Introduction
Keratins are cytoplasmic intermediate filaments
expressed by epithelial tissues and are expressed in tissue- and
differentiation-specific manners (1). They play an important role in
maintaining cell viability and structure by providing mechanical
support. In addition to their cytoprotective function, keratins are
known to modulate important signaling pathways associated with
protein synthesis, cell growth and cell differentiation (2). Studies have found that K5/14 null mice
died 2 days after birth (3) and the
loss of K14 cannot be compensated for by the ectopic expression of
any other keratin, suggesting that K14 performs an important
regulatory function (4,5). Previous research in our laboratory
demonstrated that depletion of K14 leads to an increase in Notch-1
and differentiation markers such as K1 and K10 and a decrease in
tumorigenic potential.
p63 is a member of the p53 tumor-suppressor family
of proteins (6). There are two
major isoforms of p63 dependent upon the differential promoter
usage, which are TAp63 which has a N terminal transactivation
domain and ΔNp63 which lacks the N terminal domain. Each is further
subdivided into α, β, γ depending upon the C terminal splicing.
ΔNp63 and its isoforms are known to regulate the proliferative
potential of basal cells and initiate the process of
differentiation (7), while TAp63
plays an important role in the maintenance of the basal cell
population and prevents premature ageing of the epidermis (8). TAp63 acts as a tumor suppressor and is
known to be downregulated in various types of cancers such as colon
cancer (9) and was also found to
promote epithelial-mesenchymal transition in MDCK cells (10).
In the present study, we aimed to elucidate the
molecules involved in K14-regulated cell differentiation and
decrease in tumorigenic potential. p63 is an important molecule to
study with respect to K14 as ΔNp63 is known to be a direct
regulator of K5/14 expression in basal keratinocytes (11). Furthermore, p63 is a known modulator
of epidermal homeostasis (12,13).
In the present study, we showed that TAp63 is the
isoform of p63 which undergoes upregulation after K14 depletion.
Furthermore, to understand the role of TAp63 in K14-depleted cells,
the TAp63 gene was knocked down using shRNA technology in
K14-depleted cells. We report here that TAp63 regulates cell
differentiation, cell motility and tumorigenesis in OSCC-derived
cells possibly through Notch-1.
Materials and methods
Statement of ethics
All protocols for animal studies were approved by
the Institutional Animal Ethics Committee (IAEC) (approval ID
28/2016) constituted under the guidelines of the ‘Committee for the
Purpose of Control and Supervision of Experiments on Animals
(CPCSEA)’ of the Government of India.
Housing and monitoring of experimental
animals and in vivo tumorigenicity assay
The animals were housed in a controlled environment
at 23±2°C, with 40–70% relative humidity and a dark/light cycle of
12 h each. The animals received autoclaved in-house-made food
including natural ingredients such as roasted Bengal gram, casein,
milk powder, vitamins, ground nut oil and sterile water ad
libitum. The tumorigenic potential of cells was determined by
subcutaneous injection in NOD-SCID mice. The cells were suspended
in plain medium without serum, and 6×106 cells were
injected subcutaneously into the dorsal flank of 6 to 8 week-old
mice. Four mice were injected per clone and were observed for tumor
formation over a period of approximately 2 months. Tumor volume was
determined using a digital Vernier caliper (Advance, Alandi, India)
and the volume was calculated by the modified ellipsoidal formula:
[Tumor volume = 1/2 (length × width2)]. After 6–8 weeks,
the animals were sacrificed so that the mean tumor diameter did not
exceed 1.2 cm, as described in the AAALAC guidelines for Animal
Welfare in Cancer Research (14).
The mice were euthanized using CO2 method and the tumor
was removed (18).
Plasmids and cell lines
Human tongue SCC-derived cell line AW13516 was
established in our institute (15).
This cell line was cultured in Iscove's modified Dulbecco's medium
(IMDM; Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine
serum (FBS; HyClone Thermo Scientific, Lafayette, CO, USA) and
antibiotics (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) in a
5% CO2 incubator at 37°C. The establishment and
characterization of HaCaT cells were as described previously
(16). TAp63 shRNA was designed and
cloned into the Plko1.Neo plasmid (plasmid #13425; Addgene,
Cambridge, MA, USA). Efficient knockdown was generated by
transducing a lentiviral-based Plko1.neo vector encoding shRNA and
an empty vector in K14-depleted HaCaT (D2 clone) and AW13516 (K7
clone) cell lines. The shRNA sequences for TAp63 are as follows:
TAp63A, 5′-GGAACAGCCTATATGTTCAGTTC-3′ and TAp63B,
5′-GCCCAGAGCACACAGACAAACTC-3′.
Lentiviral-mediated transduction
293FT cells were cultured until achieving 50%
confluency in complete Dulbecco's modified Eagle's medium (DMEM;
Life Technologies). Calcium phosphate precipitation method was used
for co-transfection of the lentiviral transfer and packaging
vectors (17). For transduction,
the viral supernatant along with polybrene (8 µg/ml) was added to
the 50% confluent OSCC cells. After 24 h, the supernatant was
replaced with complete media. Furthermore, the stable clones were
selected in puromycin (0.5 µg/ml) and G418 (500 µg/ml), purchased
from Sigma-Aldrich (St. Louis, MO, USA).
Western blotting
The whole cell extracts were prepared in SDS lysis
buffer with protease inhibitor cocktail (Sigma-Aldrich). For
Notch-1 inhibition, K14-depleted cells were treated with
γ-secretase inhibitor (DAPT;
N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl
ester; Merck-Millipore, Darmstadt, Germany) at concentrations of
150 and 250 nM for 72 h and then cell lysates were prepared. After
blocking, the blot was incubated with the diluted primary antibody
for 1 h at room temperature (RT) on a rocker. The blot was then
washed thrice with TBST [0.1% Tween-20 (v/v), 150 mM NaCl, 10 mM
Tris-HCl pH 8.0] followed by incubation with horseradish peroxidase
(HRP)-conjugated secondary antibody (anti-mouse; A4416 or
anti-rabbit; A-0545; Sigma-Aldrich) for 1 h at RT on a rocker
followed by TBST washes. The blots were developed using ECL
chemiluminescence reagent (WesternBright™ ECL; Advansta, East
Sussex, UK) according to the manufacturer's protocol (18). The primary antibodies for K14
(1:8,000; mouse monoclonal, clone LL002; cat. no. MCA890) and
involucrin (1:1,000 dilution ratio; mouse monoclonal, clone SY5;
cat. no. 5390-9950) were obtained from AbD Serotec (a Bio-Rad
Company, Raleigh, NC, USA). The non-phospho β-catenin (1:1,000;
rabbit monoclonal antibody D28UY; cat. no. 19807) and Notch-1
antibodies (1:1,000; rabbit monoclonal, clone D1E11; cat. no. 3608)
were purchased from Cell Signaling Technology (Danvers, MA, USA).
Vimentin (1:1,000; mouse monoclonal, clone V9; cat. no. V-6630),
β-actin (1:8,000; mouse monoclonal; cat. no. AC-74) and were
obtained from Sigma-Aldrich. Total β-catenin (1:1,000; mouse
monoclonal; cat. no. Sc7963) was purchased from Santa Cruz
Biotechnology (Dallas, TX, USA). TAp63 (1:1,500; mouse polyclonal;
cat. no. Ab53039) was obtained from Abcam (Cambridge, MA, USA).
E-cadherin antibody (1:500; mouse monoclonal; cat. no. 610182) was
purchased from BD Biosciences (Franklin Lakes, NJ, USA).
RT-PCR and RT-qPCR
For RT-PCR and RT-qPCR, RNA was isolated with the
TRI reagent and cDNA was prepared using Revert Aid First Strand
cDNA synthesis kit according to the manufacturer's protocol (MBI
Fermentas, Amherst, NY, USA). All SYBR-Green-based RT-qPCRs were
performed on the ABI 7900HT fast real-time based systems (Applied
Biosystems, Foster City, CA, USA) as previously described (17). The SYBR-Green Master Mix was
purchased from Applied Biosystems. The relative levels were
calculated using the ΔΔCt method and the relative expression fold
(2−ΔΔCt) was calculated (17). The respective gene expression was
normalized to GAPDH. The primers used for PCR amplification are as
follows: TAp63 forward, 5′-TGGTGCGACAAACAAGATTG-3′ and reverse,
5′-ATAGGGACTGGTGGACGAGG-3′; ΔNp63 forward,
5′-TGTACCTGGAAAACAATGCCCA-3′ and reverse,
5′-GACGAGGAGCCGTTCTGAATCT-3′; GAPDH forward,
5′-CTTCTTTTGCGTCGCCAGCC-3′ and reverse,
5′-GAGTTAAAAGCAGCCCTGGTGA-3′; involucrin forward,
5′-GAAACAGCCAACTCCACTGC-3′ and reverse, 5′-ATTCTTGCTCAGGCAGTCCC-3′;
vimentin forward, 5′-GTCAGCAATATGAAAGTGTGGC-3′ and reverse,
5′-GGTAGTTAGCAGCTTCAACGG-3′; K1 forward, 5′-TGACAAGGTGAGGTTCCTGG-3′
and reverse, 5′-GTTGGTCCACTCTCCTTCGG-3′; loricrin forward,
5′-GATCTGCCACCAGACCCAG-3′ and reverse 5′-CCCCTGGAAAACACCTCCAA-3′;
E-cadherin forward, 5′-CTTTGACGCCGAGAGCTACA-3′ and reverse,
5′-TTTGAATCGGGTGTCGAGG-3′; Snail forward,
5′-CCAGTGCCTCGACCACTATG-3′ and reverse,
5′-CTGCTGGAAGGTAAACTCTGGA-3′; Twist forward,
5′-TCTACCAGGTCCTCCAGAG-3′ and reverse,
5′-CTCCATCCTCCAGACCGAGA-3′.
Phenotypic assays for cell
transformation
For the invasion assay, 2×105 cells were
seeded on a Matrigel (1 mg/ml; BD Biosciences) coated insert and
incubated for 16 h (17). Transwell
migration assay was performed similar to the invasion assay but
without coating the insert with Matrigel. At the 15th h, 4 µg/ml
calcein AM (Life Technologies) was added to the lower chamber and
incubated for 1 h at 37°C. The cells on the upper chamber were
carefully removed with a cotton swab at 16th h. Fluorescence was
measured at a wavelength of 488/535 (Ex/Em) on a bottom reading
fluorescent plate reader (Berthold Technologies, Bad Wildbad,
Germany).
For soft agar assay, 1 ml of the basal layer was
made by adding equal volumes of 2X complete IMDM and 2% low melting
agarose. Cells (1,000) in complete medium containing 0.4% low
melting agarose were seeded over the basal layer. Plates were fed
with complete medium on every alternate day and incubated at 37°C
in a 5% CO2 atmosphere for 15 days. Opaque and dense
colonies were observed and counted microscopically (Zeiss Aiovert
200M; Zeiss, Oberkochen, Germany) on day 15.
Immunohistochemistry (IHC)
Mouse tumor tissues were fixed in 10% formalin
buffer and processed for histology. Immunohistochemistry was
performed as previously reported (19). As per the protocol, the tissues were
subjected to microwave treatment for antigen retrieval. The
sections were then blocked with pre-immune horse serum for 30 min
at RT followed by incubation in primary antibody K14 (1:100) and
the respective secondary antibody which was coupled with biotin
(Elite ABC Kit; Vector Laboratories, Burlingame, CA, USA). Signals
were detected by an avidin-biotin based immunoperoxidase technique.
The expression of protein in IHC staining was quantified by visual
assessment of the microscopic field by counting a total of 100
cells per field and for each section, in an upright microscope
(Leica Biosystems, Wetzlar, Germany).
Densitometric quantification and
statistical analysis
The densitometric quantification of the western
blots was performed using ImageJ software (NIH, Bethesda, MD, USA).
Band intensities were normalized with respect to their loading
controls. All the statistical analysis was performed using the
Graph Pad Prism Software (version 5.0). Two groups of data were
statistically analyzed by t-test. Two-way ANOVA using Bonferroni
test was performed for tumor volume analysis. A P-value <0.05
was considered to indicate a statistically significant result.
Results
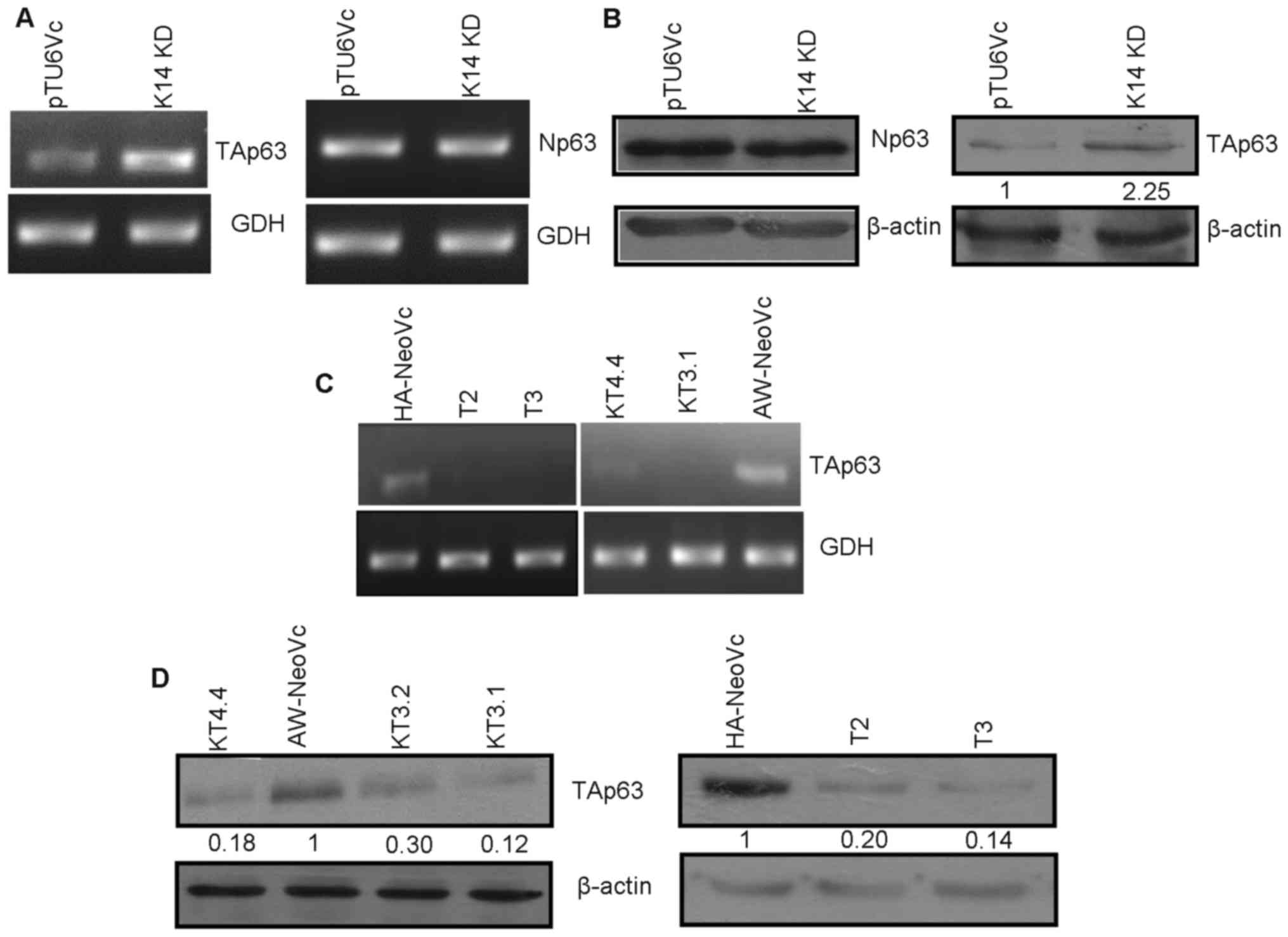
Increased expression of TAp63 in
K14-knockdown cells
p63 is known to regulate transcription of K14. In
order to assess the expression of p63 in K14-depleted cells, we
performed western blotting and reverse transcription. We observed
that there was no alteration in ΔNp63 at the mRNA and protein level
(Fig. 1A and B). However, there was
an increase in TAp63 at the transcript level (Fig. 1A). Furthermore, an 2.25-fold
increase in TAp63 protein level in the K14-knockdown (K14 KD) cells
was also observed (Fig. 1B).
Generation of TAp63 knockdown in
K14-depleted cells
TAp63 was stably downregulated in the K14-depleted
cells. Two clones (T2 and T3) obtained from HaCaT cells
demonstrated significant levels of reduction in TAp63 at the
transcript level as seen by RT-PCR (Fig. 1C). TAp63 was downregulated 80 and
86% in T2 and T3, respectively, at the protein level as compared to
the vector control (HA-NeoVc) (Fig.
1D). Ha-NeoVc stands for plko1.neo empty vector in HaCaT K14 KD
cells. TAp63 KD clones (KT3.1, 3.2 and 4.4) obtained from AW13516
cells also showed reduced TAp63 expression at the mRNA level in
RT-PCR (Fig. 1C). There was an 88,
70 and 82% reduction in TAp63 expression at the protein level in
KT3.1, 3.2 and 4.4, respectively (Fig.
1D) as compared to AW-NeoVc (vector control in AW-13516 K14 KD
cells). There were no significant changes observed in the levels of
K5, K14 and ΔNp63 respectively after depletion of TAp63 in these
cells, as seen by western blotting (Fig. 5D).
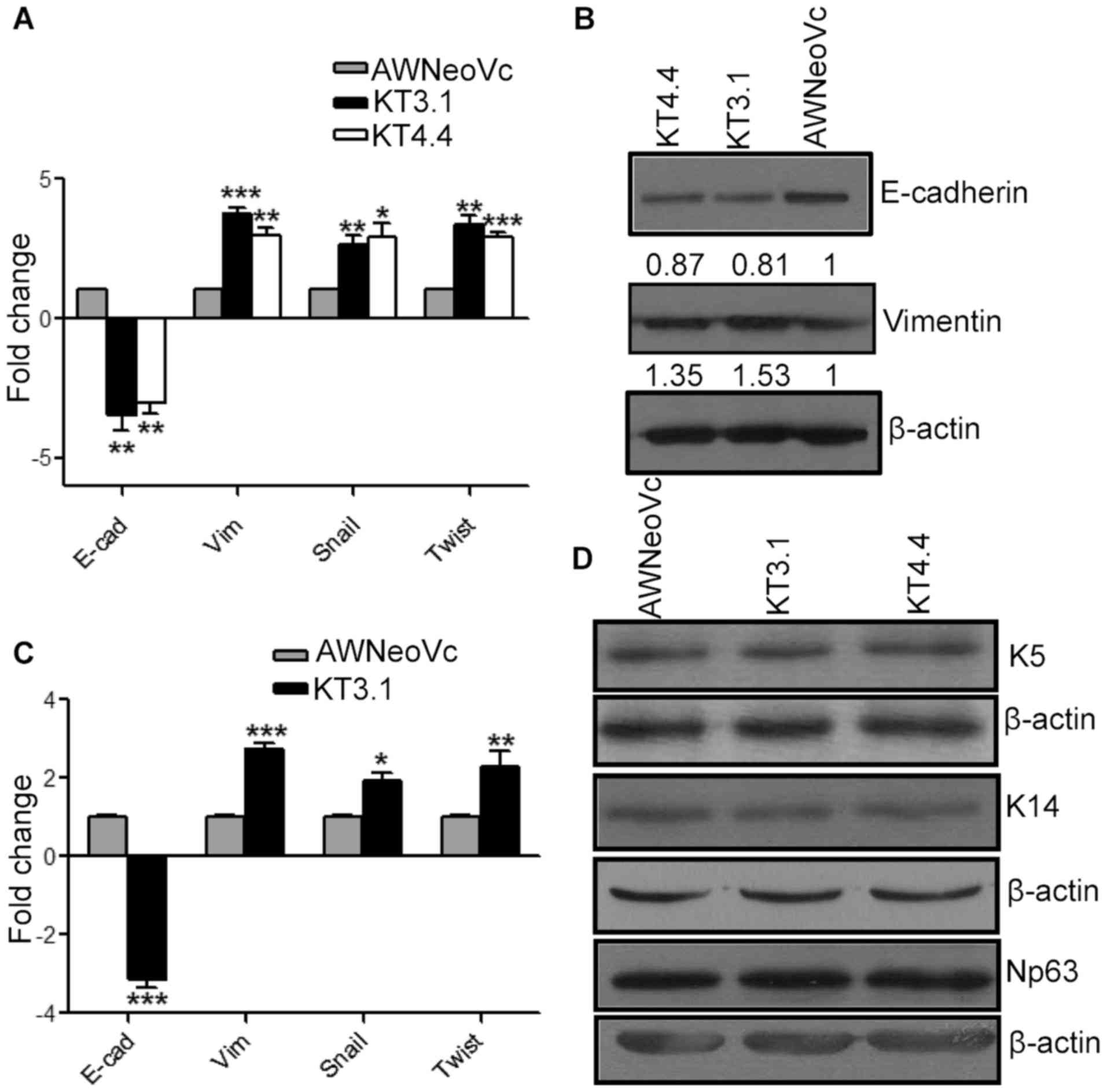 | Figure 5.TAp63 downregulation leads to an
increase in EMT. (A) The graph represents RT-qPCR analysis of
E-cadherin, vimentin, snail and twist in KT3.1, KT4.4 and AW-NeoVc.
(B) Western blot analysis of EMT markers E-cadherin and vimentin in
KT3.1, KT4.4 and AW-NeoVc. (C) The graph represents the RT-qPCR
analysis of E-cadherin, vimentin, snail and Twist in tumors
obtained from KT3.1 and AW-NeoVc NOD-SCID mice, respectively. (D)
Western blot analysis of K5, K14 and ΔNp63 in TAp63 KD clones and
AWNeoVc. Np63 stands for ΔNp63. The graph data represent the mean ±
SEM of three independent experiments. The number below the blots
indicates the relative intensity of each bands using densitometric
analysis. ***P<0.0001, **P<0.005, *P<0.05. |
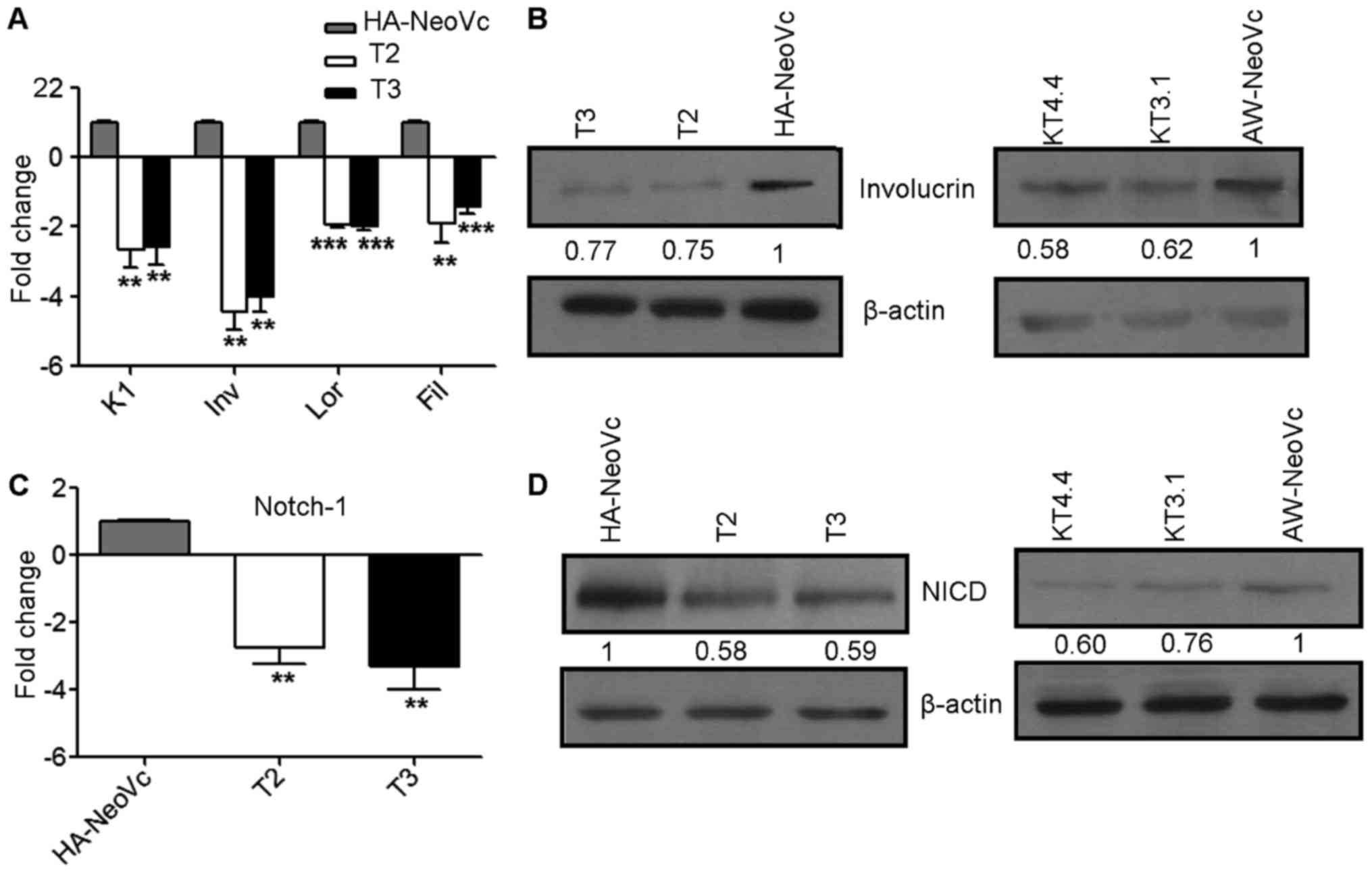
TAp63 knockdown results in
downregulation of Notch-1 and cell differentiation markers
We assessed the levels of differentiation markers by
western blotting and RT-qPCR. Involucrin, K1, loricrin and
filaggrin were downregulated 4.0-, 2.6-, 1.8- and 1.5-fold,
respectively, at the mRNA level (Fig.
2A) (P<0.005). There was an 25 and 22% decrease in the
levels of involucrin at the protein level in the T2 and T3 clones,
respectively (Fig. 2B). Similarly,
there was a 38 and 42% decrease in involucrin in KT3.1 and KT4.4,
respectively, as compared to AW-NeoVc (Fig. 2B).
p63 is known to regulate the process of
differentiation in keratinocytes through Notch-1 (20). We observed an 2.7- and 3.3-fold
decrease in Notch-1 expression in T2 and T3, respectively (Fig. 2C) (P<0.002). Consistently, a 41%
decrease in activation of Notch-1 intracellular domain (NICD) at
the protein level was observed in the TAp63KD HaCaT clones as
compared to HA-NeoVc. There was a 24 and 40% decrease in NICD in
KT3.1 and KT4.4, respectively (Fig.
2D). These observations are consistent with a previous study,
which demonstrated that TAp63 is known to activate transcription of
members of the Notch-1 pathway which in turn promotes transcription
of differentiation-related genes (21).
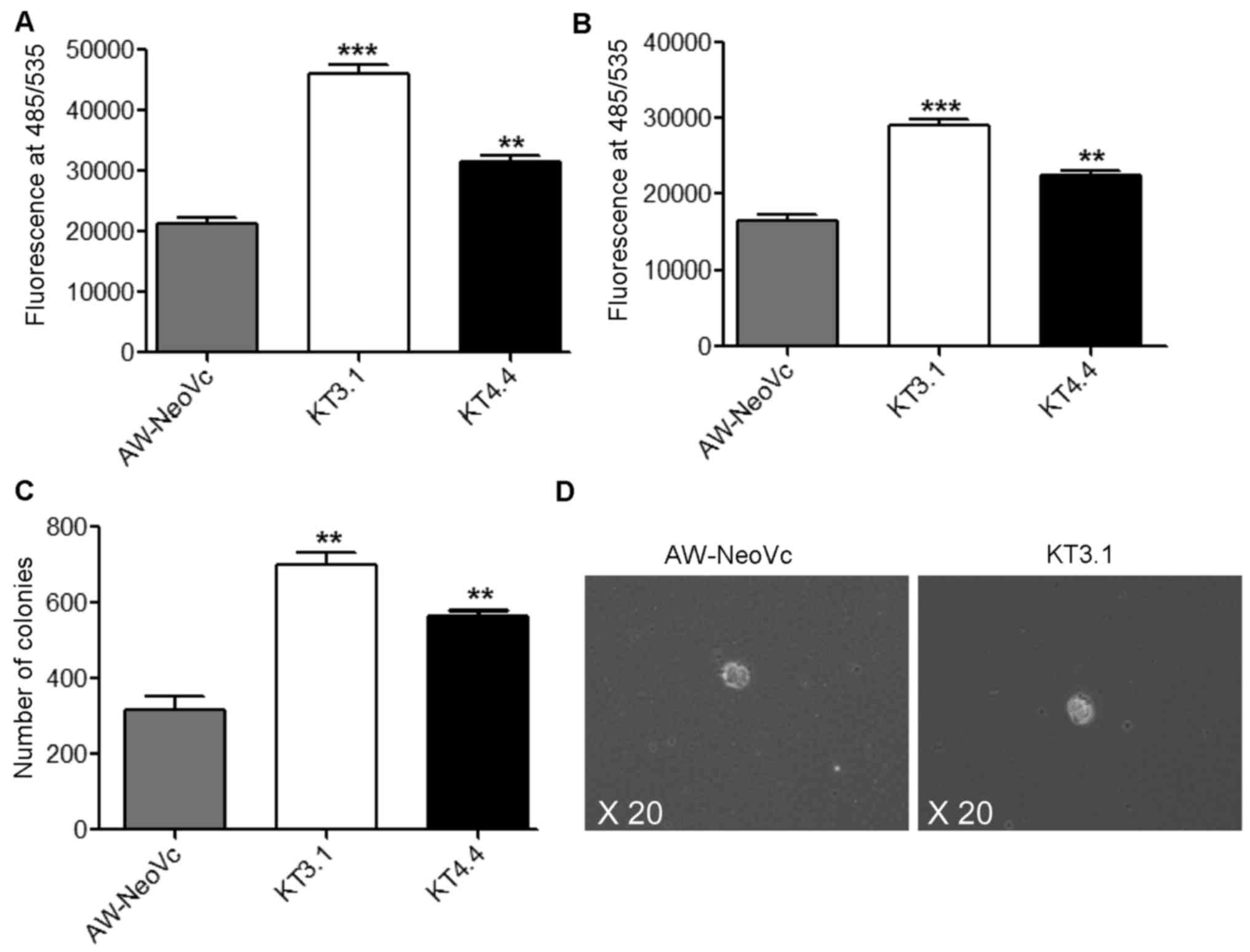
Downregulation of TAp63 leads to
changes in the transformation potential of AW13516 cells and in
vivo tumorigenicity
Knockdown of TAp63 in K14-depleted cells caused an
increase in cell migration and invasion. In the Transwell migration
assay, TAp63 KD clones KT3.1 and KT4.4 showed an 50 and 37.5%
increase in migration, respectively, as compared to the vector
control AW-NeoVc (Fig. 3A)
(P<0.001). There was an 40 and 30% increase in invasion in the
TAp63 KD clones KT3.1 and KT4.4, respectively, as compared to the
vector control AW-NeoVc (Fig. 3B)
(P<0.002). However, there were no changes in the migration and
invasion in HaCaT TAp63 KD clones as compared to HA-NeoVc (data not
shown), since HaCaT is an immortalized and not a transformed cell
line. Furthermore, there was an increase in the number of soft agar
colonies by 40% in KT3.1 and 22% in KT4.4 compared to AW-NeoVc
(Fig. 3C and D) (P<0.002). The
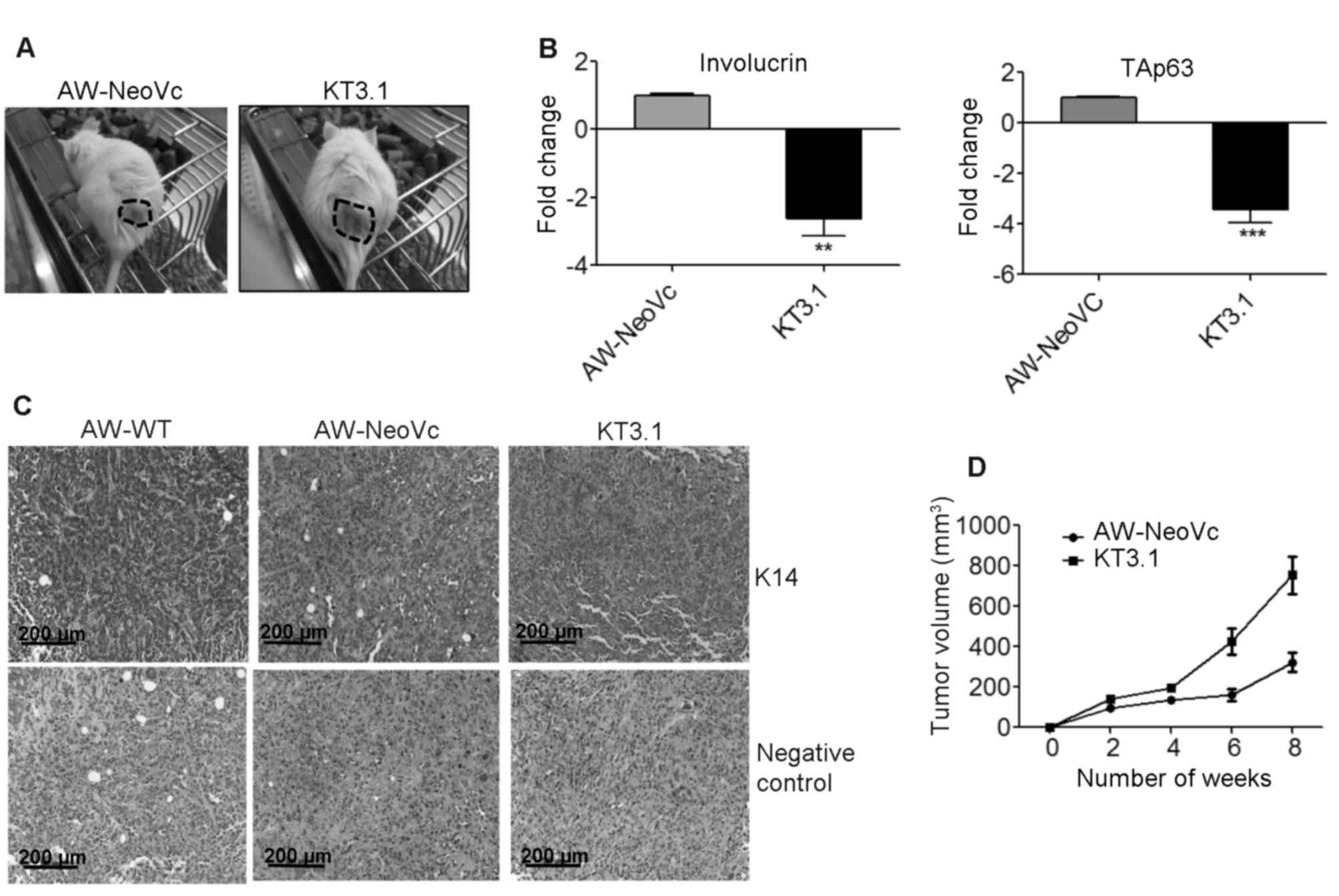
tumorigenic potential of TAp63 KD cells was determined by
subcutaneous injection into NOD-SCID mice (n=4). The volume of the
tumors formed in the TAp63 KD mice (KT3.1) was significantly higher
as compared to the vector control (AW-NeoVc) (Fig. 4A and D). IHC studies illustrated a
decrease in the expression of K14 in the tumor samples obtained
from the mice injected with AW-NeoVc and KT3.1 as compared to the
wild-type (AW-WT) tumor samples (Fig.
4C). The TAp63 antibody was not effective in IHC. Therefore,
RT-qPCR was performed to analyze the levels of TAp63 in RNA samples
derived from the respective tumors. At the mRNA level, the TAp63
level in KT3.1 was 3.4-fold less when compared to AW-NeoVc in the
tumor tissues (Fig. 4B)
(P<0.001). The involucrin mRNA level was also 2.5-fold lower in
the tumor samples derived from KT3.1 when compared to AW-NeoVc
(Fig. 4B) (P<0.002).
Downregulation of TAp63 leads to an
increase in the expression of EMT markers
We observed changes in invasion in vitro,
therefore, we assessed whether epithelial-mesenchymal transition
(EMT) was involved in the TAp63-regulated invasion. We found that
downregulation of TAp63 led to an 3.36-, 2.77- and 3.14-fold
increase in the mRNA levels of vimentin (P<0.001), snail
(P<0.05) and twist (P<0.001) in the K14-depleted cells,
respectively. A 2.91-fold downregulation of E-cadherin was noted at
the mRNA level (Fig. 5A)
(P<0.003). In addition, there was a 19 and 13% decrease in
E-cadherin protein expression in KT3.1 and KT4.4, respectively, as
compared to AW-NeoVc. Subsequently, there was an 1.53- and
1.35-fold increase in vimentin expression at the protein level in
KT3.1 and KT4.4, respectively (Fig.
5B). Likewise, we observed an increase in the expression of
vimentin, snail and twist in the KT3.1 tumor tissues at the mRNA
level (P<0.05) and subsequently, there was downregulation of
E-cadherin mRNA in the KT3.1 tumor tissues (Fig. 5C).
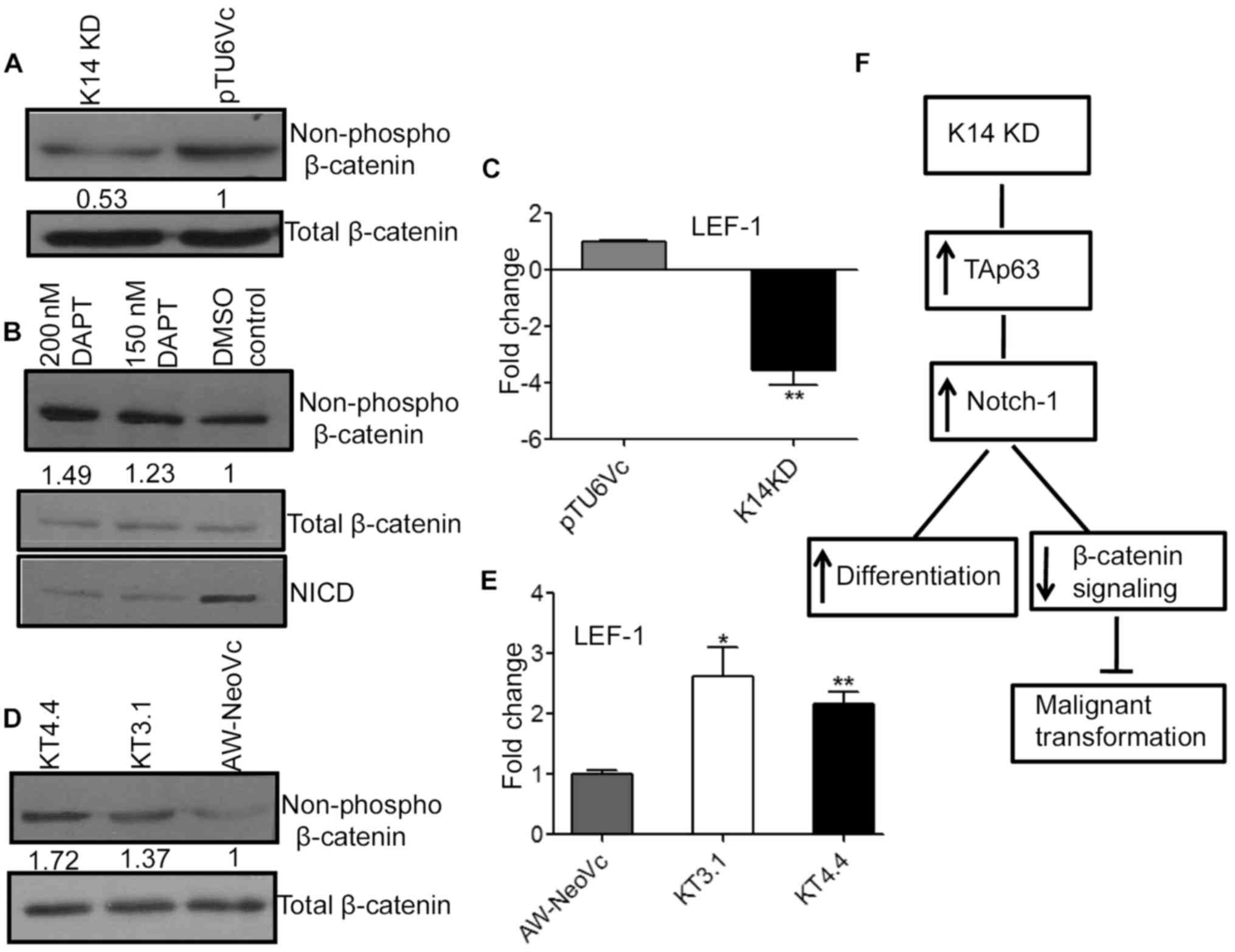
Notch-1 regulates β-catenin signaling
in the K14 KD cells
The available literature has shown that inactivation
of Notch-1 activates β-catenin signaling and its downstream target
lymphoid enhancing factor-1 (LEF-1) which leads to an increase in
tumorigenesis (22). Notch-1 was
upregulated in the K14 KD cell lines and in addition there was a
decrease in tumorigenic potential of these cells. Thus, we
hypothesized that β-catenin could be one of the targets that maybe
involved in the K14-regulated decrease in tumorigenic potential. To
assess this hypothesis, we examined the β-catenin levels in K14 KD
cells. There was a decrease in the non-phospho levels of β-catenin
(active pool) in the K14 KD cells as compared to the vector control
(Fig. 6A). The total β-catenin
levels remain unchanged. There was an 3.5-fold decrease in LEF-1
expression at the mRNA level in the K14 KD cells as compared to
pTU6Vc (Fig. 6C) (P<0.001).
LEF-1 is a downstream target of β-catenin.
To further confirm that β-catenin levels in K14 KD
cells are modulated by Notch-1, we treated the AW-K7 clone with
Notch-1 inhibitor DAPT. There was an 1.5-fold increase in
non-phospho β-catenin (active pool) levels with 200 nM of DAPT in
the AW-K7 clone as compared to the DMSO-treated AW-K7 clone
(control) (Fig. 6B). This verifies
our hypothesis that β-catenin levels are regulated by Notch-1 in
K14-depleted cells.
However, TAp63 knockdown clones KT3.1 and KT4.4
displayed an 1.37- and 1.72-fold increase in non-phospho β-catenin
(active pool) levels as compared to the vector control (Fig. 6D). Subsequently, there was an
2.3-fold increase in LEF-1 expression at the mRNA level in the
TAp63 KD clones (Fig. 6E).
Discussion
K14 is expressed in the basal cells of stratified
epithelia and as the cells move upward to the suprabasal layer, the
expression of K14 is replaced by expression of K1 and K10. This
suggests that keratins play an important regulatory role in cell
differentiation (23). A previous
study by our laboratory showed that depletion of K14 initiates the
process of cell differentiation. An increase in Notch-1 expression
and a decrease in tumorigenicity was also observed in K14 depleted
cells (24). Yet, the molecules
involved in the K14-regulated differentiation and malignant
transformation remain unknown.
p63 is a master regulator of epithelial cell
proliferation and differentiation. We showed here that
K14-downregulated cells exhibited an increase in TAp63 levels at
both the mRNA and protein levels. Upon TAp63 KD, we observed a
decrease in differentiation markers such as involucrin, K1,
filaggrin and loricrin. Furthermore, in the present study, upon
downregulation of TAp63 in K14 KD cells, there was a decrease in
the expression in the Notch-1 level, suggesting that Notch-1 is
regulated by TAp63. A previous report demonstrated that there is a
complex interplay between Notch-1 and p63 in regulating the switch
between cell differentiation and stemness. p63 plays a dual role by
suppressing Notch-1 in the basal cell compartment and then
synergizes with Notch-1 signaling in the early stages of
differentiation (20). Our data is
consistent with this report, as we showed that upon K14
downregulation, there was an increase in TAp63 expression and an
increase in intracellular levels of Notch-1, which further
regulates genes involved in differentiation.
TAp63-depleted cells further exhibited an increase
in cell migration, invasion and tumorigenicity in the
K14-downregulated cells. Our observations suggest that in
K14-downregulated cells, TAp63 modulates the process of cell
transformation and tumorigenicity. An increase in invasion could be
related to EMT. Lin et al showed that p63 regulates
metastasis in colon cancer cells by regulating EMT (9). We were curious to ascertain whether
TAp63 KD cells which show a less differentiated and more invasive
phenotype, exhibit EMT. Our results demonstrated an increase in the
expression of vimentin and a decrease in the expression of
E-cadherin at both the transcript and protein levels in the TAp63
KD AW13516 cells. Similar results were obtained in RNA samples
isolated from tumor tissues of mice injected with the TAp63 KD
cells.
Alam et al found an increase in the
expression of Notch-1 and a decrease in tumorigenicity in
vivo in K14 KD cells (24). We
were curious to ascertain whether there were any changes in
β-catenin and its downstream signaling molecules in K14 KD cells.
We found that there was a decrease in the non-phospho pool of
β-catenin (active) in K14 KD cells as seen by western blotting and
subsequent decrease in LEF-1 at the mRNA level. Furthermore, when
we treated the cells with DAPT (Notch-1 inhibitor), there was an
increase in the active pool of β-catenin. However, when we observed
β-catenin levels in TAp63 KD cells, there was an increase in
expression of non-phospho β-catenin at the protein level and LEF-1
at the mRNA level. Thus, we can infer that Notch-1 regulates
tumorigenesis in K14-depleted cells and Notch-1 expression in turn
is modulated by TAp63. Furthermore, how β-catenin levels regulate
tumorigenesis warrants further investigation. Reports available in
the literature demonstrate that inhibition of Notch-1 leads to
development of squamous cell carcinoma in which β-catenin signaling
is upregulated (25,26).
In summary, the present study demonstrated that
TAp63 plays an important role in K14-regulated cell
differentiation. Knockdown of TAp63 led to an increase in cell
motility, invasion and tumorigenesis in vivo. TAp63 may
modulate tumorigenic potential and differentiation of AW13516 cells
through Notch-1 (Fig. 6F). Romano
et al showed that gene expression of K5/14 is regulated by
p63 (11). In the present study, we
demonstrated that upon K14 downregulation, there was an increase in
the expression of TAp63. The mechanism by which K14 regulates TAp63
is yet to be discovered. Further studies are required to
investigate the interplay between p63 and K14 and to decipher the
mechanism underlying the regulation of p63 by K14.
Acknowledgements
The present study was supported by the Department of
Science and Technology (DST, India) (grant no. LS-630/2013) which
also provided a fellowship to Dr Saumya S. Srivastava. We thank Dr
Crismita D'Mello and Dr Richa Tiwari for their experimental
suggestions and Mrs. Sharda Sawant for help with the IHC analysis.
We thank Mr Sridhar Nadkar, Ms. Silvania Charles and Ms. Zinia
D'Souza for assistance in the animal experiments.
Glossary
Abbreviations
Abbreviations:
|
K14
|
keratin14
|
|
OSCC
|
oral squamous cell carcinoma
|
|
Ex/Em
|
excitation/emission
|
|
KD
|
knockdown
|
References
|
1
|
Coulombe PA and Omary MB: ‘Hard’ and
‘soft’ principles defining the structure, function and regulation
of keratin intermediate filaments. Curr Opin Cell Biol. 14:110–122.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Koch PJ and Roop DR: The role of keratins
in epidermal development and homeostasis-going beyond the obvious.
J Invest Dermatol. 123:x–xi. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan Y, Anton-Lamprecht I, Yu QC, Jäckel
A, Zabel B, Ernst JP and Fuchs E: A human keratin 14 ‘knockout’:
The absence of K14 leads to severe epidermolysis bullosa simplex
and a function for an intermediate filament protein. Genes Dev.
8:2574–2587. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hutton E, Paladini RD, Yu QC, Yen M,
Coulombe PA and Fuchs E: Functional differences between keratins of
stratified and simple epithelia. J Cell Biol. 143:487–499. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Paladini RD and Coulombe PA: Directed
expression of keratin 16 to the progenitor basal cells of
transgenic mouse skin delays skin maturation. J Cell Biol.
142:1035–1051. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Osada M, Ohba M, Kawahara C, Ishioka C,
Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M and
Ikawa S: Cloning and functional analysis of human p51, which
structurally and functionally resembles p53. Nat Med. 4:839–843.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Truong AB, Kretz M, Ridky TW, Kimmel R and
Khavari PA: p63 regulates proliferation and differentiation of
developmentally mature keratinocytes. Genes Dev. 20:3185–3197.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin
YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, et al: TAp63
prevents premature aging by promoting adult stem cell maintenance.
Cell Stem Cell. 5:64–75. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin CW, Li XR, Zhang Y, Hu G, Guo YH, Zhou
JY, Du J, Lv L, Gao K, Zhang Y and Deng H: TAp63 suppress
metastasis via miR-133b in colon cancer cells. Br J Cancer.
110:2310–2320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Yan W and Chen X: P63 regulates
tubular formation via epithelial-to-mesenchymal transition.
Oncogene. 33:1548–1557. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Romano RA, Ortt K, Birkaya B, Smalley K
and Sinha S: An active role of the DeltaN isoform of p63 in
regulating basal keratin genes K5 and K14 and directing epidermal
cell fate. PLoS One. 4:e56232009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang A, Schweitzer R, Sun D, Kaghad M,
Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C and McKeon
F: p63 is essential for regenerative proliferation in limb,
craniofacial and epithelial development. Nature. 398:714–718. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mills AA, Zheng B, Wang XJ, Vogel H, Roop
DR and Bradley A: p63 is a p53 homologue required for limb and
epidermal morphogenesis. Nature. 398:708–713. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Workman P, Aboagye EO, Balkwill F, Balmain
A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA,
Glennie MJ, et al: Guidelines for the welfare and use of animals in
cancer research. Br J Cancer. 102:1555–1577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tatake RJ, Rajaram N, Damle RN, Balsara B,
Bhisey AN and Gangal SG: Establishment and characterization of four
new squamous cell carcinoma cell lines derived from oral tumors. J
Cancer Res Clin Oncol. 116:179–186. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boukamp P, Petrussevska RT, Breitkreutz D,
Hornung J, Markham A and Fusenig NE: Normal keratinization in a
spontaneously immortalized aneuploid human keratinocyte cell line.
J Cell Biol. 106:761–771. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chaudhari PR, Charles SE, D'Souza ZC and
Vaidya MM: Hemidesmosomal linker proteins regulate cell motility,
invasion and tumorigenicity in oral squamous cell carcinoma derived
cells. Exp Cell Res. 360:125–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dmello C, Sawant S, Alam H, Gangadaran P,
Mogre S, Tiwari R, D'Souza Z, Narkar M, Thorat R, Patil K, et al:
Vimentin regulates differentiation switch via modulation of keratin
14 levels and their expression together correlates with poor
prognosis in oral cancer patients. PLoS One. 12:e01725592017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sawant SS, Vaidya Mm, Chaukar DA, Alam H,
Dmello C, Gangadaran P, Kannan S, Kane S, Dange PP, Dey N, et al:
Clinical significance of aberrant vimentin expression in oral
premalignant lesions and carcinomas. Oral Dis. 20:453–465. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blanpain C, Lowry WE, Pasolli HA and Fuchs
E: Canonical notch signaling functions as a commitment switch in
the epidermal lineage. Genes Dev. 20:3022–3035. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koh LF, Ng BK, Bertrand J and Thierry F:
Transcriptional control of late differentiation in human
keratinocytes by TAp63 and notch. Exp Dermatol. 24:754–760. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nicolas M, Wolfer A, Raj K, Kummer JA,
Mill P, van Noort M, Hui CC, Clevers H, Dotto GP and Radtke F:
Notch1 functions as a tumor suppressor in mouse skin. Nat Genet.
33:416–421. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vaidya MM and Kanojia D: Keratins: Markers
of cell differentiation or regulators of cell differentiation? J
Biosci. 32:629–634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alam H, Sehgal L, Kundu ST, Dalal SN and
Vaidya MM: Novel function of keratins 5 and 14 in proliferation and
differentiation of stratified epithelial cells. Mol Bio Cell.
22:4068–4078. 2011. View Article : Google Scholar
|
|
25
|
Proweller A, Tu L, Lepore JJ, Cheng L, Lu
MM, Seykora J, Millar SE, Pear WS and Parmacek MS: Impaired notch
signaling promotes de novo squamous cell carcinoma formation.
Cancer Res. 66:7438–7444. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duan L, Yao J, Wu X and Fan M: Growth
suppression induced by Notch1 activation involves Wnt-beta-catenin
down-regulation in human tongue carcinoma cells. Biol Cell.
98:479–490. 2006. View Article : Google Scholar : PubMed/NCBI
|