Introduction
Esophageal cancer (EC) is the eighth most common
highly aggressive gastrointestinal cancer and is the sixth leading
cause of cancer-related deaths globally (1). In 1990 EC caused 345,000 deaths and
this number rose to 400,000 in 2012 (2). The methods for diagnosing EC have
since been improved and ionizing radiation (IR) now plays an
important role in the treatment of human EC. Combining radiotherapy
with chemotherapy improves the effects of treatment for advanced
EC, however, the 5-year survival rate is still poor. Intrinsic
radioresistance accounts for the high recurrence and poor 5-year
survival rate of patients with EC (3). Thus, there is an urgent need to extend
our understanding of the molecular mechanisms underlying the
progression of EC and provide new strategies to overcome EC
radioresistance by manipulating key targets.
Multiple genetic and epigenetic alterations are
involved in the growth of ECs, including tumor suppressor gene
(TSG) mutations, loss of heterozygosity and promoter methylation,
as well as overexpression of oncogenes (4,5).
However, the underlying molecular mechanisms that cause
carcinogenesis are poorly understood. Epigenetic events, such as
aberrant de novo methylation of gene promoters, have
recently been defined as markers of human cancers and therapeutic
strategies targeting these mechanisms are currently being tested in
several clinical trials (6,7). Among the compounds being tested, the
DNA methyltransferase inhibitors (DNMTIs), such as decitabine
(5-aza-2′-deoxycytidine), have been demonstrated to reverse the
silencing of tumor-suppressor genes, such as PRKD1, TP53 and ESR1,
thus upregulating their expression (8–10).
However, the effectiveness of these compounds
depends on their incorporation into DNA, which may lead to
dose-dependent cytotoxicity (11).
Due to the potential toxicity of nucleoside analogs, there has been
a focus on discovering new compounds that may directly target
DNMTs. RG108, a non-nucleoside analog designed to target human
DNMT1, binds to the active site of DNMT and has been demonstrated
to be effective in reactivating several tumor-suppressor genes in
human colon cancer cells without affecting the methylation status
of centromeric repeats (12). In
addition, RG108 lacks the high levels of cytotoxicity associated
with 5-Aza-dCR, which inhibits DNA methyltransferase activity to
achieve DNA demethylation (12,13).
Thus, the aim of the present study was to assess the impact of
RG108 on the viability, radiosensitization, apoptosis and cell
cycle progression of EC cells.
Materials and methods
Cell culture and irradiation
treatment
The commonly used esophageal squamous cancer cell
lines Eca-109 and TE-1 which were obtained from the American Type
Culture Collection (ATCC, Manassas, VA, USA) were maintained in
Dulbecco's modified Eagle's medium (DMEM; HyClone Laboratories,
Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone
Laboratories), 100 µg/ml streptomycin and 100 U/ml penicillin in a
humidified incubator at 37°C with 5% CO2. RG108 was
purchased from Selleck Chemicals (Houston, TX, USA), dissolved in
dimethyl sulfoxide (DMSO) and stored at −20°C until use. The cells
were pretreated with different concentrations of RG108 diluted in
DMEM, followed by irradiation with a single dose of X-ray
irradiation using a linear accelerator (Rad Source Technologies
Inc., Suwanee, GA, USA) at a dose rate of 1.15 Gy/min and 160 kV
X-ray energy.
Cell viability assay
The viability of cells was evaluated using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. The cells were seeded at a density of
2.5×103/well in 96-well flat bottom plates 24 h before
treatment with RG108 and/or irradiation. The cells were then
incubated at 37°C in a 5% CO2 environment for 48 h.
Twenty microliters of a 5 mg/ml MTT solution were added into each
well and the cells were incubated for another 4 h. Then the
supernatant was removed and the cells were lysed by adding 100 µl
of DMSO. The absorbance at 490 nm was assessed by an enzyme-linked
immunosorbent assay reader. All tests were repeated three times
independently.
Colony formation assay
The cells were seeded in triplicate into 6-well flat
bottom plates at a density of 200–6,000 cells/well depending on the
dose of radiation. Subsequently, the cells were treated with or
without 25 µM RG108 for 6 h and the supernatant was removed. Then,
the cells were irradiated with 0, 2, 4, 6 or 8 Gy X-ray radiation
and incubated for 12 days to form cell clones. Subsequently, the
cells were fixed with methanol and stained with 0.1% crystal
violet. The cells were manually counted under a dissecting
microscope and clones were defined as groups of more than 50 cells.
The radiation sensitivity enhancement ratios (SER) were calculated
according to the multi-target single hit model. Each experiment was
repeated three times.
Cell apoptosis and cell cycle
analysis
The apoptotic cells were quantified using an Annexin
V/7-aminoactinomycin D (7-AAD) double staining kit according to the
manufacturer's instructions (BD Biosciences, San Jose, CA, USA).
Briefly, the cells were exposed to 25 µM RG108 for 6 h and then
harvested 48 h after treatment with either 6 Gy of X-ray
irradiation or sham treatment. The percentage of apoptotic cells
was assessed with a FACSCalibur system (BD Biosciences). The
percentage of both Annexin V+/7AAD− cells,
early in the apoptotic process, and Annexin
V+/7AAD+ cells, in late apoptosis, was
quantified.
For the cell cycle analysis, the cells were
collected 24 h after treatment with RG108 or RG108 combined with 6
Gy of X-ray irradiation and fixed with 70% precooled ethanol
overnight. After staining with propidium iodide (PI, 10 µg/ml;
Sigma-Aldrich, St. Louis, MO, USA) in the dark for 30 min, flow
cytometry was performed with a FACSCalibur system using ModFit LT
software (Verity Software House Inc., Topsham, ME, USA).
Western blot analysis
The cells were harvested 48 h after treatment with
RG108 and/or 6 Gy irradiation. Then they were incubated for 40 min
on ice with ice-cold RIPA buffer (50 mM Tris pH 7.2, 150 mM NaCl,
1% NP-40, 1% sodium deoxycholate, 0.05% SDS and 1 mM PMSF). After
centrifugation at 4°C for 5 min at 12,000 × g, supernatants were
analyzed with an enhanced BCA protein assay kit (Beyotime Institute
of Biotechnology, Nantong, China). The proteins were separated by
sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE) with 12% gels and transferred onto polyvinylidene
fluoride (PVDF) membranes. The gels were run and transferred under
the same experimental conditions. The membranes were blocked with
5% skim milk in TBS containing 0.1% Tween-20 (TBST) for 1 h at room
temperature, followed by incubation overnight at 4°C with either
Bax (1:200; mouse; cat. no. sc-20067) or Bcl-2 primary antibodies
(1:1,000; mouse; cat. no. sc-7382; both from Santa Cruz
Biotechnology, Santa Cruz, CA, USA). The membranes were washed
three times in TBST and incubated with horseradish
peroxidase-conjugated anti-mouse or anti-rabbit secondary
antibodies (1:1,000; cat. no. A0192) for 2 h at room temperature.
β-actin (Beyotime Institute of Biotechnology) was used as a loading
control.
Tumor xenografts
We established a tumor transplantation model in
BALB/c mice (SLAC Laboratory Animal Center, Shanghai, China) with
Eca-109 cells. The mice were kept under specific pathogen-free
conditions on a 12-h light-dark cycle. To produce tumors,
1×107 Eca-109 cells were suspended in 150 µl of PBS and
subcutaneously injected into each posterior flank region of
5-week-old male BALB/c nude mice (~18 g). When the tumors reached
150–200 mm3, the mice were randomly divided into four
groups as follows (5 mice/group): i) control (no radiation,
injection of 100 µl of sterile DMSO daily for 6 days); ii) RG108
alone (no radiation, injection of RG108 daily for 6 days, 50 mg/kg
total in a volume of 100 µl); iii) IR plus DMSO (injection of 100
µl of 0.01% sterile DMSO daily for 6 days, followed by 8 Gy
radiation 1 h after the last DMSO injection); and iv) IR plus RG108
treatment (injection of RG108 daily for 6 days, 50 mg/kg total in a
volume of 100 µl, followed by 8 Gy radiation after the last RG108
treatment). Both DMSO and RG108 were intraperitoneally injected.
The tumor sizes were assessed with digital calipers at regular
intervals and their volumes were calculated according to the
following formula: tumor volume = 0.52 × length × width2
(14). Tumor growth curves were
produced and data are presented as the mean ± SEM. The animals were
sacrificed 30 days after the first inoculation and their tumors
were frozen at −80°C or fixed in 10% formalin overnight and
subjected to IHC analysis. The animal experiments were approved by
the Institutional Animal Care and Use Committee of Soochow
University.
RNA-seq analysis of gene
expression
Total RNA was extracted from cells with TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). RNA
purity was checked using a Nano Photometer®
spectrophotometer (Implen Inc., Westlake Village, CA, USA) and RNA
concentrations were assessed using a Qubit® RNA Assay
kit with a Qubit® 2.0 Fluorometer (Invitrogen Life
Technologies). The transcriptome library for sequencing was
generated using a VAHTSTM mRNA-seq v2 Library Prep Kit for
Illumina® (Vazyme Biotech Co., Ltd, Nanjing, China) and
the index-coded samples were clustered using VAHTS RNA Adapters
set1/set2 for Illumina (Vazyme Biotech) according to the
manufacturer's instructions. After clustering, the libraries were
sequenced on an Illumina Hiseq X Ten platform using a (2×150 bp)
paired-end module. The raw images were transformed into raw reads
by base calling using CASAVA 1.8 (http://www.illumina.com/support/documentation.ilmn).
In addition, cluster analysis, Gene Ontology (GO) and pathway
enrichment analysis (KOBAS 2.0) of differentially expressed genes
were implemented.
Statistical analysis
Data are presented as the mean ± SEM from three
independent experiments. Differences were analyzed using Student's
t-test when only two groups were present, while differences among
more than two groups were tested by one-way analysis of variance
(ANOVA). The interaction between RG108 and radiation was assessed
using two-way ANOVA for both in vitro and in vivo
efficacy assays. SPSS 19.0 (SPSS, Inc., Chicago, IL, USA) was used
for all statistical analyses. Differences were considered
statistically significant when P<0.05.
Results
Growth inhibition effects of RG108 in
EC cells
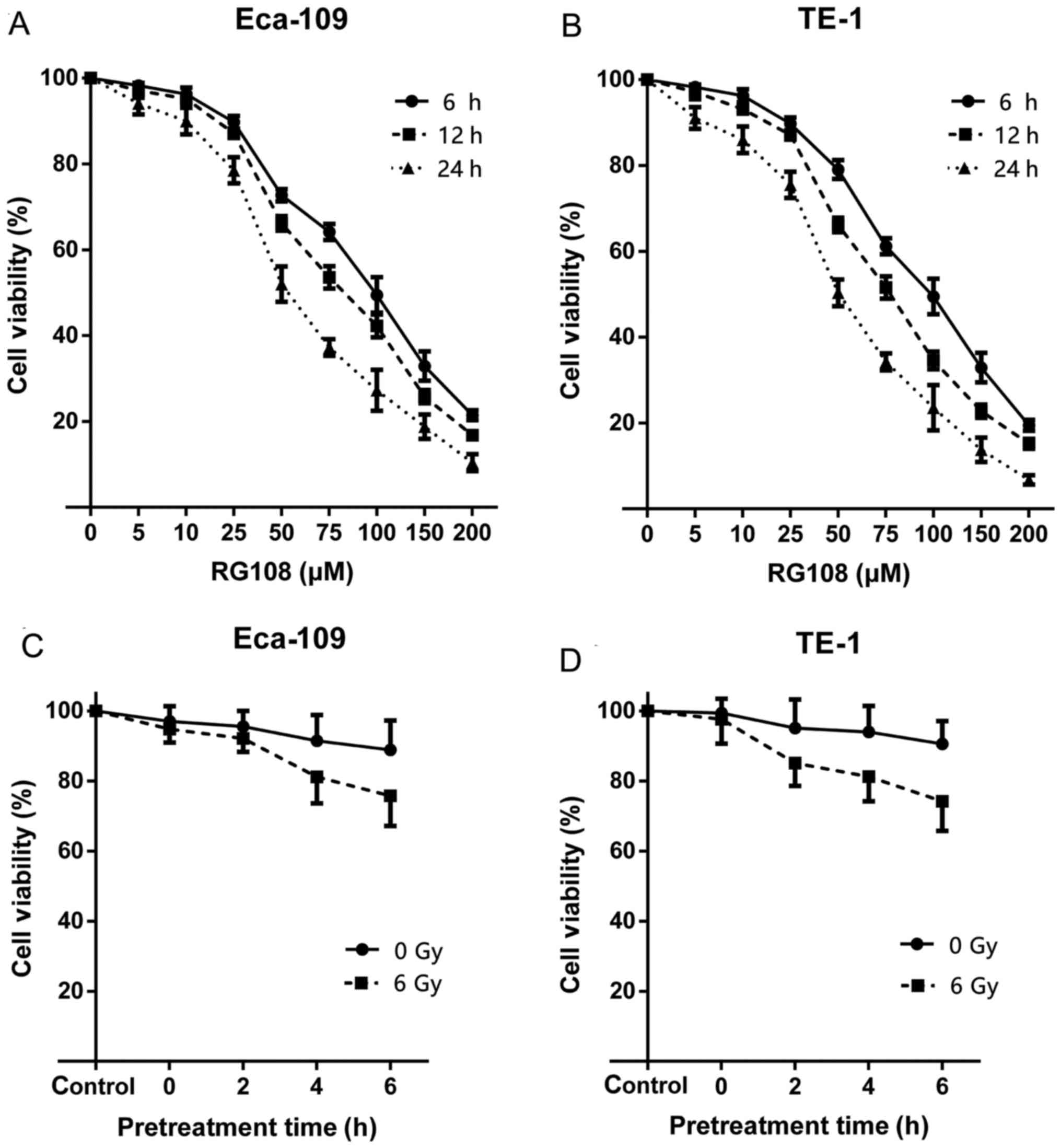
To evaluate the inhibitory effects of RG108 on the
growth of the cultured human EC cell lines Eca-109 and TE-1, we
first tested whether RG108 alone, at a range of different
concentrations, inhibited the proliferation of human EC cells. The
results indicated that RG108 inhibited cell proliferation in both
Eca-109 and TE-1 cells in a dose- and time-dependent manner
(Fig. 1A and B). The 50% inhibitory
concentrations (IC50) of RG108 against Eca-109 and TE-1
cells were 70 and 75 µM, respectively. We selected 25 µM RG108,
which resulted in ~90% cell viability, as the non-toxic dose for
the subsequent experiments.
We next assessed whether pretreatment with RG108
enhanced the anti-proliferative effect of irradiation on EC cells.
The results revealed that pretreatment with 25 µM of RG108
inhibited proliferation of both cell lines in a time-dependent
manner (Fig. 1C and D). Given that
6 h of RG108 pretreatment before IR sensitized the EC cells to
IR-induced cell death, this pretreatment time was used for further
studies.
RG108 at a non-toxic dose reduces
focus formation and enhances the anti-growth effect of radiation in
both Eca-109 and TE-1 cells
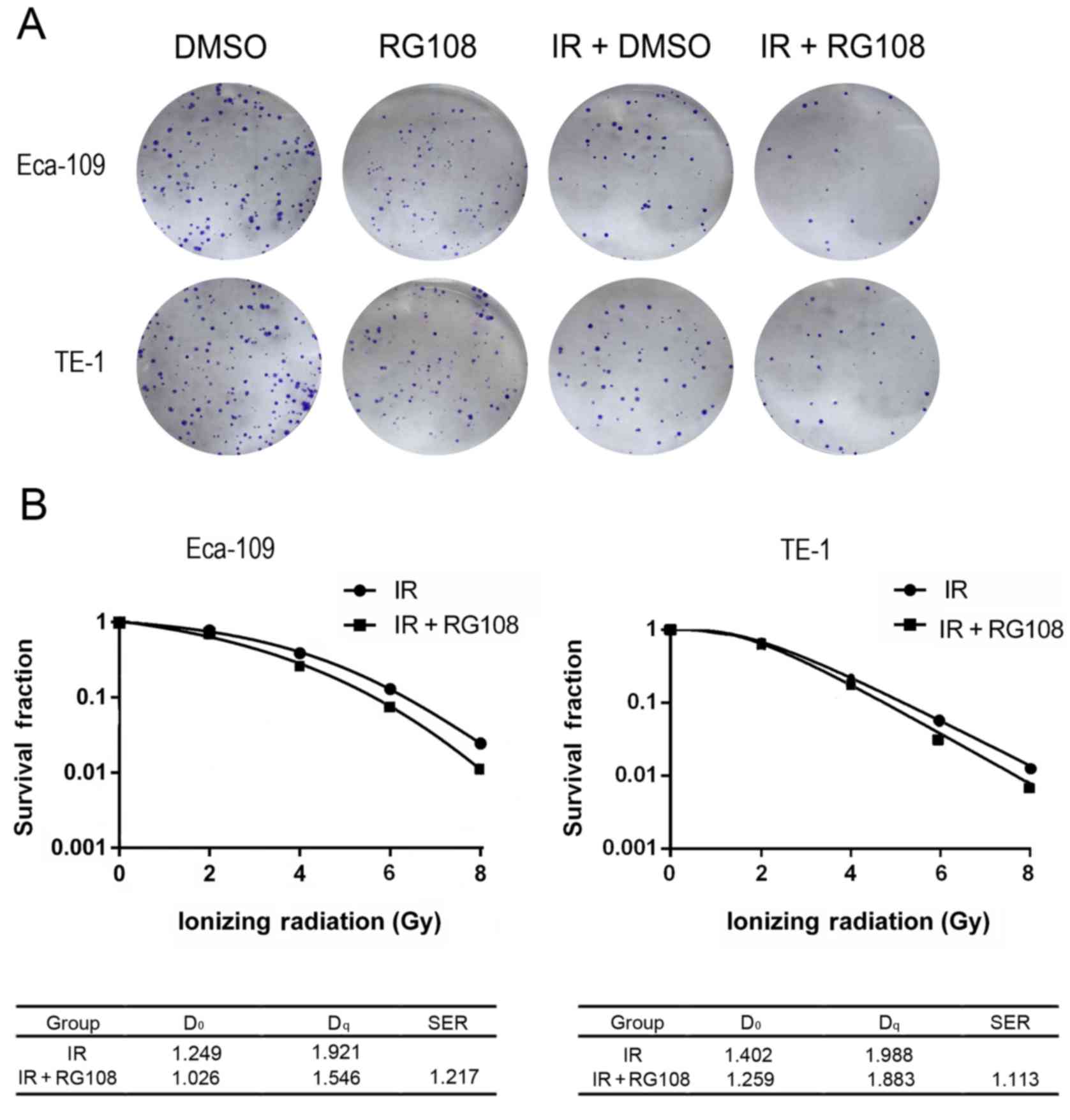
Clonogenic assays were performed to investigate the
effect of a low RG108 dose on the radiosensitivity of EC cell
lines. Firstly, we examined the clonogenicity of cells after
treatment with 25 µM RG108 or an equivalent volume of DMSO
(control) for 6 h with or without IR. As shown in Fig. 2A, the number of colonies after
treatment with 25 µM RG108 and 6 Gy of X-ray irradiation was
significantly decreased compared with the control (P<0.05) for
both Eca-109 and TE-1 cells. Thus, RG108 reduced the
radioresistance of both Eca-109 and TE-1 cells (SER = 1.217,
Fig. 2B). These results
demonstrated that a non-toxic dose of RG108 (25 µM) decreased focus
formation and significantly reduced the radioresistance of EC
cells.
RG108 modulates apoptosis of human EC
cells after IR
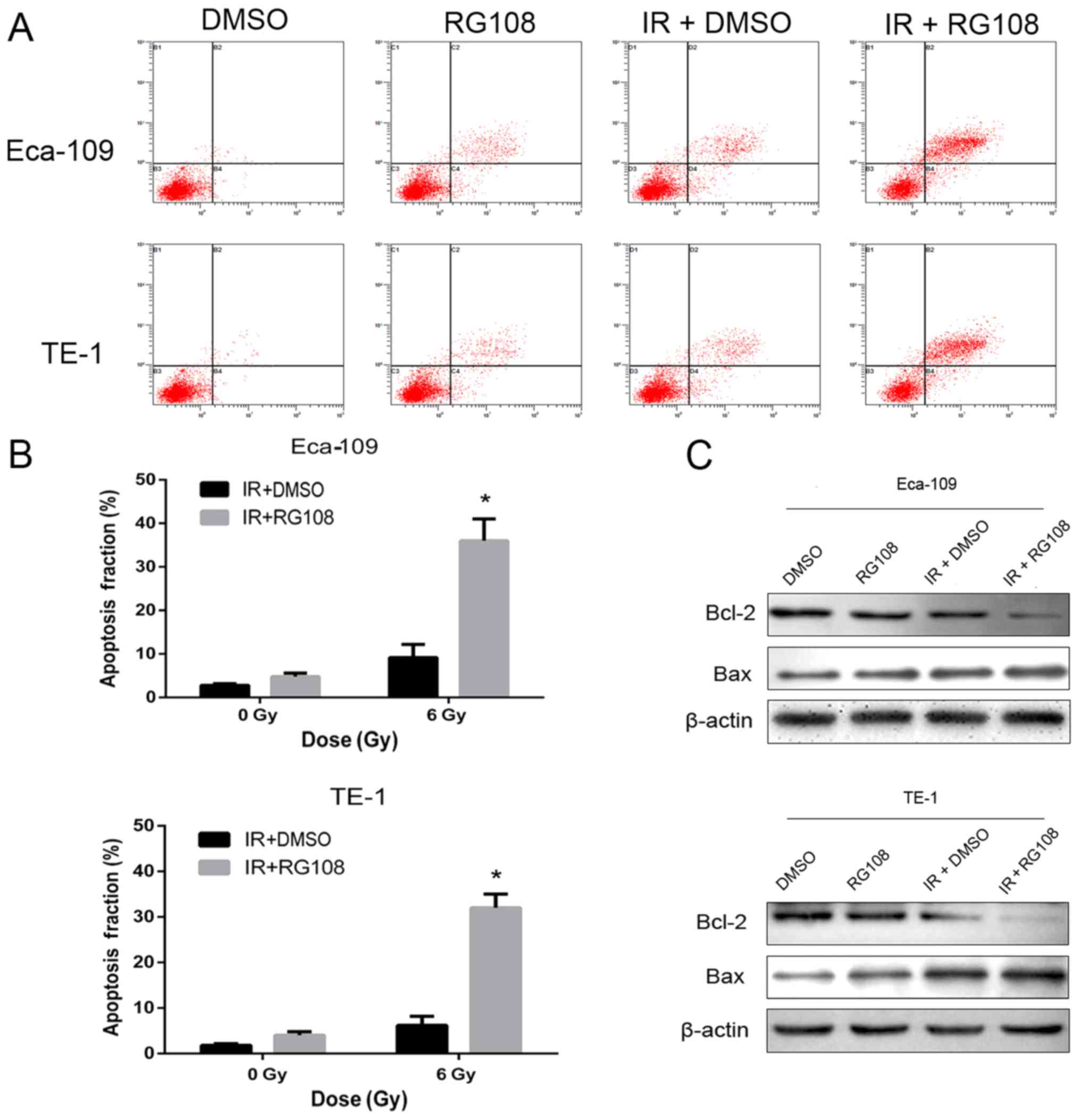
To investigate whether RG108 at a non-toxic dose
could enhance radiation-induced apoptosis in EC cells, we performed
Annexin V/7-AAD double-staining assays to analyze cellular
apoptosis. As depicted in Fig. 3A and
B, compared with control cells, apoptosis was modestly
increased in both cell lines by treatment with RG108 or IR alone.
However, apoptosis was significantly increased by treating cells
with 25 µM RG108 for 6 h before exposure to IR in both cell
lines.
Subsequently, we examined whether RG108 can regulate
the expression of genes associated with apoptosis by western blot
analysis. The results revealed that pretreatment with 25 µM RG108
for 6 h before 6 Gy irradiation significantly inhibited the
expression of Bcl-2, while it upregulated the expression of Bax, in
both Eca-109 and TE-1 cells (Fig.
3C). Collectivelly, these results indicated that apoptosis of
EC cells was significantly increased by RG108 at a range of
non-toxic doses in combination with IR.
RG108 combined with IR enhances G2/M
cell cycle arrest in EC cells
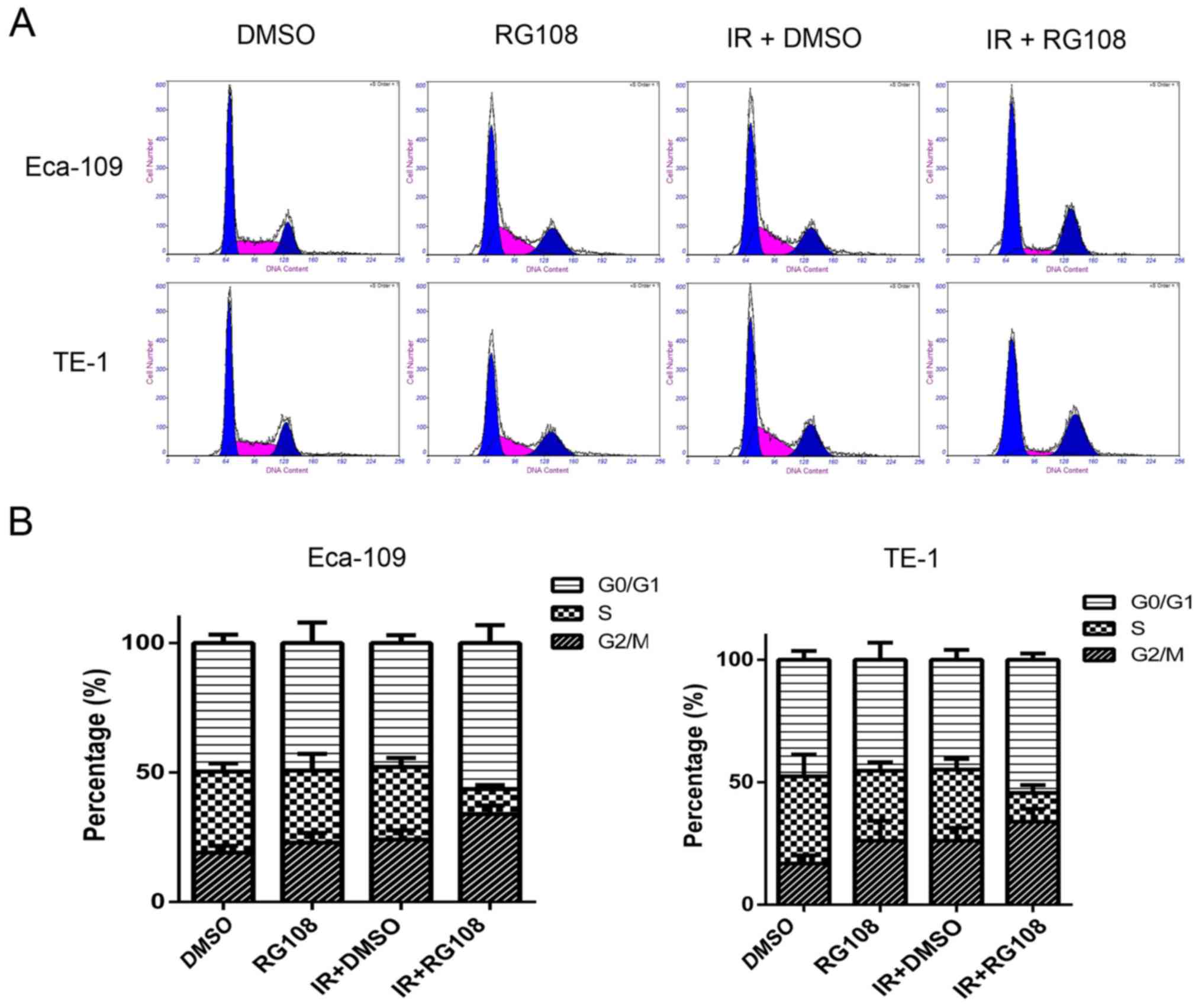
Eca-109 and TE-1 cells were mock treated or
pretreated with 25 µM RG108 for 6 h. Subsequently, the cells were
exposed to IR and the cell cycle was analyzed after 48 h. As
demonstrated in Fig. 4, compared
with the control group, pretreatment with 25 µM RG108 alone did not
change the distribution of the cell cycle phases in either of the
two cell lines. While 6 Gy of X-ray irradiation alone induced a
slight increase of Eca-109 and TE-1 cells in G2/M phase,
pretreatment with 25 µM RG108 for 6 h in combination with IR
significantly increased this radiation-induced arrest inthe G2/M
phase by 9.77% in Eca-109 cells and 7.97% in TE-1 cells (both
P<0.05). These results indicated that combined treatment with
RG108 and IR increased the G2/M arrest of the EC cells induced by
radiation.
RG108 inhibits tumor growth and
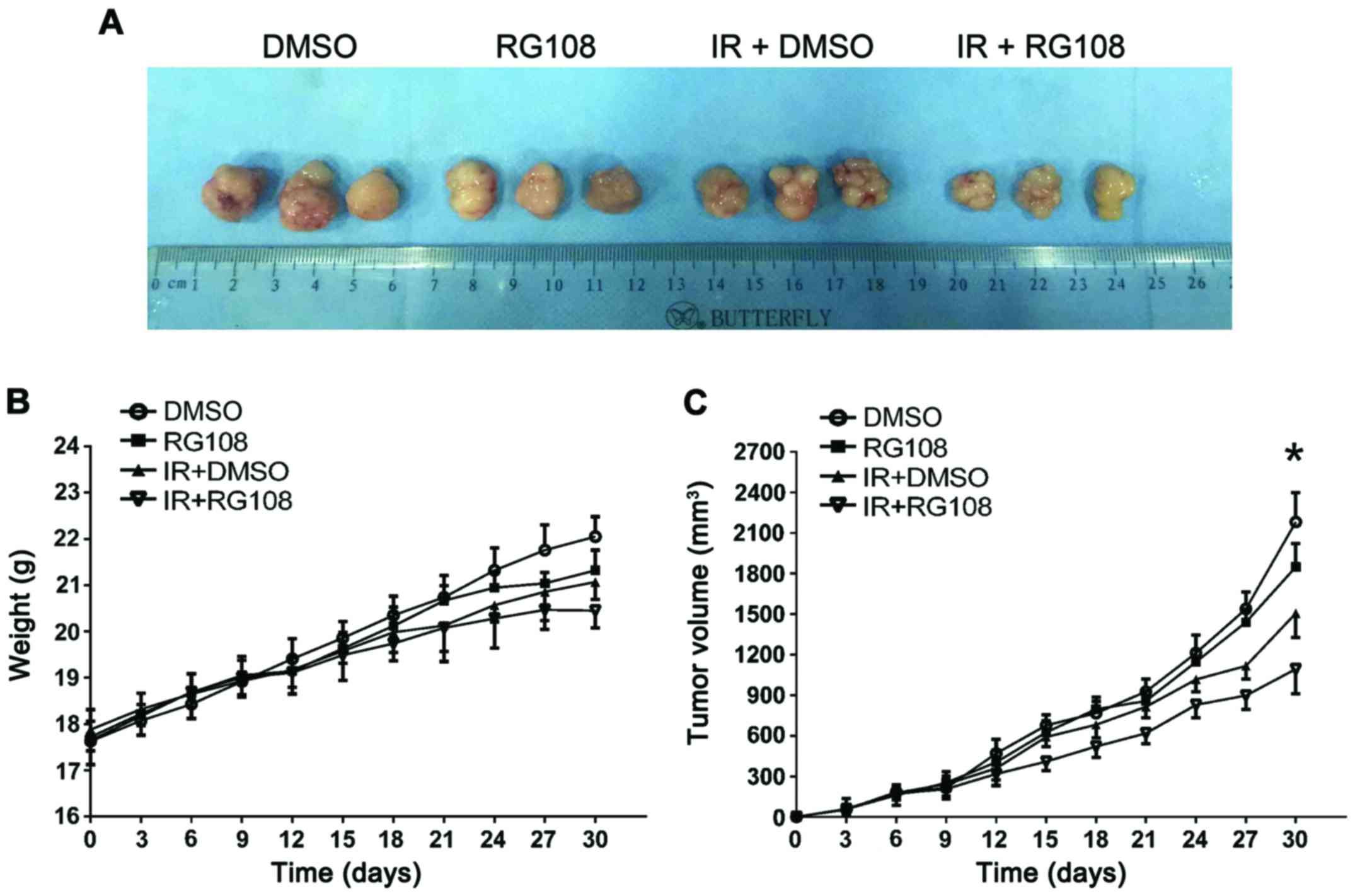
enhances radiosensitivity in a mouse xenograft model
To examine the inhibitory effect of RG108 on EC-cell
growth in vivo, the Eca-109 cells were subcutaneously
inoculated into the right posterior flank region of 5-week-old male
BALB/c nude mice. When tumors reached 150–200 mm3 in
volume, the mice were randomly divided into four groups. Either
DMSO or RG108 was intraperitoneally injected into mice daily for 6
days. Mice in each group exhibited no differences in body weight
and no observable pathological abnormalities (data not shown),
indicating no gross toxicity. As demonstrated in Fig. 5A and B, treatment with RG108 or IR
alone slightly decreased the xenograft size, whereas the
combination of RG108 and IR significantly decreased the volume of
tumors compared with the control treatment (57% reduction,
P<0.05).
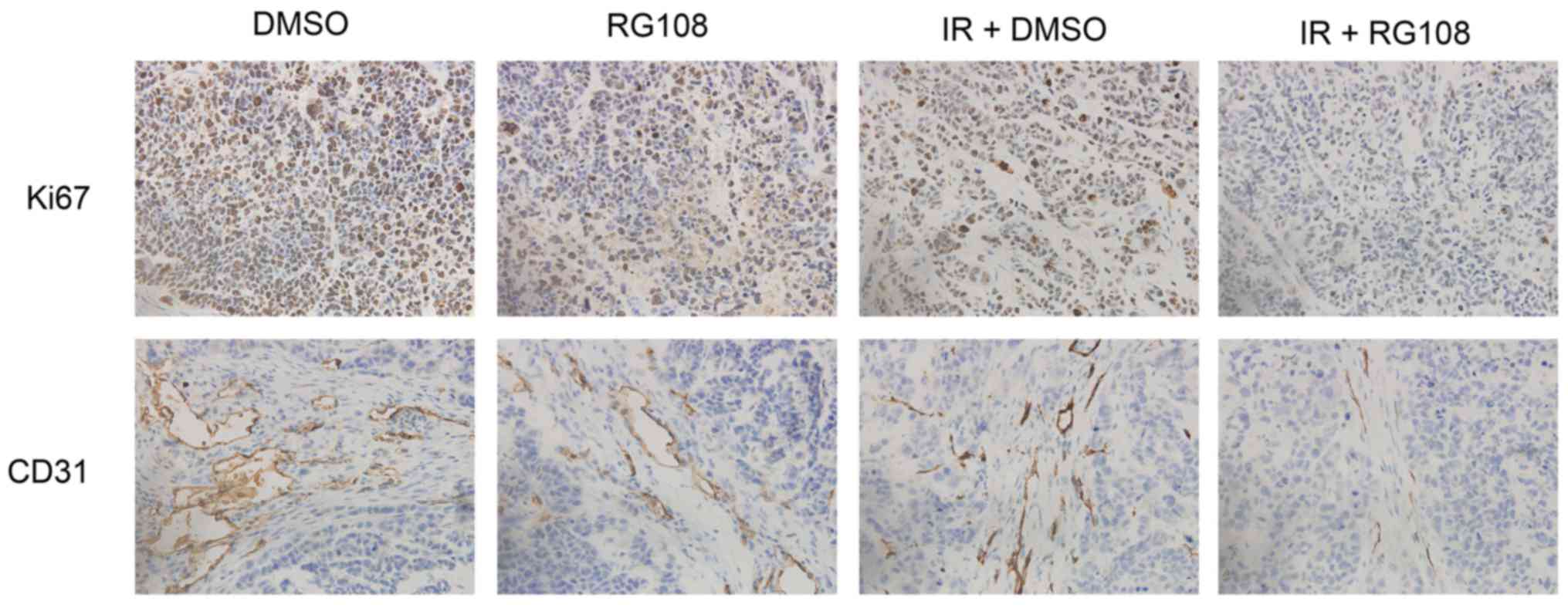
RG108 inhibits angiogenesis and cell
proliferation in response to X-ray irradiation in vivo
To characterize the molecular changes of the
xenografts, the proliferation marker Ki67 and angiogenesis marker
CD31 were examined. As depicted in Fig.
6 (top), RG108 or IR alone modestly reduced the expression of
Ki67, whereas the number of Ki67-positive cells was significantly
decreased in Eca-109 tumors by combined treatment with both RG108
and IR (P<0.05). Similarly, compared to treatment with RG108 or
IR alone, the combination therapy significantly reduced tumor
angiogenesis (P<0.05). As shown in Fig. 6 (bottom), treatment with RG108 or IR
alone modestly reduced the expression of CD31, but combined
treatment with both RG108 and IR significantly decreased the
expression of CD31.
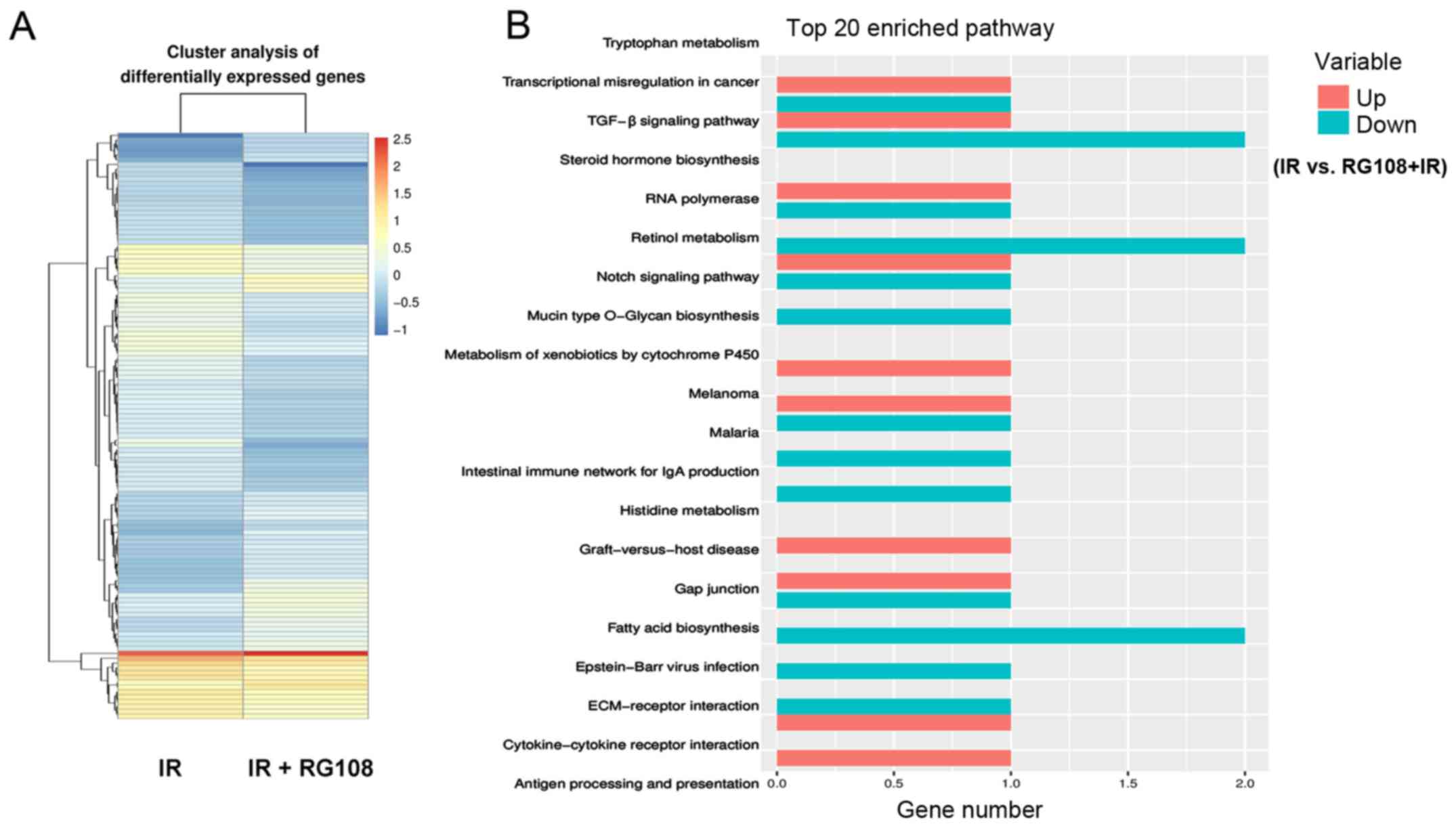
RG108 increases the radiosensitivity
of Eca-109 cells via a complex mechanism
We profiled the gene expression of Eca-109 cells
after either 6 Gy of X-ray irradiation or a combination of 25 µM
RG108 and irradiation to further analyze the underlying mechanisms
responsible for RG108-mediated radiosensitivity. RNA-seq analysis
identified a total of 121 genes (45 upregulated and 76
downregulated) with significantly altered expression levels (|log2
ratio| ≥1 and P-value <0.05) between the two treatments
(Tables I and II). RG108 appeared to modulate the
radiosensitivity of Eca-109 cells via a complex mechanism. Pathway
analysis revealed that RG108 treatment affected multiple pathways,
including the TGF-β signaling pathway and Epstein-Barr virus
infection pathway (Fig. 7).
 | Table I.Microarray analysis of upregulated
genes in Eca-109 cells (6 Gy of X-ray irradiation plus RG108 vs. 6
Gy of X-ray irradiation alone). |
Table I.
Microarray analysis of upregulated
genes in Eca-109 cells (6 Gy of X-ray irradiation plus RG108 vs. 6
Gy of X-ray irradiation alone).
| ID | Gene name | Locus | log2 (RG108 +
IR/ctr + IR) | P-value | Description |
|---|
| 1 |
ABHD14A-ACY1 | chr3 | 1.79769e+308 | 0.0000000328 | ABHD14A-ACY1
read-through |
| 2 |
LOC388780 | chr20 | 1.79769e+308 | 0.0007827010 | Uncharacterized
LOC388780 |
| 3 |
AGAP1-IT1 | chr2 | 1.79769e+308 | 0.0037878500 | AGAP1 intronic
transcript 1 |
| 4 |
RNF103-CHMP3 | chr2 | 3.0834500000 | 0.0008902720 | RNF103-CHMP3
read-through |
| 5 | CNFN | chr19 | 2.4991900000 | 0.0015629000 | Cornifelin |
| 6 |
PIK3CD-AS2 | chr1 | 2.4472000000 | 0.0048119700 | PIK3CD antisense
RNA 2 |
| 7 | ID2-AS1 | chr2 | 2.3098600000 | 0.0037836900 | ID2 antisense RNA 1
(head to head) |
| 8 |
SLMO2-ATP5E | chr20 | 2.2677500000 | 0.0002843230 | SLMO2-ATP5E
read-through |
| 9 | AMTN | chr4 | 2.2395500000 | 0.0057826300 | Amelotin |
| 10 | F2RL2 | chr5 | 2.0639000000 | 0.0003827790 | Coagulation factor
II thrombin receptor like 2 |
| 11 |
ZNF559-ZNF177 | chr19 | 2.0248700000 | 0.0050070200 | ZNF559-ZNF177
read-through |
| 12 | GCOM1,
MYZAP | chr15 | 1.8443900000 | 0.0000197514 | GRINL1A complex
locus 1 |
| 13 | C1orf53 | chr1 | 1.7266700000 | 0.0000011706 | Chromosome 1 open
reading frame 53 |
| 14 | MMP13 | chr11 | 1.6353900000 | 0.0021975000 | Matrix
metallopeptidase 13 |
| 15 |
TOPORS-AS1 | chr9 | 1.6260200000 | 0.0043661900 | TOPORS antisense
RNA 1 |
| 16 |
RPS10-NUDT3 | chr6 | 1.5762500000 | 0.0000082438 | RPS10-NUDT3
read-through |
| 17 |
C7orf55-LUC7L2 | chr7 | 1.5491400000 | 0.0000433101 | C7orf55-LUC7L2
read-through |
| 18 | FAM9B | chrX | 1.4650900000 | 0.0002975920 | Family with
sequence similarity 9 member B |
| 19 | GGACT | chr13 | 1.4237200000 | 0.0003725270 | γ-glutamylamine
cyclotransferase |
| 20 | EVA1A | chr2 | 1.3947700000 | 0.0071589000 | Eva-1 homolog A,
regulator of programmed cell death |
 | Table II.Microarray analysis of downregulated
genes in Eca-109 cells (6 Gy of X-ray irradiation plus RG108 vs. 6
Gy of X-ray irradiation alone). |
Table II.
Microarray analysis of downregulated
genes in Eca-109 cells (6 Gy of X-ray irradiation plus RG108 vs. 6
Gy of X-ray irradiation alone).
| ID | Gene name | Locus | log2 (RG108 +
IR/ctr + IR) | P-value | Description |
|---|
| 1 | FOXB1 | chr15 | −3.2752 | 0.0021565800 | Forkhead box
B1 |
| 2 | RDH5 | chr12 | −3.0106 | 0.0003470500 | Retinol
dehydrogenase 5 |
| 3 |
C19orf71 | chr19 | −2.85166 | 0.0001936530 | Chromosome 19 open
reading frame 71 |
| 4 | RHAG | chr6 | −2.17175 | 0.0020515500 | Rh-associated
glycoprotein |
| 5 | CNTD2 | chr19 | −1.83579 | 0.0006623760 | Cyclin N-terminal
domain containing 2 |
| 6 | MAFB | chr20 | −1.76114 | 0.0000580816 | MAF bZIP
transcription factor B |
| 7 | GATSL3 | chr22 | −1.62313 | 0.0030739500 | GATS protein-like
3 |
| 8 |
INO80B-WBP1 | chr2 | −1.60966 | 0.0000335464 | INO80B-WBP1
read-through (NMD candidate) |
| 9 | YPEL1 | chr22 | −1.57025 | 0.0002142530 | Yippee like 1 |
| 10 |
POC1B-GALNT4 | chr12 | −1.46739 | 0.0000269476 | POC1B-GALNT4
read-through |
| 11 |
LOC100507472 | chr15 | −1.45729 | 0.0000060501 | Uncharacterized
LOC100507472 |
| 12 |
LOC389199 | chr4 | −1.45482 | 0.0010464800 | Uncharacterized
LOC389199 |
| 13 | AMDHD1 | chr12 | −1.45002 | 0.0000036856 | Amidohydrolase
domain containing 1 |
| 14 | SOX7 | chr8 | −1.42999 | 0.0002313520 | SRY-box 7 |
| 15 | CHRD | chr3 | −1.42985 | 0.0015947400 | Chordin |
| 16 | DFNB59 | chr2 | −1.41377 | 0.0070473000 | Deafness, autosomal
recessive 59 |
| 17 | ICOSLG | chr21 | −1.40911 | 0.0000000006 | Inducible T-cell
costimulator ligand |
| 18 | CRB2 | chr9 | −1.39949 | 0.0000000000 | Crumbs 2, cell
polarity complex component |
| 19 | CYP26B1 | chr2 | −1.3987 | 0.0000000793 | Cytochrome P450
family 26 subfamily B member 1 |
| 20 | EIF3CL | chr16 | −1.35891 | 0.0017581100 | Eukaryotic
translation initiationfactor 3 subunit C-like |
Discussion
Radiotherapy is an important treatment for cancer
and plays a critical role in the management of human EC. However,
radioresistance significantly decreases the efficacy of
radiotherapy in the treatment of EC (15). Therefore, the efficacy of
radiotherapy is limited both by the total dosage of radiation that
can be administeredwithout damaging normal tissues and
radioresistance of EC. The resistance of malignant tumor cells to
anticancer agents remains the major cause of failure in treating
patients with EC.
Tumorigenesis and tumor progression are connected
with genetic and epigenetic changes and one of these epigenetic
factors is DNA methylation (16).
Previous studies have revealed that the methylation statuses of
specific genes may potentially be molecular markers of thyroid
(17), breast (18), prostate (19), gastric (20) and colon carcinomas (21). The reversion of epigenetic mutations
by DNMTIs, a promising class of novel drugs, represents an
experimental strategy with great promise for epigenetic cancer
therapy (22). In fact, two
nucleoside analogs, 5-azacytidine and 5-aza-2′-deoxycytidine, have
already been approved by the US Food and Drug Administration
(USFDA) for the treatment of myelodysplastic syndrome (23).
Epigenetics and DNA methylation have recently become
one of the most exciting frontiers for research on the
radioresistance of cancer cells. By integrating mRNA and
methylation profiles, Luo et al (24) found that decreased expression of the
transcription factor Sall2, with a corresponding increase in
methylation of the Sall2 gene, was associated with the aggressive
phenotypes acquired by EC cells after radiotherapy (24). RG108, the first DNMTi discovered by
rational drug design, functions without being integrated into DNA
and effectively blocks DNMTs at their active sites. This leads to
reactivation of tumor-suppressor genes and demethylation of genomic
DNA, while exhibiting little toxicity in human cancer cell lines
(12,25). The present study first illustrated
the role of RG108 in EC cell proliferation and
radiosensitivity.
To date, apoptosis has been the most widely studied
mechanism in anticancer therapy (26,27).
In the present study, we found that pretreatment with RG108 prior
to IR increased apoptosis of EC cells via the overexpression of
Bax, accompanied by a reduction of Bcl-2. As demonstrated in a
previous study (28), Bcl-2 and Bax
are two members of the Bcl-2 family, which is composed of both
apoptosis-promoting and anti-apoptotic proteins that exert opposing
effects on mitochondria. Increased expression of Bcl-2 (an
anti-apoptotic protein) is involved in the development and
progression of many tumor types. The present study demonstrated
that RG108 increased the radiosensitivity and promoted apoptosis of
EC cells both in vitro and in vivo. However, in the
in vivo experiments, RG108 was administered at a
concentration of 50 mg/kg to achieve effective mouse blood
concentrations. Further investigation with other levels of RG108 is
needed. The degradation of cyclin B1 has been reported to be a
novel phenomenon that is caused by high-dose radiation, leading to
G2/M cell-cycle arrest and sensitivity to radiotherapy, although
the mechanism of this cell-cycle suspension is still under
investigation (29). Zheng et
al (30) and Liu et al
(31) have reported that treatment
with either miRNA-200c or MG132 promoted G2/M arrest in cancer
cells, leading to the conclusion that strategies that target G2/M
arrest may be effective for promoting radiosensitivity in cancer
therapy. These previously mentioned results support the conclusion
of the present study that RG108 holds promise as an effective
radiosensitizer in EC therapy by increasing G2/M arrest.
Analysis of mRNA expression indicated that RG108
combined with IR treatment increased the expression of EVA1A, which
in most cancer tissues has reduced or undetectable expression
compared to normal tissues (32,33).
The restoration of EVA1A expression induces death in some cancer
cell lines through both autophagy and apoptosis, indicating that
EVA1A is an effective tumor-suppressing molecule. RG108 combined
with radiotherapy increased the expression of EVA1A, which may be
one of the reasons why RG108 inhibited tumor growth. The present
study also revealed that RG108 modulated the radiosensitivity of
Eca-109 cells via a complex mechanism by affecting multiple
pathways, including the TGF-β signaling pathway and Epstein-Barr
virus infection pathway.
In the present study, we elucidated the effects of
RG108 and IR on the growth of EC cells. The results revealed that
RG108 inhibited the growth of EC cells by increasing cell apoptosis
and G2/M arrest. The results of tumor xenograft experiments
revealed that RG108, combined with IR, significantly inhibited the
proliferation of Eca-109 cells in vivo. RNA-seq analysis
demonstrated that, compared with radiation treatment alone, X-ray
irradiation plus RG108 altered the expression of 121 genes that
function in multiple pathways, including the TGF-β signaling
pathway and Epstein-Barr virus infection pathway. In conclusion,
RG108 enhanced radiation-induced apoptosis and increased G2/M
arrest in EC, thus showing promise as an effective radiosensitizer
in EC therapy. However, the effects of RG108 on tumorigenesis and
the value of its application for treating other cancers require
further study.
Acknowledgements
This study was supported by the Jiangsu Provincial
Special Program of Medical Science (nos. BE2015631and BK20161152),
the Medicine and Health and Scientific Development Program of
Shandong Province (no. 2015WSB30011), the Scientific Research of
Changzhou (nos. QN201503 and CJ20160015), the Scientific Research
of Jintan (no. JT2016065) and the Changzhou High Level Medical
Talents Training Project (no. 2016CZLJ026).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the Global
Burden of Disease Study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang C, Chen X, Li L, Zhou Y, Wang C and
Hou S: The Association between telomere length and cancer
prognosis: Evidence from a meta-analysis. PLoS One.
10:e01331742015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ito S, Ohga T, Saeki H, Nakamura T,
Watanabe M, Tanaka S, Kakeji Y and Maehara Y: p53 mutation
profiling of multiple esophageal carcinoma using laser capture
microdissection to demonstrate field carcinogenesis. Int J Cancer.
113:22–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuwano H, Kato H, Miyazaki T, Fukuchi M,
Masuda N, Nakajima M, Fukai Y, Sohda M, Kimura H and Faried A:
Genetic alterations in esophageal cancer. Surg Today. 35:7–18.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karahoca M and Momparler RL:
Pharmacokinetic and pharmacodynamic analysis of
5-aza-2′-deoxycytidine (decitabine) in the design of its
dose-schedule for cancer therapy. Clin Epigenetics. 5:32013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh V, Sharma P and Capalash N: DNA
methyltransferase-1 inhibitors as epigenetic therapy for cancer.
Curr Cancer Drug Targets. 13:379–399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Borges S, Döppler H, Perez EA, Andorfer
CA, Sun Z, Anastasiadis PZ, Thompson E, Geiger XJ and Storz P:
Pharmacologic reversion of epigenetic silencing of the PRKD1
promoter blocks breast tumor cell invasion and metastasis. Breast
Cancer Res. 15:R662013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karpf AR, Moore BC, Ririe TO and Jones DA:
Activation of the p53 DNA damage response pathway after inhibition
of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol.
59:751–757. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eiseler T, Döppler H, Yan IK, Goodison S
and Storz P: Protein kinase D1 regulates matrix metalloproteinase
expression and inhibits breast cancer cell invasion. Breast Cancer
Res. 11:R132009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jüttermann R, Li E and Jaenisch R:
Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated
primarily by covalent trapping of DNA methyltransferase rather than
DNA demethylation. Proc Natl Acad Sci USA. 91:pp. 11797–11801.
1994; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brueckner B, Garcia Boy R, Siedlecki P,
Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M and Lyko F:
Epigenetic reactivation of tumor suppressor genes by a novel
small-molecule inhibitor of human DNA methyltransferases. Cancer
Res. 65:6305–6311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bardenheuer W, Lehmberg K, Rattmann I,
Brueckner A, Schneider A, Sorg UR, Seeber S, Moritz T and Flasshove
M: Resistance to cytarabine and gemcitabine and in vitro selection
of transduced cells after retroviral expression of cytidine
deaminase in human hematopoietic progenitor cells. Leukemia.
19:2281–2288. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Z, Hou J, Lu L, Qi Z, Sun J, Gao W,
Meng J, Wang Y, Sun H, Gu H, et al: Small ribosomal protein subunit
S7 suppresses ovarian tumorigenesis through regulation of the
PI3K/AKT and MAPK pathways. PLoS One. 8:e791172013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shridhar R, Almhanna K, Meredith KL,
Biagioli MC, Chuong MD, Cruz A and Hoffe SE: Radiation therapy and
esophageal cancer. Cancer Contr. 20:97–110. 2013. View Article : Google Scholar
|
|
16
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hoque MO, Rosenbaum E, Westra WH, Xing M,
Ladenson P, Zeiger MA, Sidransky D and Umbricht CB: Quantitative
assessment of promoter methylation profiles in thyroid neoplasms. J
Clin Endocrinol Metab. 90:4011–4018. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miyamoto K, Fukutomi T, Akashi-Tanaka S,
Hasegawa T, Asahara T, Sugimura T and Ushijima T: Identification of
20 genes aberrantly methylated in human breast cancers. Int J
Cancer. 116:407–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McKie AB, Douglas DA, Olijslagers S,
Graham J, Omar MM, Heer R, Gnanapragasam VJ, Robson CN and Leung
HY: Epigenetic inactivation of the human sprouty2 (hSPRY2)
homologue in prostate cancer. Oncogene. 24:2166–2174. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
An C, Choi IS, Yao JC, Worah S, Xie K,
Mansfield PF, Ajani JA, Rashid A, Hamilton SR and Wu TT: Prognostic
significance of CpG island methylator phenotype and microsatellite
instability in gastric carcinoma. Clin Cancer Res. 11:656–663.
2005.PubMed/NCBI
|
|
21
|
Feinberg AP, Cui H and Ohlsson R: DNA
methylation and genomic imprinting: Insights from cancer into
epigenetic mechanisms. Semin Cancer Biol. 12:389–398. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abdulhaq H and Rossetti JM: The role of
azacitidine in the treatment of myelodysplastic syndromes. Expert
Opin Investig Drugs. 16:1967–1975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo J, Wang W, Tang Y, Zhou D, Gao Y,
Zhang Q, Zhou X, Zhu H, Xing L and Yu J: mRNA and methylation
profiling of radioresistant esophageal cancer cells: The
involvement of Sall2 in acquired aggressive phenotypes. J Cancer.
8:646–656. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stresemann C, Brueckner B, Musch T,
Stopper H and Lyko F: Functional diversity of DNA methyltransferase
inhibitors in human cancer cell lines. Cancer Res. 66:2794–2800.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang H, Tian ST, Wu RY, Chen Y, Mei ZN,
Wang CY and Yang GZ: Glycoborinine induces apoptosis through
mitochondrial pathway in HepG2 cells. J Asian Nat Prod Res.
16:991–999. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wimardhani YS, Suniarti DF, Freisleben HJ,
Wanandi SI, Siregar NC and Ikeda MA: Chitosan exerts anticancer
activity through induction of apoptosis and cell cycle arrest in
oral cancer cells. J Oral Sci. 56:119–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oh KJ, Barbuto S, Pitter K, Morash J,
Walensky LD and Korsmeyer SJ: A membrane-targeted BID BCL-2
homology 3 peptide is sufficient for high potency activation of BAX
in vitro. J Biol Chem. 281:36999–37008. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He J, Li J, Ye C, Zhou L, Zhu J, Wang J,
Mizota A, Furusawa Y and Zhou G: Cell cycle suspension: A novel
process lurking in G2 arrest. Cell Cycle. 10:1468–1476.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng R, Liu Y, Zhang X, Zhao P and Deng
Q: miRNA-200c enhances radiosensitivity of esophageal cancer by
cell cycle arrest and targeting P21. Biomed Pharmacother.
90:517–523. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu J, Shen W, Tang Y, Zhou J, Li M, Zhu
W, Yang H, Wu J, Zhang S and Cao J: Proteasome inhibitor MG132
enhances the antigrowth and antimetastasis effects of radiation in
human nonsmall cell lung cancer cells. Tumour Biol. 35:7531–7539.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu D, Yang F, He H, Hu J, Lv X, Ma D and
Chen YY: Expression of TMEM166 protein in human normal and tumor
tissues. Appl Immunohistochem Mol Morphol. 21:543–552. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun W, Ma XM, Bai JP, Zhang GQ, Zhu YJ, Ma
HM, Guo H, Chen YY and Ding JB: Transmembrane protein 166
expression in esophageal squamous cell carcinoma in Xinjiang,
China. Asian Pac J Cancer Prev. 13:3713–3716. 2012. View Article : Google Scholar : PubMed/NCBI
|