Introduction
Colon cancer remains the most commonly diagnosed
cancer and the leading cause of cancer-related deaths in the world
(1,2). Current therapeutic strategies for
colon cancer include surgery, radiotherapy, chemotherapy or a
combination of these therapies. Drug resistance is often regarded
as the cause of the failure of chemotherapeutic drug treatments in
patients with colon cancer. Oxaliplatin is a third-generation
platinum drug that is currently used as the front line treatment of
colon cancer (3). To date, there
have been extensive studies on the mechanisms of oxaliplatin
resistance. Several factors including alterations in transport,
detoxification, DNA damage response and repair, cell death, and
epigenetic processes have been described to induce oxaliplatin
resistance (4). However, little is
known about the effects of glycosylation on the development of
oxaliplatin resistance in colon cancer.
As the most common post-translational modifications
of proteins, glycosylation is an effective way to induce diversity
due to the structural variations of glycans. Glycans are more
diverse in terms of chemical structure and information density than
are DNA and proteins (5). The
attachment of glycans usually occurs during or after the process of
protein synthesis, which involves several glycogenes and isoforms
of these enzymes. Cancer cells frequently express glycans at
atypical levels or with different structural attributes than those
found in normal cells (6).
Correlations between drug resistance and glycosylation changes have
been extensively studied. For example, Zhang et al
demonstrated that N-glycans were associated with adriamycin
resistance in human leukemia cells (7). Ma et al found that the
alterations of glycogenes in breast cancer cells was correlated
with tumor sensitivity to chemotherapeutic drugs (8). Kudo et al revealed that
N-glycan alterations contributed to the drug-resistant phenotype in
human hepatocellular carcinoma (9).
Wu et al reported that N-glycosites and site-specific
glycoforms of secreted proteins in drug-resistant cell lines were
distinctly different from those in the parental cell lines
(10). Therefore, it is of interest
to clarify the relationship between aberrant glycosylation and
oxaliplatin resistance in colon cancer and to find therapeutic
targets for effective medical treatment.
Lectin microarray has been used as a robust,
high-throughput, exceedingly sensitive, and reliable quantitative
method for obtaining whole-cell glycan signatures (11,12).
In the present study, we utilized this high-throughput glycomics
technology to profile specific glycans from an
oxaliplatin-resistant colon cancer cell line and its parental cell
line. We also investigated whether glycogenes participated in the
regulation of oxaliplatin resistance. In addition, the possible
pathways underlying glycosylation-mediated oxaliplatin resistance
were further explored.
Materials and methods
Cell culture and establishment of an
oxaliplatin-resistant cell line
The human colon cancer cell line SW620 was purchased
from the cell library of the Chinese Academy of Sciences (Shanghai,
China). An oxaliplatin-resistant cell line SW620R was established
by continuous exposure of its parental cell line SW620 to gradually
increasing concentrations of oxaliplatin (Sigma-Aldrich, St. Louis,
MO, USA). Both cell lines were cultured in RPMI-1640 medium
containing 10% fetal bovine serum (Gibco-BRL, Carlsbad, CA, USA).
The SW620R cell line was incubated in the presence of 0.1 µg/ml
oxaliplatin to maintain the drug-resistant phenotype.
Drug sensitivity assay of various
anticancer drugs using CCK-8
A total of 1×103 cells/well were seeded
into 96-well plates and incubated overnight to allow for full
adherence. Then, varied concentrations of anticancer drugs
including oxaliplatin (0–0.5 µg/ml), etoposide (0–1 µg/ml),
vincristine (0–0.1 µg/ml) and cisplatin (0–1 µg/ml) were added to
each well. After 72 h, cell proliferation was determined by Cell
Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology,
Jiangsu, China). Then, the absorbance at 450 nm was assessed using
a microplate reader (Molecular Devices, Sunnyvale, CA, USA). The
50% inhibition concentration (IC50) was calculated based
on individual cytotoxicity plots.
Lectin microarray scanning
A total of 1×106 cells were collected,
washed twice with ice-cold PBS, and labeled with CFDA-SE cell
tracking dye (Life Technologies, Carlsbad, CA, USA) at 10 µM for 30
min at 4°C. Then, the cells were resuspended in PBS with 1% BSA
(Sigma-Aldrich), 0.5 mM CaCl2 and 0.1 mM
MnCl2. The lectin microarrays with 30 lectins (BC
Biotechnology, Guangdong, China) were blocked for 1 h at room
temperature with 1% BSA. Subsequently, the cells were allowed to
bind on lectin microarrays in the dark for 1 h. The excess and/or
unbound cells were gently removed with washing buffer (PBS with
0.5% Tween). GenePix 5.0 (Molecular Devices) was used for
extracting binding signals from the scanned images (13).
Lectin flow cytometry
A total of 1×106 cells were collected and
washed with fluorescence-activated cell sorting (FACS) buffer (PBS
with 20 mg/ml bovine serum). Then, cells were stained with one of
the 6 FITC-lectins (Vector Laboratories, Inc., Burlingame, CA, USA)
at a final concentration of 10 µg/ml for 1 h at 4°C in the dark.
After three washes with FACS buffer, the cells were centrifuged at
1,000 × g for 5 min and resuspended in 200 µl of PBS. The samples
were analyzed using FACScan flow cytometry (Becton-Dickinson,
Mountain View, CA, USA). Ten thousand cells were analyzed for each
condition.
Analysis of glycogenes
A total of 1×106 cells were harvested for
real time RT-PCR analysis. Total mRNA was extracted using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) and then reversed
transcribed to cDNA with the Superscript First Strand synthesis
system (Invitrogen) according to the manufacturer's instructions.
All PCR reactions were performed on an ABI PRISM® 7500
Sequence Detection System (Applied Biosystems, Foster City, CA,
USA) for 40 cycles (5 sec at 95°C, 30 sec at 60°C and 30 sec at
72°C). The SYBR-Green Real-Time PCR Master Mix kit (Toyobo, Osaka,
Japan) was used. Primer sequences are listed in Table I. The expression level of each
glycogen was normalized to GAPDH. The results were calculated using
the 2−ΔΔCt method.
 | Table I.Sequences of the primers used for
real-time RT-PCR. |
Table I.
Sequences of the primers used for
real-time RT-PCR.
| Primer name | Sequences
(5′-3′) |
|---|
| β3GnT8 | F:
GTCGCTACAGTGACCTGCTG |
|
| R:
GTCTTTGAGCGTCTGGTTGA-3 |
| GnT-III | F:
ATGAAGATGAGACGCTACAAGC |
|
| R:
GCTGGACACCAGGTTAGGG |
| GALNT1 | F:
TTCCCAGCGACTCCAGAAACAC |
|
| R:
TGGGATAACCTGCATCCACGGA |
| ST6GaL1 | F:
GTGGGCACAAAAACTACCAT |
|
| R:
GGCTCTGGGCTCATAAACTG |
| FUT8 | F:
CCTGGCGTTGGATTATGCTCA |
|
| R:
CCCTGATCAATAGGGCCTTC |
| GnT-V | F:
CTTCACTCCGTGGAAGTTGTC |
|
| R:
TGGATGGTAAAGTGCAGAAGC |
| GADPH | F:
CCAACCGCGAGAAGATGA |
|
| R:
CCAGAGGCGTACAGGGATAG |
Small interference RNA (siRNA)
transfection
The siRNA directed against β3GnT8 and nontargeting
negative siRNA (NC) were designed and synthesized by GenePharma
(Shanghai, China). The following sequences were used: β3GnT8 sense,
5′-CAUUCGGCUCUGGAAACAAdT dT-3′ and antisense,
5′-UUGUUUCCAGAGCCGAAUGCTT-3′; NC sense, 5′-UUCUCCGAACGUGUCACGUdT
dT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT3′ (14). SW620R cells were transfected with
100 nM siRNA using Lipofectamine 3000 (Invitrogen) as suggested by
the manufacturer.
Plasmid construction and
transfection
The full coding sequence of β3GnT8 was cloned into
the pEGFP-C1 eukaryotic expression vector (Clontech, Heidelberg,
Germany). Primers 5′ and 3′ for PCR were
5′-AATCTCGAGTAATGCGCTGCCCCAAG-3′ and
5′-GCGGAATTCTCAGCACTGGAGCCTT3-3′. After being identified by
digestion with restriction enzymes XhoI and EcoRI
(MBI, Fermentas, Vilnius, Lithuania), the pEGFP-c1-β3GnT8 plasmid
was transfected into SW620 cells using Lipofectamine 3000 reagent.
The empty vector pEGFP-C1 was also transfected into SW620 cells as
a mock control.
Western blotting
Proteins were extracted and quantified with a BCA
protein assay kit (Beyotime Institute of Biotechnology). Equal
amounts of the protein samples were separated by 10% SDS-PAGE and
transferred onto a PVDF membrane (Millipore, Bedford, MA, USA). The
membrane was blocked with 5% non-fat milk solution at room
temperature for 1 h. After being incubated with primary antibodies
at 4°C overnight, the blots were incubated with HRP-conjugated
corresponding secondary antibodies at room temperature for 1 h,
followed by detection with an ECL kit (Beyotime Institute of
Biotechnology). All the antibodies were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). The antibodies and dilutions
were as follows: pY397 FAK (1:800; mouse monoclonal; cat. no.
sc-81493), FAK (1:1,000; mouse monoclonal; cat. no. sc-271126),
pY118 paxillin (1:800; mouse monoclonal; cat. no. sc-365020) and
paxillin (1:1,000; mouse monoclonal; cat. no. sc-365059).
Lectin blotting
Proteins were separated by 10% SDS-PAGE and
transferred onto a PVDF membrane as aforementioned for western
blotting. The membrane was blocked with Carbo-Free Blocking
Solution (Vector Laboratories, Inc.) at room temperature for 1 h.
After incubation with 2 µg/ml biotinylated LEA (Vector
Laboratories, Inc.) at 4°C overnight, the blots were incubated with
HRP-conjugated streptavidin (Beyotime Institute of Biotechnology)
at room temperature for 1 h, followed by detection with an ECL
kit.
Lectin immunoprecipitation (IP)
A total of 1×106 cells were collected and
lysed. Then, cell lysates were incubated with 10 µg/ml of the LEA
overnight at 4°C on a rotating shaker. Protein A agarose beads
(Thermo Fisher Scientific, Waltham, MA, USA) were added to the
aforementioned complex and incubated for 4 h at 4°C. Finally, the
beads were washed, boiled, centrifuged and subjected to western
blotting as previously described. Precipitated proteins were
immunoblotted to detect integrin β1 with the anti-integrin β1
antibody (1:1,000; mouse monoclonal; cat. no. sc-13590; Santa Cruz
Biotechnology).
Statistical analysis
Each assay was performed at least three times. The
data were expressed as the mean ± SD. All analyses for
statistically significant differences were determined by the
Student's t-test. P<0.05 was considered to indicate a
statistically significant result.
Results
Differential glycan composition is
defined by lectin microarray in SW620 and SW620R cell lines
First, a CCK-8 assay was performed to determine the
IC50 value of oxaliplatin in sensitive SW620 and
resistant SW620R cells. As shown in Table II, the IC50 of
oxaliplatin in SW620 and SW620R cells was 0.015±0.002 and
0.183±0.024 µg/ml, respectively (P<0.05). In addition to
oxaliplatin, SW620R cells also exhibited cross-resistance to other
chemotherapeutic drugs including etoposide, vincristine and
cisplatin. The IC50 values for these drugs were greater
in SW620R cells than those in SW620 cells. It suggested that the
oxaliplatin-resistant cell line SW620R was successfully established
and could be used for the successive experiments.
 | Table II.Sensitivity and cross-resistance of
SW620 and SW620R cells to anticancer drugs. |
Table II.
Sensitivity and cross-resistance of
SW620 and SW620R cells to anticancer drugs.
|
| IC50
(µg/ml) |
|
|---|
|
|
|
|
|---|
| Drug | SW620 | SW620R | P-value |
|---|
| Oxaliplatin | 0.015±0.002 | 0.183±0.024 | 0.009 |
| Etoposide | 0.117±0.003 | 0.302±0.015 | 0.005 |
| Vincristine | 0.014±0.005 | 0.033±0.006 | 0.021 |
| Cisplatin | 0.109±0.027 | 0.577±0.062 | 0.018 |
Then, lectin microarray assay was performed to
detect the glycan signatures in SW620 and SW620R cell lines. Using
this microarray containing 27 lectins, two negative controls (0.5%
BSA and Cy5), and one positive control (Cy3), a cell binding map
was generated (Fig. 1). SW620R
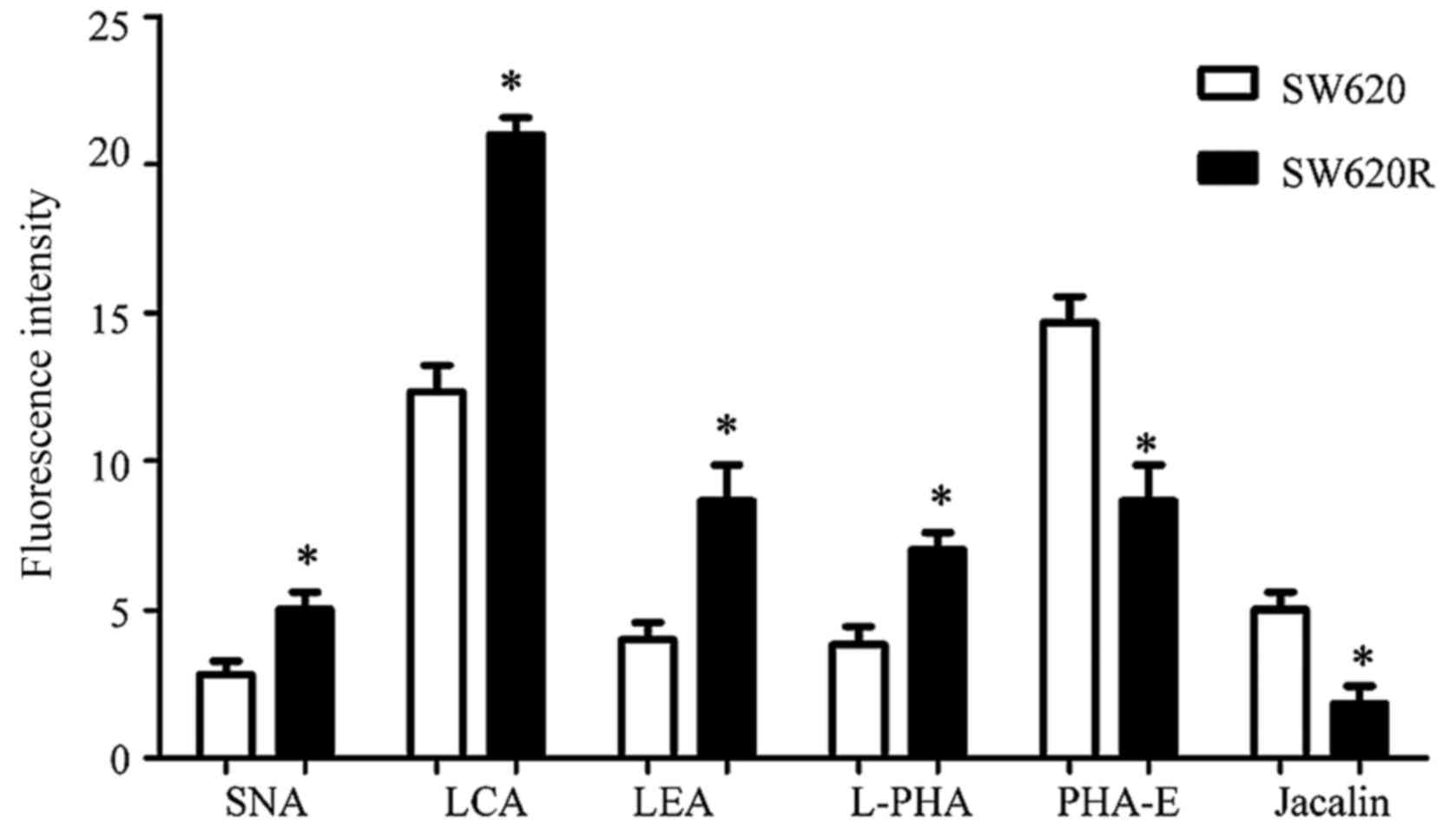
cells had higher expression of four glycan structures recognized by
lectins SNA, LCA, LEA, and L-PHA, and lower expression of two
glycan structures recognized by PHA-E and Jacalin. Increased
staining by SNA, LCA, LEA, and L-PHA indicated elevated levels of
α2-6 sialylation, core fucosylation, polylactosamine-type N-glycans
and β1,6-GlcNAc branched N-glycans in SW620R cells. Reduced
staining by PHA-E and Jacalin indicated downregulation of bisecting
GlcNAc and Galβ1-3GalNAcα-Ser/Thr (T antigen) in SW620R cells.
These data revealed that differential glycan composition may be
associated with colon cancer drug resistance.
Differential glycan profiles are
defined by FITC-lectin flow cytometry in SW620 and SW620R cell
lines
To analyze the glycan profiles of SW620 and SW620R
cells, an FITC-lectin flow cytometric assay was performed.
Differences in fluorescence intensity for SNA, LCA, LEA, L-PHA,
PHA-E and Jacalin were evident when SW620 cells were compared to
SW620R cells (Fig. 2). The SW620R
cells exhibited higher fluorescence signal intensities of SNA, LCA,
LEA and L-PHA but lower intensities of PHA-E and Jacalin, which
were consistent with the results of the lectin microarray
analysis.
Differential expression of glycogenes
in SW620 and SW620R cell lines
Differences in glycan structures highlight the
importance of glycosyltransferases encoded by glycogenes. A
real-time RT-PCR analysis was performed to analyze the expression
of glycogenes in colon cancer cells and paired
oxaliplatin-resistant cells. Six genes were differentially
expressed between the two cell lines, SW620 and SW620R. Two
glycogenes, GnT-III and GALNT1 were expressed at an elevated level
(i.e., >3-fold higher) in the SW620 cells compared with those in
SW620R cells. Conversely, four glycogenes, ST6GaL1, FUT8, β3GnT8
and GnT-V were expressed at a higher level in the SW620R cells
compared with those in the SW620 cells (i.e., >3-fold higher,
Table III). Since it was revealed
that knockdown of β3GnT8 could increase 5-fluorouracil sensitivity
in SW620 cells, we then tested whether β3GnT8 affected the
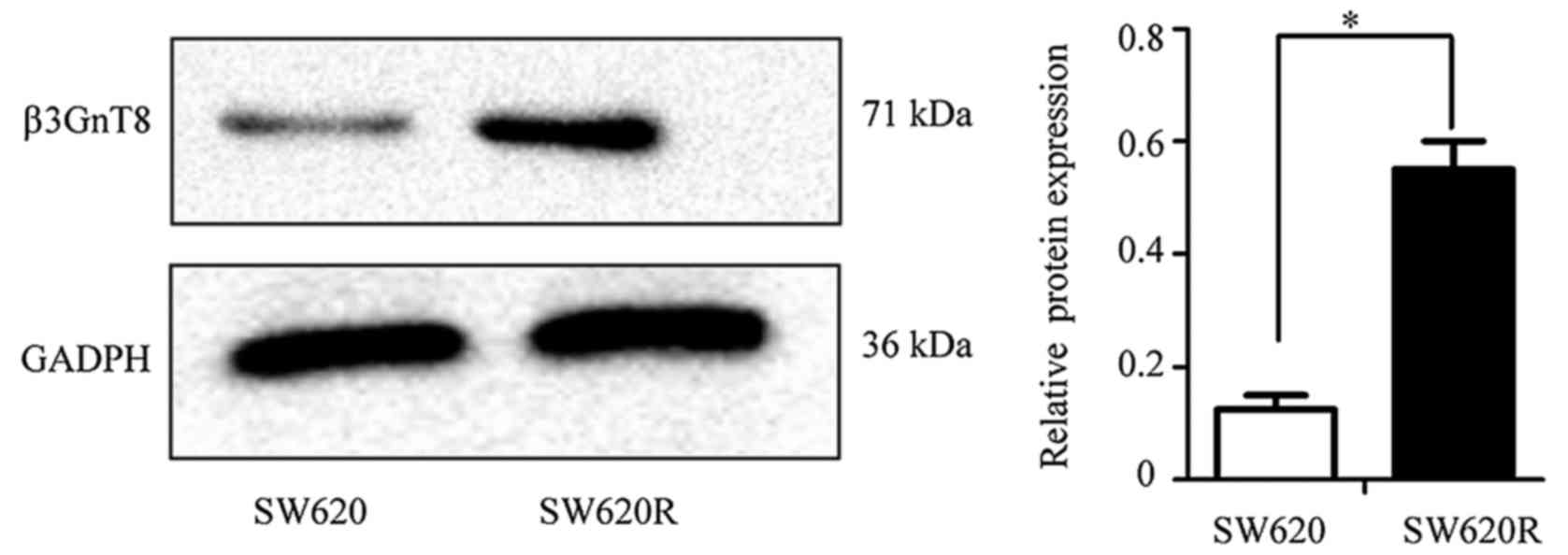
oxaliplatin resistance in the same cancer cells (15). Western blot analysis was performed
to detect the expression of the β3GnT8 protein. The relative level
of the β3GnT8 protein expression in the SW620 cell line was
significantly lower than that in the SW620R cell line (Fig. 3). High expression of β3GnT8 also
corresponded with high fluorescence intensity of LEA in SW620R
cells (data not shown).
 | Table III.Differential expression of glycogenes
in SW620 and SW620R cell lines. |
Table III.
Differential expression of glycogenes
in SW620 and SW620R cell lines.
|
| Ratio
(>3-fold) |
|
|---|
|
|
|
|
|---|
| Glycogene | SW620R/SW620 | SW620/SW620R | Enzyme |
|---|
| ST6GaL1 | 4.61±0.25 |
| GMP-NeuAc:
Galactoside α-2, 6-sialyltransferase |
| FUT8 | 3.92±0.19 |
| α1,
6-Fucosyltransferase 8 |
| β3GnT8 | 5.57±0.14 |
| β1,
3-N-Acetylglucosaminyltransferase 8 |
| GnT-V | 3.89±0.35 |
| α-3-D-Mannoside-β1,
6-N-acetylglucosaminyltransferase V |
| GnT-III |
|
4.16±0.12 | α-3-D-Mannoside-β1,
4-N-acetylglucosaminyltransferase V |
| GALNT1 |
|
4.29±0.33 | Polypeptide
N-acetylgalactosaminyltransferase 1 |
Silencing of β3GnT8 in SW620R cells
results in increased sensitivity to oxaliplatin
To elucidate the direct effect of β3GnT8 expression
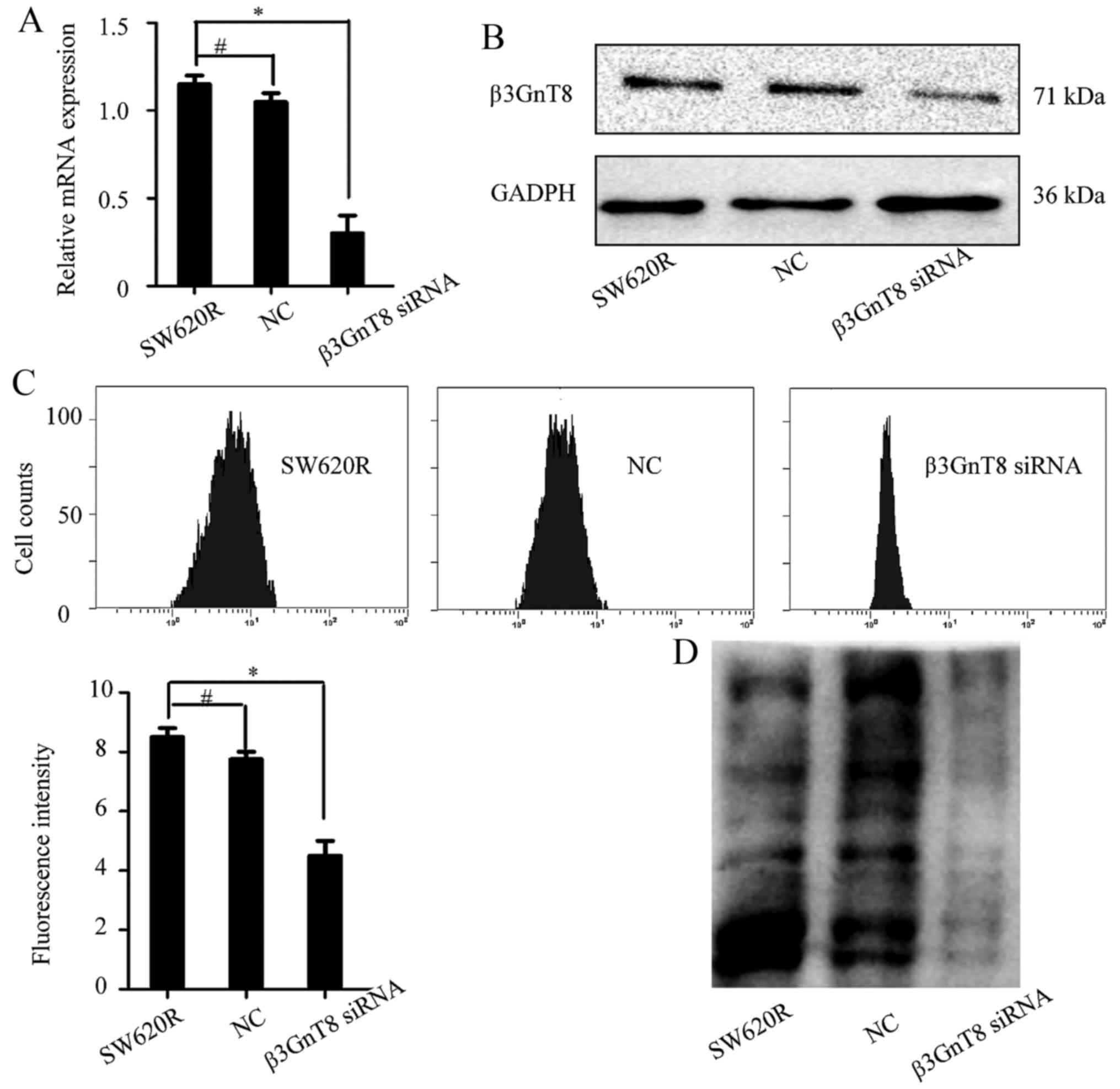
on the sensitivity of SW620R cells, we silenced β3GnT8 with siRNA.
Compared with the negative siRNA-transfected cells, β3GnT8
expression at the mRNA and protein levels was downregulated in
β3GnT8-siRNA transfected SW620R cells (Fig. 4A and B). To further evaluate the
effect of β3GnT8 silencing on cell chemosensitivity, a CCK-8 assay
was performed. The IC50 values of oxaliplatin in SW620R
cells transfected with negative or β3GnT8 siRNA were 0.176±0.019
and 0.042±0.007 µg/ml, respectively (P<0.05) (data not shown).
LEA lectin, which specifically recognizes polylactosamine chains
(product of β3GnT8), was used to analyze the alteration of glycan
structures. Fig. 4C revealed that
β3GnT8 knockdown resulted in a decrease of fluorescence intensity
compared with the negative control cells. The results of lectin
blotting were consistent with the FITC-lectin binding analysis
(Fig. 4D). Thus, downregulation of
β3GnT8 could enhance the sensitivity of SW620R cells to oxaliplatin
by alteration of polylactosamine structures.
Overexpression of β3GnT8 in SW620
cells resulting in increased resistance to oxaliplatin
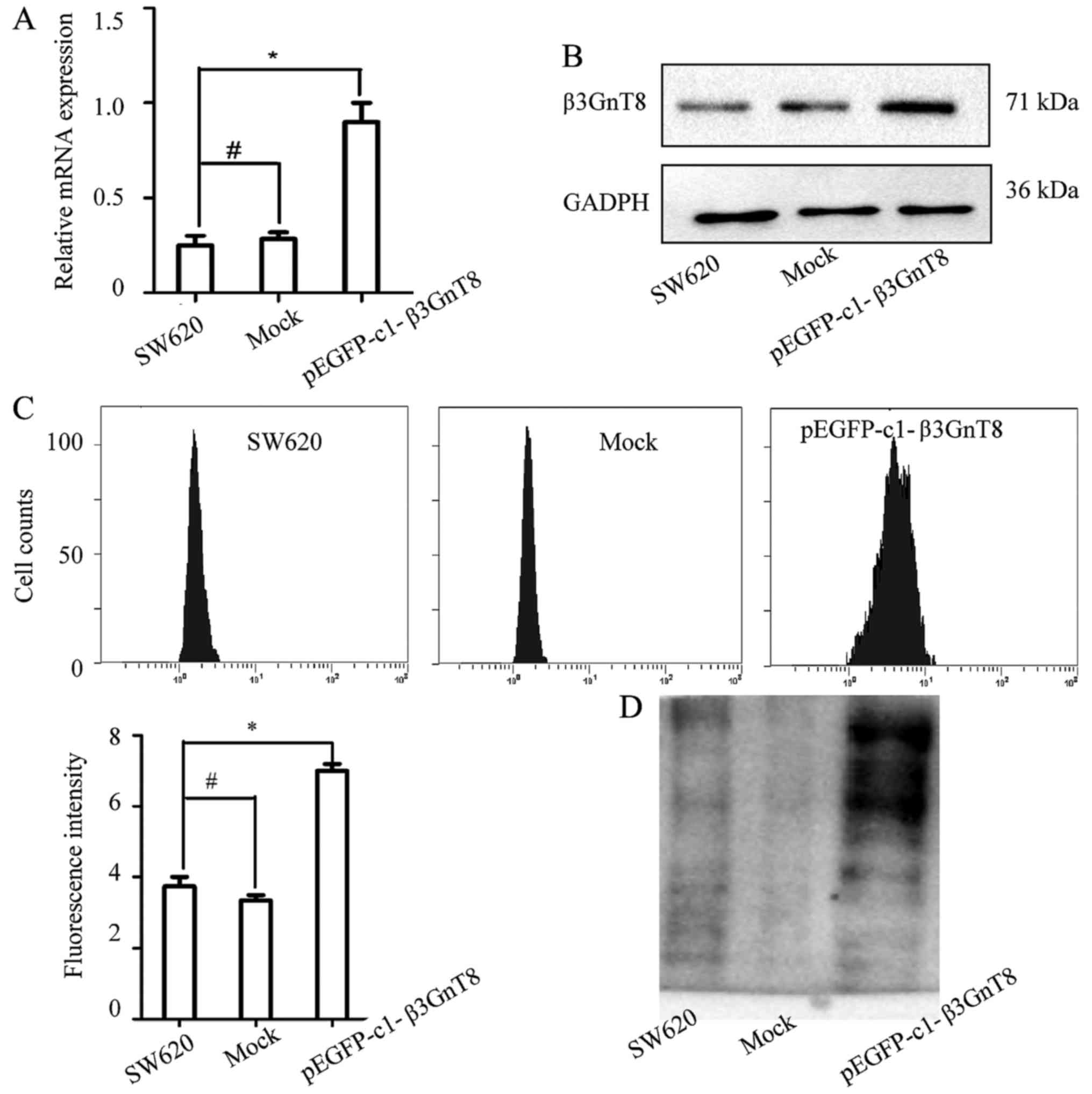
To further investigate the effect of β3GnT8
expression on the sensitivity of SW620 cells to oxaliplatin, a
SW620 cell line stably expressing β3GnT8 was established. As shown
in Fig. 5A and B, β3GnT8 expression
at the mRNA and protein levels was increased in the pEGFP-c1-β3GnT8
plasmid-transfected SW620 cells. The IC50 values of
oxaliplatin in SW620 cells transfected with the pEGFP-c1-β3GnT8
plasmid or empty plasmid were 0.122±0.058 and 0.013±0.008 µg/ml,
respectively (P<0.05) (data not shown). Fig. 5C revealed that β3GnT8 overexpression
resulted in an increase of fluorescence intensity compared with the
mock cells. The results of lectin blotting were consistent with the
FITC-lectin binding analysis (Fig.
5D). These data clearly confirmed that β3GnT8 contributed to
the development of oxaliplatin resistance via the regulation of
polylactosamine chains.
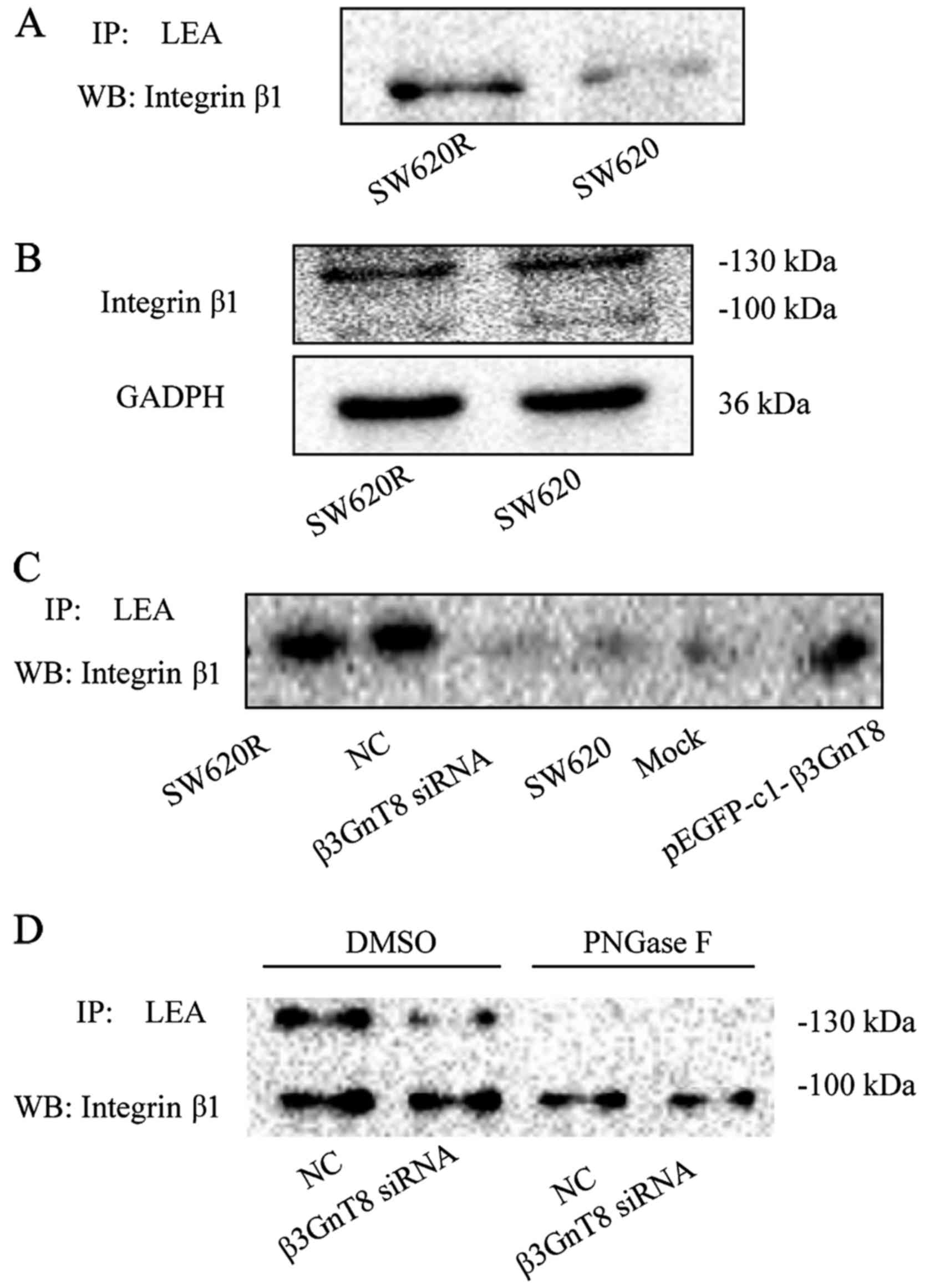
Integrin β1 is a target of β3GnT8 in
SW620R cells
Since integrin β1 is an extensively glycosylated
glycoprotein, we further evaluated whether β3GnT8 modified glycan
structures on integrin β1. Cell lysates were first incubated with
LEA and LEA-bound glycoproteins were then immunoblotted for
integrin β1. In line with the relative expression of β3GnT8, the
binding of LEA to integrin β1 in SW620R cells was stronger than
that in SW620 cells (Fig. 6A).
However, there was no significant difference in the expression of
the integrin β1 protein between SW620 and SW620R cells (Fig. 6B). These results revealed that
integrin β1 was post-translationally modified by β3GnT8. Moreover,
β3GnT8 overexpression in SW620 cells increased LEA binding to
integrin β1, whereas β3GnT8 knockdown in SW620R cells decreased LEA
binding to integrin β1 (Fig. 6C).
To further confirm the existence of LEA-recognized carbohydrate
structures on integrin β1, PNGase F (25 units; Sigma-Aldrich) was
used to remove N-glycans from glycoproteins. As shown in Fig. 6D, the binding of LEA to integrin β1
was almost completely eliminated by PNGase F treatment in SW620R
cells. All these data indicated that integrin β1 could be a target
glycoprotein affected by β3GnT8.
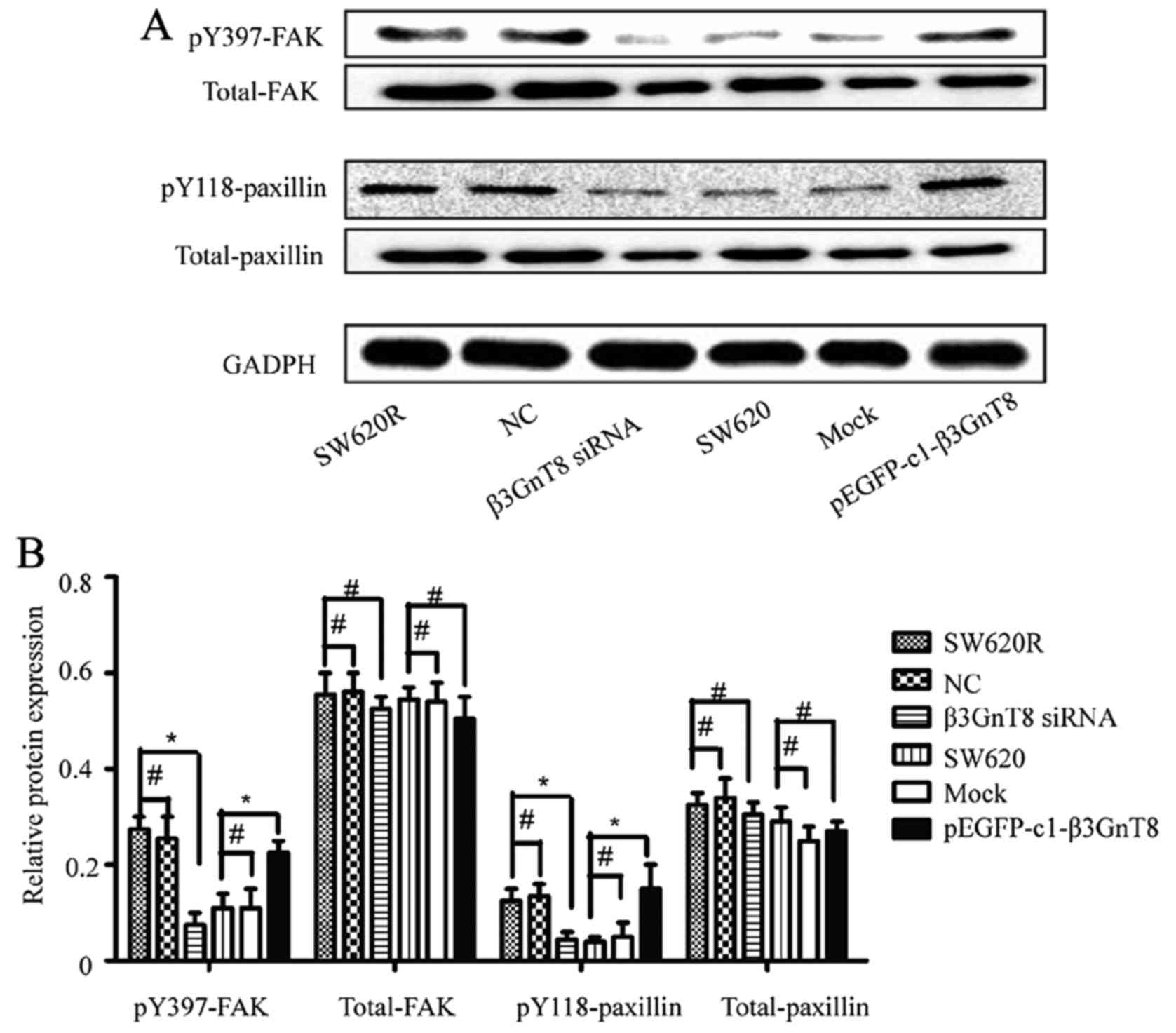
β3GnT8 regulates the downstream
signaling pathway of integrin β1
Since integrin β1 was involved in β3GnT8-mediated
oxaliplatin resistance and β3GnT8 regulated the glycosylation of β1
integrin, we next examined whether β3GnT8 knockdown could regulate
the expression of integrin β1 downstream signaling molecules. As
shown in Fig. 7, β3GnT8 knockdown
in SW620R cells reduced the phosphorylation of focal adhesion
kinase (FAK) and paxillin. Conversely, β3GnT8 overexpression in
SW620 cells enhanced FAK and paxillin phosphorylation.
Discussion
In the present study, we investigated the possible
correlations between oxaliplatin resistance and glycosylation
changes in parental and oxaliplatin-resistant human colon cancer
cell lines using lectin microarray, FITC-lectin binding and real
time RT-PCR. Additionally, we confirmed that the glycogene β3GnT8
affected oxaliplatin resistance through modification of the
polylactosamine structures on integrin β1.
Lectin microarray has been demonstrated to be useful
in revealing glycan components and glycome structures of a
biological sample (16). It can be
exploited to identify aberrant glycosylation patterns, which in
turn would help in enhancing the specificity of cancer diagnosis
(17). For example, using lectin
microarray analysis, Wu et al revealed that gastric cancer
cells exhibited elevated levels of α2-6 sialylation, core
fucosylation, and the Tn antigen (10). Zhou et al discovered that
lectin RCA-I specifically bound to metastasis-associated cell
surface glycans in triple-negative breast cancer (11). Moreover, the FITC-lectin flow
cytometric analysis is an effective approach to validate the
results of lectin microarray not only qualitatively but also
quantitatively. Combined use of these two methods ensured
identification of correct glycosylation status and led to several
important findings. The integrated strategy employed in the present
study revealed differential expression of α2-6 sialylation, core
fucosylation, polylactosamine-type N-glycans, β1,6-GlcNAc branched
N-glycans, bisecting GlcNAc and the T antigen (recognized by SNA,
LCA, LEA, L-PHA, PHA-E, and Jacalin, respectively) between colon
cancer cells SW620 and its oxaliplatin-resistant cells SW620R. In
previous studies, polylactosamine structure alterations were
reported to be involved in drug resistance of human colon cancer,
breast cancer and leukemia (7,8,15). Our
results further support that increasing polylactosamine chain
expression may enhance colon cell oxaliplatin resistance.
Glycogens play important roles in glycan synthesis
and modification. In the present study, the expression profiles of
glycogenes were analyzed using real-time PCR analysis. We
determined that the glycogenes including ST6GaL1, FUT8, β3GnT8,
GnT-V, GnT-III and GALNT1 were differentially expressed between the
SW620 and SW620R cells. These data were consistent with the lectin
microarray and FITC-lectin flow cytometric analysis. The glycogene
β3GnT8 belongs to the family of β1,
3-N-acetylglucosaminyltransferase (β3GnT), which catalyze the
biosynthesis of polylactosamine-type N-glycans (18). In the present study, we clearly
demonstrated that the expression of β3GnT8 was increased in the
oxaliplatin-resistant cells SW620R as compared to the parental
cells SW620. In addition, we further demonstrated that the
silencing of β3GnT8 in SW620R cells resulted in increased
chemosensitivity to oxaliplatin. The product of β3GnT8 also
decreased in β3GnT8-siRNA transfected SW620R cells labeled with LEA
lectin. Conversely, a stable high expression of β3GnT8 in SW620
cells could increase resistance to oxaliplatin. The β3GnT8 product
was also increased in pEGFP-c1-β3GnT8 plasmid-transfected SW620
cells. All these features suggest that β3GnT8 contributed to the
development of oxaliplatin resistance by catalyzing the formation
of polylactosamine-type N-glycans.
Integrin β1 is a highly N-glycosylated transmembrane
protein that is composed of 12 potential N-glycosylation sites on
its polypeptide backbone (19).
Most studies of altered integrin β1 function have focused on either
changes in integrin β1 expression or regulation of activity through
‘inside-out’ signaling mechanisms (20,21).
However, there is growing evidence for the role of glycosylation in
the regulation of integrin β1 function. For example,
ST6Gal-I-mediated sialylation of integrin β1 enhanced cancer cell
adhesion to, and migration on collagen I (22). Aberrant expression of β4GalT3
through alteration of integrin β1 activation and glycan structures
may enhance cancer cell invasiveness (23). It has been reported that
polylactosamine chains mainly appear on the N-glycans of integrin
β1 in colon cancer cells (23). In
the present study, altered glycosylation of integrin β1 was
reveales in SW620R cells. We observed decreased and increased
polylactosamine carried on integrin β1 with β3GnT8 knockdown and
overexpression, respectively. Exogenous β3GnT8 induced marked
alteration of the glycosylated forms on integrin β1. Integrin β1
could therefore be a target of β3GnT8 in SW620R cells.
Activation of FAK/paxillin is considered to be an
important step in integrin β1 signaling (24,25).
The present study confirmed that the resistant cell line SW620R
exhibited higher FAK/paxillin activity than the sensitive one.
Altered expression of β3GnT8 markedly regulated the activity of
FAK/paxillin in colon cancer cells.
Collectively, our findings reveal new insights into
the regulation of oxaliplatin resistance by aberrant expression of
β3GnT8 through alteration of integrin β1 glycosylation in colon
cancer. It also provides a potentially new biomarker for prediction
of drug resistance in colon cancer patients.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (81502666), the Initial Project
for Post-Graduates of Hubei University of Medicine (2016QDJZR10),
the Natural Science Foundation of the Hubei Provincial Department
of Education (Q20162115), and the Scientific and Technological
Project of Shiyan City of Hubei Province (15K65).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen WQ, Zheng RS, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer Statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dey H and Liu ZR: Phosphorylation of P68
RNA Helicase by P38 MAP kinase contributes to colon cancer cells
apoptosis induced by oxaliplatin. BMC Cell Biol. 13:272012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martinez-Balibrea E, Martínez-Cardús A,
Ginés A, Ruiz de Porras V, Moutinho C, Layos L, Manzano JL, Bugés
C, Bystrup S, Esteller M and Abad A: Tumor-related molecular
mechanisms of oxaliplatin resistance. Mol Cancer Ther.
14:1767–1776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Raman R, Raguram S, Venkataraman G,
Paulson JC and Sasisekharan R: Glycomics: An integrated systems
approach to structure-function relationships of glycans. Nat
Methods. 2:817–824. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Freire-de-Lima L: Sweet and sour: The
impact of differential glycosylation in cancer cells undergoing
epithelial-mesenchymal transition. Front Oncol. 4:592014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Z, Zhao Y, Jiang L, Miao X, Zhou H
and Jia L: Glycomic alterations are associated with multidrug
resistance in human leukemia. Int J Biochem Cell Biol.
44:1244–1253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma H, Miao X, Ma Q, Zheng W, Zhou H and
Jia L: Functional roles of glycogene and N-glycan in multidrug
resistance of human breast cancer cells. IUBMB Life. 65:409–422.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kudo T, Nakagawa H, Takahashi M, Hamaguchi
J, Kamiyama N, Yokoo H, Nakanishi K, Nakagawa T, Kamiyama T,
Deguchi K, et al: N-glycan alterations are associated with drug
resistance in human hepatocellular carcinoma. Mol Cancer. 6:322007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu J, Qin H, Li T, Cheng K, Dong J, Tian
M, Chai N, Guo H, Li J, You X, et al: Characterization of
site-specific glycosylation of secreted proteins associated with
multi-drug resistance of gastric cancer. Oncotarget. 7:25315–25327.
2016.PubMed/NCBI
|
|
11
|
Zhou SM, Cheng L, Guo SJ, Wang Y,
Czajkowsky DM, Gao H, Hu XF and Tao SC: Lectin RCA-I specifically
binds to metastasis-associated cell surface glycans in
triple-negative breast cancer. Breast Cancer Res. 17:362015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fry SA, Afrough B, Lomax-Browne HJ, Timms
JF, Velentzis LS and Leathem AJ: Lectin microarray profiling of
metastatic breast cancers. Glycobiology. 21:1060–1070. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tao SC, Li Y, Zhou J, Qian J, Schnaar RL,
Zhang Y, Goldstein IJ, Zhu H and Schneck JP: Lectin microarrays
identify cell-specific and functionally significant cell surface
glycan markers. Glycobiology. 18:761–769. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Shen L, Xu L, Sun X, Zhou J and Wu
S: Down-regulation of β-1,3-N-acetylglucosaminyltransferase-8 by
siRNA inhibits the growth of human gastric cancer. Mol Med Report.
4:497–503. 2011.
|
|
15
|
Shen L, Yu M, Xu X, Gao L, Ni J, Luo Z and
Wu S: Knockdown of β3GnT8 reverses 5-fluorouracil resistance in
human colorectal cancer cells via inhibition the biosynthesis of
polylactosamine-type N-glycans. Int J Oncol. 45:2560–2568. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
An HJ, Kronewitter SR, de Leoz ML and
Lebrilla CB: Glycomics and disease markers. Curr Opin Chem Biol.
13:601–607. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Syed P, Gidwani K, Kekki H, Leivo J,
Pettersson K and Lamminmäki U: Role of lectin microarrays in cancer
diagnosis. Proteomics. 16:1257–1265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishida H, Togayachi A, Sakai T, Iwai T,
Hiruma T, Sato T, Okubo R, Inaba N, Kudo T, Gotoh M, et al: A novel
β1,3-N-acetylglucosaminyltransferase (β3Gn-T8), which synthesizes
poly-N-acetyllactosamine, is dramatically upregulated in colon
cancer. FEBS Lett. 579:71–78. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Janik ME, Litynska A and Vereecken P: Cell
migration-the role of integrin glycosylation. Biochim Biophys Acta.
1800:545–555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Christie DR, Shaikh FM, Lucas JA IV, Lucas
JA III and Bellis SL: ST6Gal-I expression in ovarian cancer cells
promotes an invasive phenotype by altering integrin glycosylation
and function. J Ovarian Res. 1:32008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hou S, Hang Q, Isaji T, Lu J, Fukuda T and
Gu J: Importance of membrane-proximal N-glycosylation on integrin
β1 in its activation and complex formation. FASEB J. 30:4120–4131.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seales EC, Jurado GA, Brunson BA,
Wakefield JK, Frost AR and Bellis SL: Hypersialylation of beta1
integrins, observed in colon adenocarcinoma, may contribute to
cancer progression by up-regulating cell motility. Cancer Res.
65:4645–4652. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen CH, Wang SH, Liu CH, Wu YL, Wang WJ,
Huang J, Hung JS, Lai IR, Liang JT and Huang MC:
β-1,4-Galactosyltransferase III suppresses β1 integrin-mediated
invasive phenotypes and negatively correlates with metastasis in
colorectal cancer. Carcinogenesis. 35:1258–1266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nalla AK, Asuthkar S, Bhoopathi P, Gujrati
M, Dinh DH and Rao JS: Suppression of uPAR retards
radiation-induced invasion and migration mediated by integrin
β1/FAK signaling in medulloblastoma. PLoS One. 5:e130062010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yun SP, Ryu JM and Han HJ: Involvement of
β1-integrin via PIP complex and FAK/paxillin in
dexamethasone-induced human mesenchymal stem cells migration. J
Cell Physiol. 226:683–692. 2011. View Article : Google Scholar : PubMed/NCBI
|