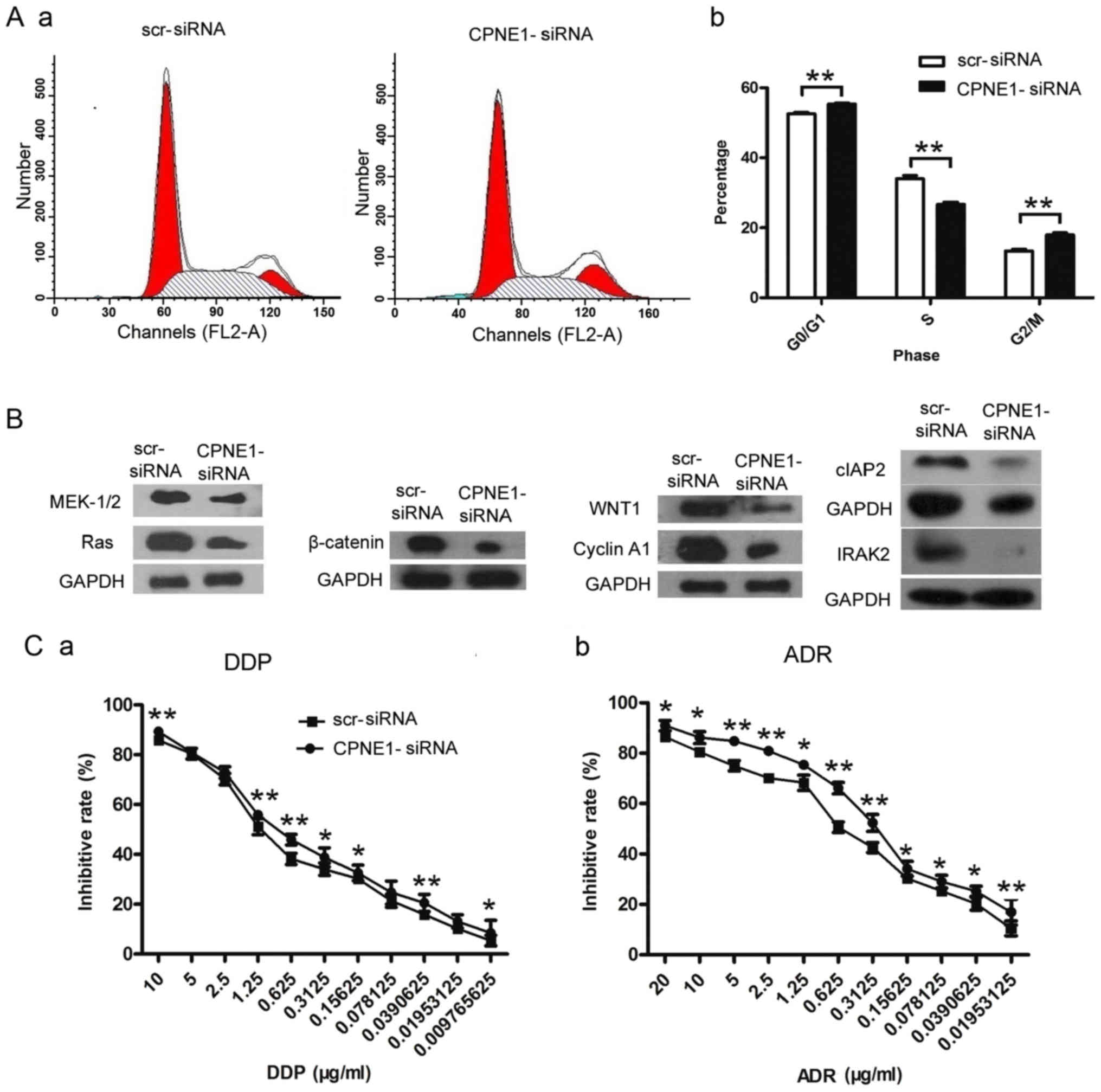
used to detect the changes in the cell cycle. As shown in Fig. 4A-a and -b, CPNE1-siRNA treatment
resulted in an increase in the percentage of Saos-2 cells in the
G2/M phase from 13.38±0.51 to 18.01±0.63% (P<0.01). In
accordance with this increase in the percentage of cells in the
G2/M phase, there was a significant decrease in the percentage of
cells in the S phase from 34.04±0.89 to 26.67±0.66% (P<0.01).
CPNE1-siRNA treatment also resulted in an increase in the
percentage of cells in the G0/G1 phase from 52.57±0.42 to
55.32±0.27% (P<0.01). These results suggested that CPNE1
depletion inhibited the proliferation of osteosarcoma cells through
G2/M and G0/G1 phase arrest of the cell cycle in Saos-2 cells.
Suppression of CPNE1 affects the
expression of related proteins
To test the possible mechanisms underlying the
inhibitory effects of CPNE1 knockdown on the biological
functions of Saos-2 cells, the expression of various related
proteins was examined. The results revealed that CPNE1
knockdown downregulated Ras, MEK-1/2, WNT1, β-catenin, cyclin A1,
cIAP2 and IRAK2 in the Saos-2 cells. These results suggest that the
inhibitory effects on the biological functions associated with
CPNE1 downregulation may be partly mediated by these related
proteins in Saos-2 cells (Fig. 4B).
Certainly, it is not clear whether these results are due to actions
at the transcriptional or translational or both levels. Thus,
further research is needed to elucidate the underlying molecular
mechanism.
CPNE1 silencing sensitizes Saos-2
cells to chemotherapeutic agents
The effect of CPNE1 silencing on the
sensitivity of Saos-2 cells towards DDP and ADR was determined. As
shown in Fig. 4C (a and b), the
IC50 for DDP in the CPNE1-siRNA group was 0.494±0.008
µg/ml at 72 h, which was significantly less than that in the
scr-siRNA group (0.832±0.030 µg/ml; P<0.01); the IC50
for ADR in the scr-siRNA group was 0.265±0.025 µg/ml at 72 h, which
was significantly less than that in the CPNE1-siRNA group
(0.619±0.066 µg/ml; P<0.01). These results suggested that Saos-2
cells could be effectively chemosensitized by siRNA-mediated
CPNE1 silencing.
Discussion
CPNE1 is a highly conserved protein and is
ubiquitously expressed in various tissues (6), but its biological function is not well
known. In the present study, we observed that CPNE1 is highly
expressed in osteosarcoma tissue, which indicates that CPNE1 may
play an important role in the development of osteosarcoma.
We also found that CPNE1 silencing inhibited
the proliferation, migration and invasion of Saos-2 and HOS cells.
In addition, CPNE1 knockdown caused cell cycle arrest in the
G2/M and G0/G1 phases in the Saos-2 cells. Yet, not only cell cycle
analysis, but also other important mechanisms (apoptotic or
necrotic cell death) are needed to be investigated in the future.
All these results provide direct evidence that CPNE1 may serve as a
target for osteosarcoma treatment.
To elucidate the mechanisms underlying the function
of CPNE1 in osteosarcoma, the expression of several related
proteins was examined. Ras is a membrane-bound GTP-binding protein
that functions as a molecular switche to transduce various signals
from the cell membrane to the nucleus. Activated Ras recruits and
activates Raf kinase at the plasma membrane. Activated Raf
subsequently activates mitogen-activated protein kinase/ERK kinase
MEK-1/2, which in turn phosphorylates and activates ERK1/2. Next,
activated ERK1/2 regulates various cellular processes (13,14).
The RAS/RAF/MEK/ERK signaling pathway is one of the most important
oncogenic pathways, which plays a central role in the regulation of
cell proliferation and survival. This pathway is aberrantly
activated in various malignancies (15). Oncoprotein β-catenin, which is
normally localized in the cytoplasm, funtions as an important
transcriptional co-activator and transmits extracellular signals
for the activation of certain target genes in the canonical Wnt
pathway (16,17). WNT1 binds to the target cell surface
receptors of the Frizzled (Fzd) family to activate several
different intracellular signal transduction pathways, resulting in
β-catenin accumulation and nuclear translocation. Nuclear β-catenin
induces the expression of downstream target genes, such as
E-cadherin, c-Myc, and cyclin D1 (18–20).
Cyclin A1 is highly expressed in cancers of the ovary, breast, lung
and prostate (21–24). cyclin A1 is associated with the
enhanced proliferation and invasiveness of various types of cancers
(25–28). Inhibitor of apoptosis proteins
(IAPs) are widely expressed in human tumor tissues and play an
important role in cell apoptosis (29). Among these, cellular IAP2 (cIAP2)
indirectly regulates apoptosis by preventing the formation of
caspase-8-activating platform and blocking Smac-mediated
XIAP-caspase interaction (30,31).
Interleukin 1 receptor-associated kinase 2 (IRAK2) plays a critical
role in sustaining NF-κB activation during TLR-mediated signaling,
and IRAK2 overexpression activates NF-κB (32–34).
Notably, all the above factors, including Ras,
MEK-1/2, WNT1, β-catenin, cyclin A1, cIAP2 and IRAK2, were
downregulated after CPNE1 knockdown, which indicated that
the inhibition of the proliferation, migration, and invasion of
human osteosarcoma cells after CPNE1 silencing is through
downregulation of these factors. Further experiments may be
essentially performed in future research.
Cisplatin (DDP) is a first-line chemotherapeutic
agent that is widely used in various types of cancers, including
those of the lung, bladder, cervix, ovary, endometrium and
testicles (35,36). Its cytotoxic effects are mediated by
interaction with cellular DNA to form DNA adducts, which activates
several signal transduction pathways, and culminates in the
activation of cell apoptosis (37).
Adriamycin (ADR) is also a first-line chemotherapeutic drug in the
treatment of various types of cancers and induces DNA damage by
topoisomerase II inhibition and free radical generation as an
anticancer mechanism (38,39).
To further confirm the synergistic effects of CPNE1
inhibition with typical cancer chemotherapeutic drugs on the
suppression of osteosarcoma cell proliferation, we treated Saos-2
cells with different doses of the two chemotherapeutic drugs (DDP
and ADR) combined with CPNE1 knockdown. The results showed
that CPNE1 knockdown enhanced the suppression of effects of
these two drugs on Saos-2 cell proliferation. This is the direct
evidence that CPNE1 depletion could be combined with
first-line chemotherapeutic drugs for treating osteosarcoma.
In conclusion, the present study firstly
demonstrated that RNAi-mediated CPNE1 downregulation
inhibited the proliferation and metastatic potential of
osteosarcoma cells, and reduced expression of various tumor-related
genes. In addition, blocking CPNE1 expression enhanced the
chemosensitivity of osteosarcoma cells. However, the molecular
mechanism of CPNE1 is complex, and further detailed studies and
clinical trials are required.
Acknowledgements
The present study was supported by a grant from the
Natural Science Foundation for the Youth (Beijing, China) (no.
81402220).
References
|
1
|
Ando K, Heymann MF, Stresing V, Mori K,
Rédini F and Heymann D: Current therapeutic strategies and novel
approaches in osteosarcoma. Cancers. 5:591–616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perry JA, Kiezun A, Tonzi P, Van Allen EM,
Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS,
et al: Complementary genomic approaches highlight the PI3K/mTOR
pathway as a common vulnerability in osteosarcoma. Proc Natl Acad
Sci USA. 111:pp. E5564–E5573. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maugg D, Rothenaigner I, Schorpp K,
Potukuchi HK, Korsching E, Baumhoer D, Hadian K, Smida J and
Nathrath M: New small molecules targeting apoptosis and cell
viability in osteosarcoma. PLoS One. 10:e01290582015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tomsig JL and Creutz CE: Copines: A
ubiquitous family of Ca(2+)-dependent phospholipid-binding
proteins. Cell Mol Life Sci. 59:1467–1477. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maitra R, Grigoryev DN, Bera TK, Pastan IH
and Lee B: Cloning, molecular characterization, and expression
analysis of Copine 8. Biochem Biophys Res Commun. 303:842–847.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Creutz CE, Tomsig JL, Snyder SL, Gautier
MC, Skouri F, Beisson J and Cohen J: The copines, a novel class of
C2 domain-containing, calcium-dependent, phospholipid-binding
proteins conserved from Paramecium to humans. J Biol Chem.
273:1393–1402. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perestenko PV, Pooler AM, Noorbakhshnia M,
Gray A, Bauccio C and Jeffrey McIlhinney RA: Copines-1, −2, −3, −6
and −7 show different calcium-dependent intracellular membrane
translocation and targeting. FEBS J. 277:5174–5189. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomsig JL, Snyder SL and Creutz CE:
Identification of targets for calcium signaling through the copine
family of proteins. Characterization of a coiled-coil
copine-binding motif. J Biol Chem. 278:10048–10054. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomsig JL, Sohma H and Creutz CE:
Calcium-dependent regulation of tumour necrosis factor-alpha
receptor signalling by copine. Biochem J. 378:1089–1094. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hwang LH: Gene therapy strategies for
hepatocellular carcinoma. J Biomed Sci. 13:453–468. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Follenzi A and Gupta S: The promise of
lentiviral gene therapy for liver cancer. J Hepatol. 40:337–340.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zielske SP and Stevenson M: Importin 7 may
be dispensable for human immunodeficiency virus type 1 and simian
immunodeficiency virus infection of primary macrophages. J Virol.
79:11541–11546. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rocks O, Peyker A and Bastiaens PI:
Spatio-temporal segregation of Ras signals: One ship, three
anchors, many harbors. Curr Opin Cell Biol. 18:351–357. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Campbell SL, Khosravi-Far R, Rossman KL,
Clark GJ and Der CJ: Increasing complexity of Ras signaling.
Oncogene. 17(11 Reviews): 1–1413. 1998.PubMed/NCBI
|
|
15
|
Prior IA, Lewis PD and Mattos C: A
comprehensive survey of Ras mutations in cancer. Cancer Res.
72:2457–2467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim W, Kim M and Jho EH: Wnt/β-catenin
signalling: From plasma membrane to nucleus. Biochem J. 450:9–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sasaya K, Sudo H, Maeda G, Kawashiri S and
Imai K: Concomitant loss of p120-catenin and β-catenin membrane
expression and oral carcinoma progression with E-cadherin
reduction. PLoS One. 8:e697772013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LT, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arsenic R, Braicu EI, Letsch A, Dietel M,
Sehouli J, Keilholz U and Ochsenreither S: Cancer-testis antigen
cyclin A1 is broadly expressed in ovarian cancer and is associated
with prolonged time to tumor progression after platinum-based
therapy. BMC Cancer. 15:7842015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shames DS, Girard L, Gao B, Sato M, Lewis
CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, et al: A
genome-wide screen for promoter methylation in lung cancer
identifies novel methylation markers for multiple malignancies.
PLoS Med. 3:e4862006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Syed Khaja AS, Dizeyi N, Kopparapu PK,
Anagnostaki L, Härkönen P and Persson JL: Cyclin A1 modulates the
expression of vascular endothelial growth factor and promotes
hormone-dependent growth and angiogenesis of breast cancer. PLoS
One. 8:e722102013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wegiel B, Bjartell A, Ekberg J, Gadaleanu
V, Brunhoff C and Persson JL: A role for cyclin A1 in mediating the
autocrine expression of vascular endothelial growth factor in
prostate cancer. Oncogene. 24:6385–6393. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji P, Agrawal S, Diederichs S, Bäumer N,
Becker A, Cauvet T, Kowski S, Beger C, Welte K, Berdel WE, et al:
Cyclin A1, the alternative A-type cyclin, contributes to G1/S cell
cycle progression in somatic cells. Oncogene. 24:2739–2744. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim J, Kim WJ, Liu Z, Loda M and Freeman
MR: The ubiquitin-specific protease USP2a enhances tumor
progression by targeting cyclin A1 in bladder cancer. Cell Cycle.
11:1123–1130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marlow LA, von Roemeling CA, Cooper SJ,
Zhang Y, Rohl SD, Arora S, Gonzales IM, Azorsa DO, Reddi HV, Tun
HW, et al: Foxo3a drives proliferation in anaplastic thyroid
carcinoma through transcriptional regulation of cyclin A1: A
paradigm shift that impacts current therapeutic strategies. J Cell
Sci. 125:4253–4263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wegiel B, Bjartell A, Culig Z and Persson
JL: Interleukin-6 activates PI3K/Akt pathway and regulates cyclin
A1 to promote prostate cancer cell survival. Int J Cancer.
122:1521–1529. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hoeller D and Dikic I: Targeting the
ubiquitin system in cancer therapy. Nature. 458:438–444. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vucic D, Dixit VM and Wertz IE:
Ubiquitylation in apoptosis: A post-translational modification at
the edge of life and death. Nat Rev Mol Cell Biol. 12:439–452.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vandenabeele P and Bertrand MJ: The role
of the IAP E3 ubiquitin ligases in regulating pattern-recognition
receptor signalling. Nat Rev Immunol. 12:833–844. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rhyasen GW and Starczynowski DT: IRAK
signalling in cancer. Br J Cancer. 112:232–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin SC, Lo YC and Wu H: Helical assembly
in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature.
465:885–890. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kawagoe T, Sato S, Matsushita K, Kato H,
Matsui K, Kumagai Y, Saitoh T, Kawai T, Takeuchi O and Akira S:
Sequential control of Toll-like receptor-dependent responses by
IRAK1 and IRAK2. Nat Immunol. 9:684–691. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang D and Lippard SJ: Cellular processing
of platinum anticancer drugs. Nat Rev Drug Discov. 4:307–320. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Teng ZY, Cheng XL, Cai XT, Yang Y, Sun XY,
Xu JD, Lu WG, Chen J, Hu CP, Zhou Q, et al: Ancient Chinese formula
Qiong-Yu-Gao protects against cisplatin-induced nephrotoxicity
without reducing anti-tumor activity. Sci Rep. 5:155922015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Siddik ZH: Cisplatin: Mode of cytotoxic
action and molecular basis of resistance. Oncogene. 22:7265–7279.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee K, Qian DZ, Rey S, Wei H, Liu JO and
Semenza GL: Anthracycline chemotherapy inhibits HIF-1
transcriptional activity and tumor-induced mobilization of
circulating angiogenic cells. Proc Natl Acad Sci USA. 106:pp.
2353–2358. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mizutani H, Tada-Oikawa S, Hiraku Y,
Kojima M and Kawanishi S: Mechanism of apoptosis induced by
doxorubicin through the generation of hydrogen peroxide. Life Sci.
76:1439–1453. 2005. View Article : Google Scholar : PubMed/NCBI
|