Introduction
Since colorectal cancer (CRC) is the third most
common cause of cancer-related deaths worldwide it has attracted
attention in recent years (1).
Research has revealed that stresses including oxidative stress and
low nutrient availability are intrinsic of the tumor
microenvironment and have significant roles in CRC development,
progression and therapy (2,3).
FoxO3a as a transcription factor is evolutionarily
conserved. It is involved in many pivotal biological processes,
such as cell cycle regulation, DNA damage repair, vascular
development, reactive oxygen species detoxification pathways,
longevity and regulation of immune responses (4–9). In
cancer, previous studies have revealed that FoxO3a is a
tumor suppressor gene and controls diverse genetic pathways
(10,11). For example, it has been found that
decreased expression of FoxO3a predicted advanced recurrence and
poor survival in CRC (12),
indicating a tumor-suppressor role for this transcription factor.
In addition, recent data indicated that FoxO3a may also function as
a metastasis inductor and promote tumor progression in CRC through
interaction with β-catenin (13).
These studies indicated that the function of FoxO3a in CRC is
highly context dependent, relying on the upstream regulation.
However, little research has been conducted concerning the
regulation of FoxO3a gene transcription.
SP1 is a zinc finger transcription factor that binds
to GC-rich motifs of many promoters. SP1 is involved in many
cellular processes, including cell growth, apoptosis, immune
response, chromatin remodeling and DNA damage response. It is
reported that inhibition of SP1 suppresses colon cancer stem cell
growth and induces apoptosis in vitro and in vivo
(14). SP1 also has
anti-proliferative effects and promotes apoptosis in oral squamous
cell carcinoma (15,16). The activity of SP1 is modulated by
growth factors, cytokines and tumor promoters (17).
Therefore, in the present study, we performed a
functional analysis of FoxO3a promoter and identified the core
sequences within the 5′ regulatory region that critically affected
FoxO3a transcriptional activity. Furthermore, plenty of SP1
transcription factor binding sites were found in the core promoter
regulatory region and we verified that SP1 played an important
function in FoxO3a transcription. Collectively, our findings
identified SP1 as a critical transcription factor in regulating
FoxO3a expression, which may offer new insights in CRC progression
and treatment.
Materials and methods
Cell culture and treatment
Caco2 and HT29 cell lines were obtained from the
Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
Caco2 cells were maintained in Dulbecco's modified Eagle's medium
(DMEM; HyClone Laboratories; GE Healthcare, Logan, UT, USA)
containing 10% fetal bovine serum (FBS). The HT29 cells were
cultured in RMPI-1640 medium (HyClone Laboratories; GE Healthcare)
supplemented with 10% FBS. Both cell lines were cultured in a 5%
CO2 humidified atmosphere at 37°C. The cells were
treated with CoCl2 (200 µM/l) or
H2O2 (200 µmol/l) for 24 h.
cDNA synthesis and quantitative
real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from Caco2 and HT29 cell
lines using TRIzol reagent (Life Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Subsequently, mRNA was reverse
transcribed using SuperScript First-Strand Synthesis System
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's instructions and then qRT-PCR was
performed using SYBR Green reagents (Takara Bio, Inc., Otsu, Japan)
containing 100 µM final concentration of FoxO3a primers (primer
sequences are listed in Table I).
PCR conditions were as follows: 95°C for 15 sec, followed by 40
cycles of 95°C for 5 sec, 60°C for 30 sec. PCRs were performed in
triplicates using the Roche LightCycler 480 II RT-PCR system (Roche
diagnostics Nederland BV, Almere, The Netherlands). The expression
levels of the target genes were normalized to β-actin. The relative
levels were calculated by the comparative Ct (ΔΔCt) method and the
relative expression fold (2−ΔΔCt) was calculated
(18).
 | Table I.Sequences of primers used for PCR in
the present study. |
Table I.
Sequences of primers used for PCR in
the present study.
| Primer name | Primer sequences |
|---|
| FoxO3a primer-F |
GGTGCTGTATAGGTGCTTTCT |
| FoxO3a primer-R |
AAAGGTGGTCCCAACTATTCC |
| FoxO3a
promoter-WT-F |
CCGGAATTCATAAGGACTTGTGCAGATGTTTTT |
| FoxO3a
promoter-WT-R |
CGACGCGTCAGGAGGACCTGAAGACGTG |
| FoxO3a
promoter-1037-F |
CCGCTCGAGTGAACTAGTGTGTAGACTTTTGGTGTG |
| FoxO3a
promoter-513-F |
CCGCTCGAGACACCGGGGCTGGCCCAGA |
| FoxO3a
promoter-62-F |
TCGAGCGCCCGCCGTCAGCCTAGGTTGAGGCGCCCTGCGTGTGTCTATAACTTTGTGCTGCTGCCGCA |
| FoxO3a
promoter-62-R |
AGCTTGCGGCAGCAGCACAAAGTTATAGACACACGCAGGGCGCCTCAACCTAGGCTGACGGCGGGCGC |
| Human-SP1-F |
GAGCGACCAAGATCACTCCATG |
| Human-SP1-R |
CCGCTCGAGTCAGAAGCCATTGCCACTGAT |
| Human-TFAP2A-F |
ACTTTGGAAATTGACGGATAATATCA |
| Human-TFAP2A-R |
CCGCTCGAGTCACTTTCTGTGCTTCTCCTCTTT |
Western blot analysis
Cell samples were collected and lysed in RIPA buffer
with protease inhibitor cocktails (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Total cell protein extracts (20 µg/lane) were
subjected to SDS-PAGE analysis. The membrane was blocked with 5%
milk in TBST before being incubated with FoxO3a antibody (cat. no.
12829; Cell Signaling Technology, Inc., Danvers, MA, USA), SP1
antibody (cat. no. 07-645; EMD Millipore, Billerica, MA, USA) or
AP2 antibody (cat. no. H00000160-K; Abnova, Taipei, Taiwan)
overnight at 4°C. The membranes were washed with TBST and incubated
with the secondary antibodies (cat. no. sc-2357; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The immunoreactive proteins
were visualized by chemiluminescence reagents (ECL; Amersham
Biosciences; GE Healthcare, Chicago, IL, USA).
Lentiviral-vector mediated
FoxO3a-knockdown stable cells
The target sequences for FoxO3a were
5′-GCTCTTGGTGGATCATCAA-3′ (FoxO3a-KD1) and
5′-GCATGTTCAATGGGAGCTTGGA-3′ (FoxO3a-KD2). Lentiviral vectors with
green fluorescent protein (GFP) sequence containing human
FoxO3a-shRNA were constructed. All of the constructs were validated
by DNA sequencing. The recombinant FoxO3a knockdown and negative
control (NC) lentivirus were prepared. Caco2 or HT29 cells were
seeded in six-well dishes in antibiotic-free medium 1 day before
infection. The cells reached 80–90% confluency at the time of
transfection. The expression of GFP was confirmed using a
fluorescence microscope after the lentivirus infection. The culture
medium was added with 4 µg/ml puromycin for at least 14 days. The
puromycin-resistant cell clones were then amplified in medium
containing 2 µg/ml puromycin for 7–9 days. The clones were
designated as FoxO3a-KD or NC cells.
Cell proliferation assay, cell
migration and cell apoptosis assay
The viability of CRC cells was determined using Cell
Counting Kit-8 (cat. no. C0039; Beyotime Institute of
Biotechnology, Shanghai, China). Briefly, 2,000 cells/well were
plated in 96-well plates. CCK-8 (10 µl) was added to the culture
medium. After 2 h, the plates were assessed using a microplate
reader (Biotek ELx800; BioTek Instruments, Inc., Winooski, VT,
USA).
Cell migration was examined by a scratch assay.
After culturing for 2 days, the cells were deprived of serum for 16
h and then artificially injured using a 200 µl plastic pipette tip.
Cells migrating to the front of the wound were imaged after 48 and
72 h. The migration capacity was quantified by determining the
percentage of open area using the following formula: (1-current
wound size/initial wound size) ×100.
The Annexin V-PE Apoptosis Detection kit (BD
Pharmingen; BD Biosciences, Franklin Lakes, NJ, USA) was used to
analyze apoptosis according to the manufacturer's instructions.
Stained cells underwent flow cytometry.
Animal studies
BALB/c nude mouse (six- to 8-week-old) weighing
~18–22 g, were obtained from SLAC Laboratory Animal Co., Ltd.
(Shanghai, China). The animals were sacrificed when tumor reached
1,500 mm3 in size. HT29 cells were subcutaneously
injected into the nude mice. The xenograft size was assessed twice
a week and the volume was calculated using the following formula: a
× b2/2 (a represents length and b represents width).
After one month, all mice were euthanized using CO2. The
tumor tissues were weighed. All animal studies were approved by the
Institutional Animal Care and Use Committee of the Shanghai
Institutes for Biological Sciences.
Construction of luciferase reporter
vectors
The FoxO3a gene promoter-driven luciferase
reporter construct was generated by inserting a 2,000 bp fragment
containing the 5′ flanking region of the FoxO3a gene from positions
−2,000 to 0 into a pGL3-basic vector (Promega, Fitchburg, WI, USA).
Subsequently, to identify the critical transcription activation
region of the FoxO3a promoter, a series of 5′ deletion fragments
[-1,037 bp/0, −513 bp/0 and −62 bp/0] luciferase reporter
constructs were generated by PCR using the full length FoxO3a
promoter fragment as a template. All the constructs were verified
by sequencing to rule out the possibility of any PCR error. The
primers used to generate all constructs are listed in Table I. The putative transcription factor
binding sites within FoxO3a gene promoter were screened using the
TFsearch program (Kyoto University, Kyoto, Japan).
Transient transfections and luciferase
assays
HT29 cells were seeded in 96-well plates and grew to
a density of 80% confluency before transfection. One hundred
nanograms of each luciferase reporter construct and 0.3 µl
Lipofectamine 2000 (Invitrogen) reagent in Opti-MEM were
transfected according to the manufacturer's instructions. In order
to validate the transfection efficiency, an additional 4 ng of
pRL-TK vector which contained the Renilla luciferase gene
(hRluc) under the control of the Herpes simplex virus thymidine
kinase promoter was co-transfected in each well. The cells were
harvested after 48-h transfection and then lysed with 20 µl passive
lysis buffer for each well (Promega). Luciferase activity was
determined using Dual-Luciferase Reporter Assay system (Promega) on
a luminescence reader (AccuFLEX Lumi 400; Aloka, Tokyo, Japan).
Each expression vector was mixed with the luciferase vectors at 1:2
concentration ratio. All the experiments were repeated at least
three times and the results were expressed as the mean ± standard
deviation (SD).
Statistical analysis
In the present study, data are presented as the mean
± SD of three independent experiments. Two-tailed Student's t-test
was used to analyze the differences between groups using GraphPad
Prism version 5.0 (GraphPad Software Inc., La Jolla, CA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
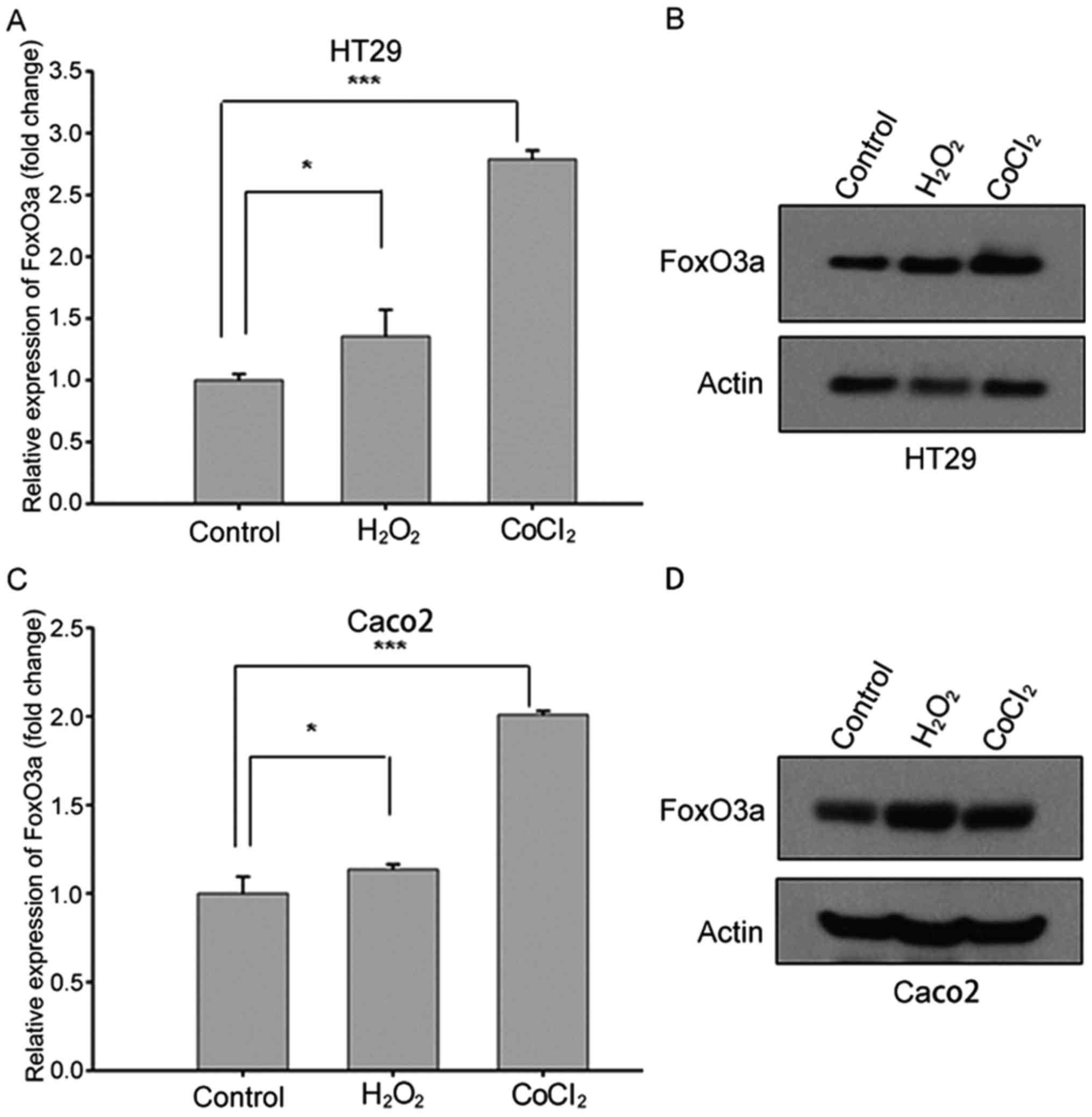
Upregulated FoxO3a expression in CRC
cell lines after stress stimuli
In order to elucidate the function of FoxO3a in CRC
under stress stimuli, we investigated the expression levels of
FoxO3a in HT29 and Caco2 cell lines treated with
H2O2 or CoCl2 using qRT-PCR and
western blot analysis. Both the qRT-PCR and western blotting
results revealed that the expression of FoxO3a increased after
oxidative stress stimuli treatment (Fig. 1), indicating that FoxO3a
transcription was upregulated in response to hypoxia and oxidative
stress.
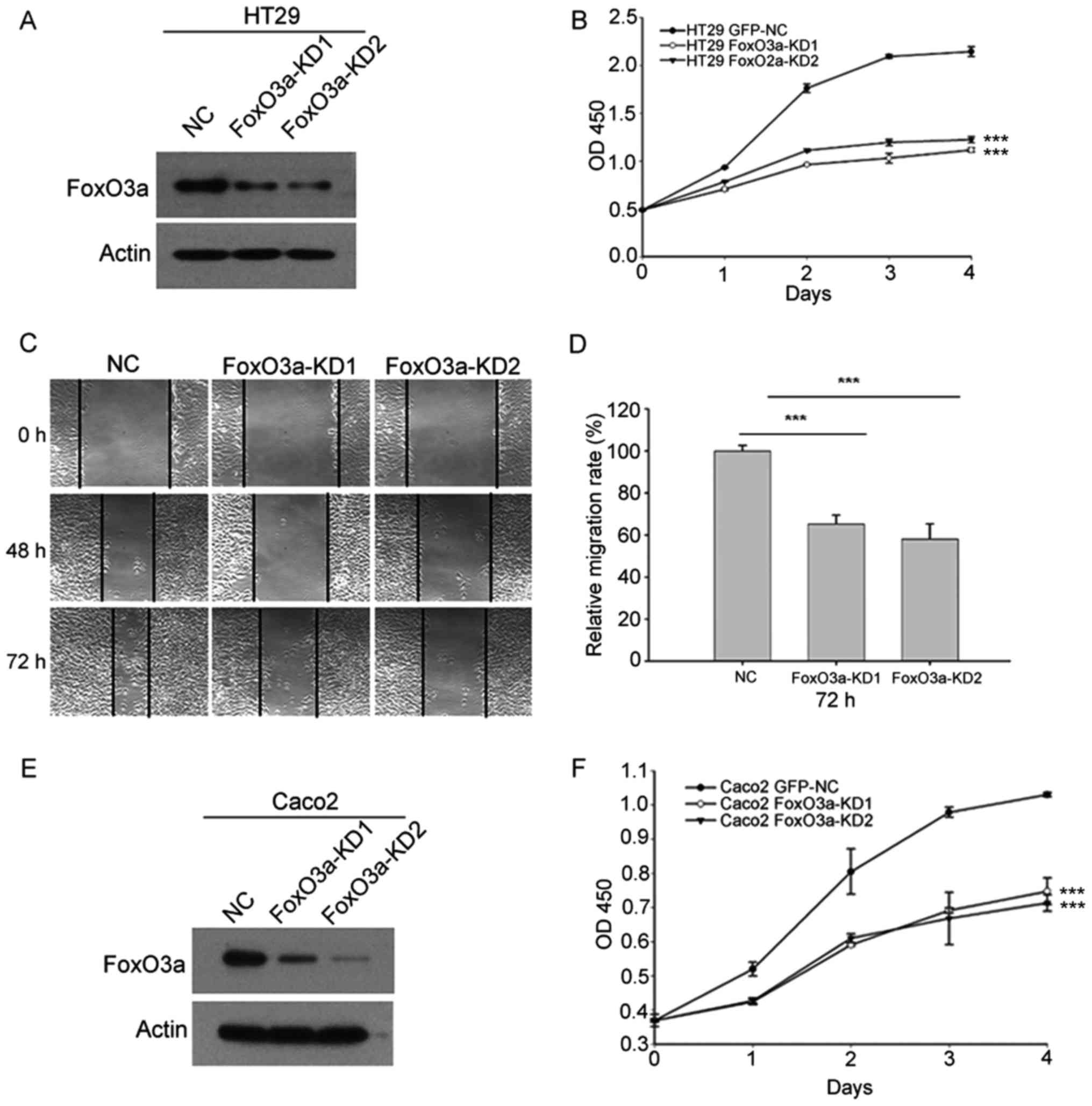
Knockdown of FoxO3a reduces CRC cell
proliferation and migration ability
We stably downregulated FoxO3a using
lentivirus-mediated shRNA. The effect of FoxO3a-knockdown in HT29
cells was verified by western blot analysis (Fig. 2A). CCK-8 analysis revealed that
knockdown of FoxO3a in HT29 cells reduced cell proliferation
(Fig. 2B). Scratch analysis
indicated that inhibition of FoxO3a also suppressed cell migration
in HT29 cells (Fig. 2C and D).
Subsequently we knocked down FoxO3a in Caco2 cells (Fig. 2E). Consistently, CCK-8 analysis
revealed that FoxO3a-knockdown suppressed cell proliferation in
Caco2 cells (Fig. 2F). These
results indicated that FoxO3a was important for CRC cell
proliferation and migration.
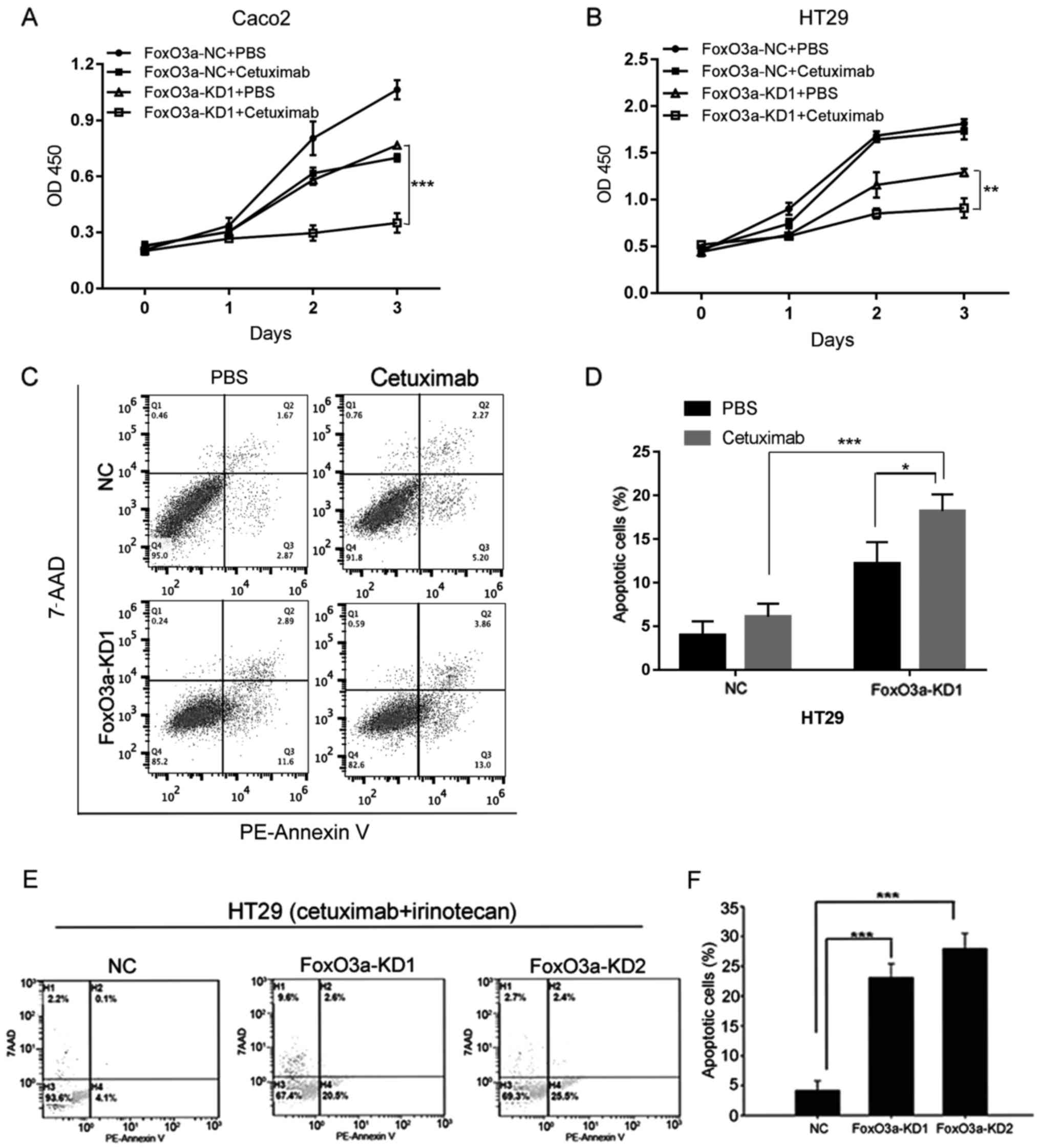
Knockdown of FoxO3a sensitizes CRC
cells to cetuximab and combination treatment
In order to elucidate the function of FoxO3a in CRC
therapy, we treated these cells with cetuximab alone or combined
with irinotecan (Fig. 3). CCK-8
analysis indicated that knockdown of FoxO3a in Caco2 and HT29 cells
reduced cell proliferation after treatment with cetuximab (Fig. 3A and B). Apoptotic analysis using
Annexin V/7-AAD revealed that knockdown of FoxO3a promoted the
apoptosis of HT29 cell lines treated with cetuximab (Fig. 3C and D). Knockdown of FoxO3a also
increased cell apoptosis after treatment of cetuximab combined with
irinotecan (Fig. 3E and F). In
conclusion, our data indicated that FoxO3a may participate in CRC
response to cetuximab treatment.
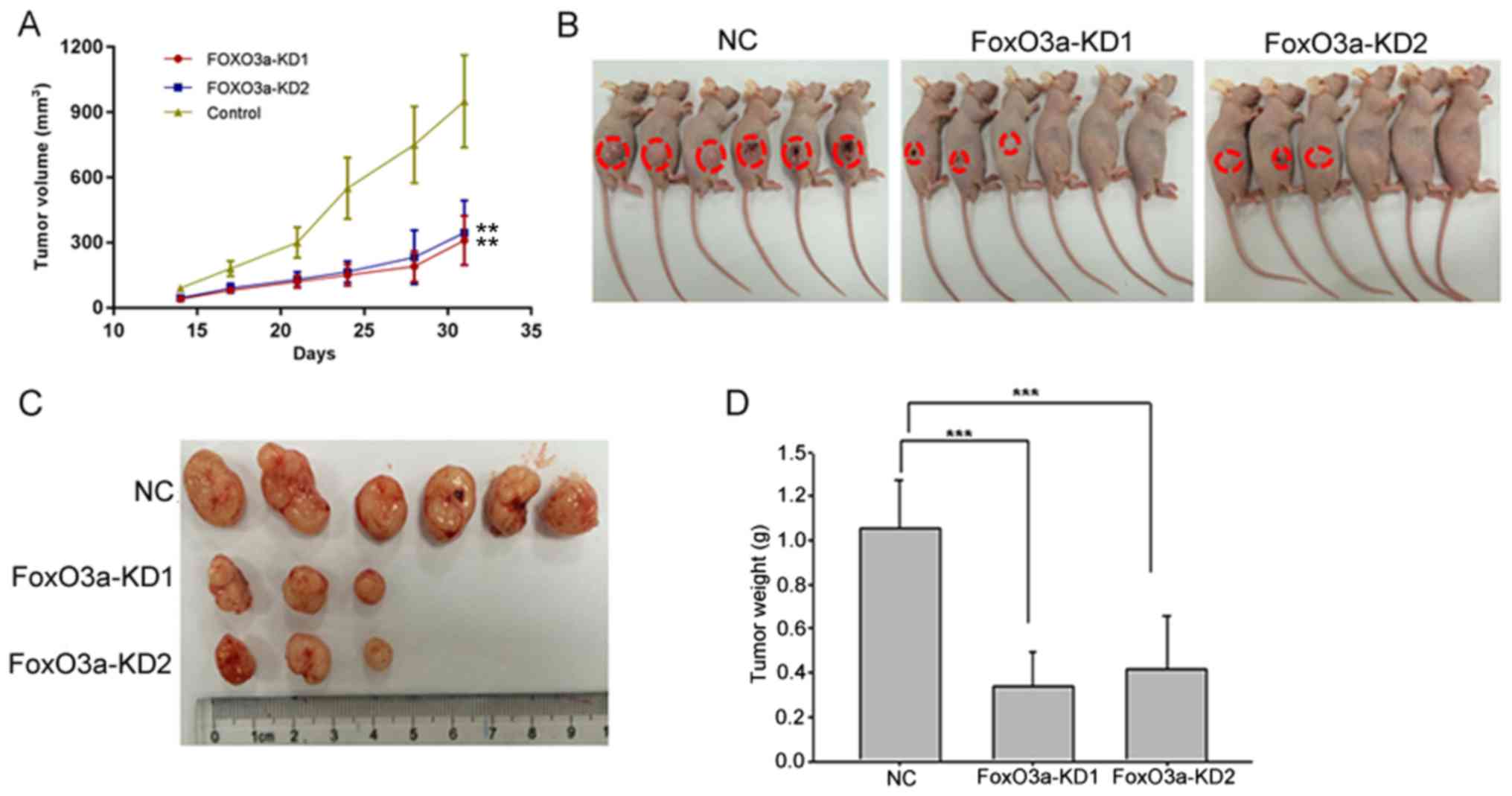
Inhibition of FoxO3a reduces CRC
progression in vivo
After subcutaneously inoculating HT29 cells with or
without knockdown of FoxO3a in a xenograft animal model, we
observed that the tumorigenesis ability of the FoxO3a-knockdown
group was significantly reduced compared with the NC group. Both
tumor volume and weight were significantly decreased in
FoxO3a-knockdown group compared with NC group (Fig. 4). In conclusion, these findings
indicated an important role of FoxO3a in CRC progression.
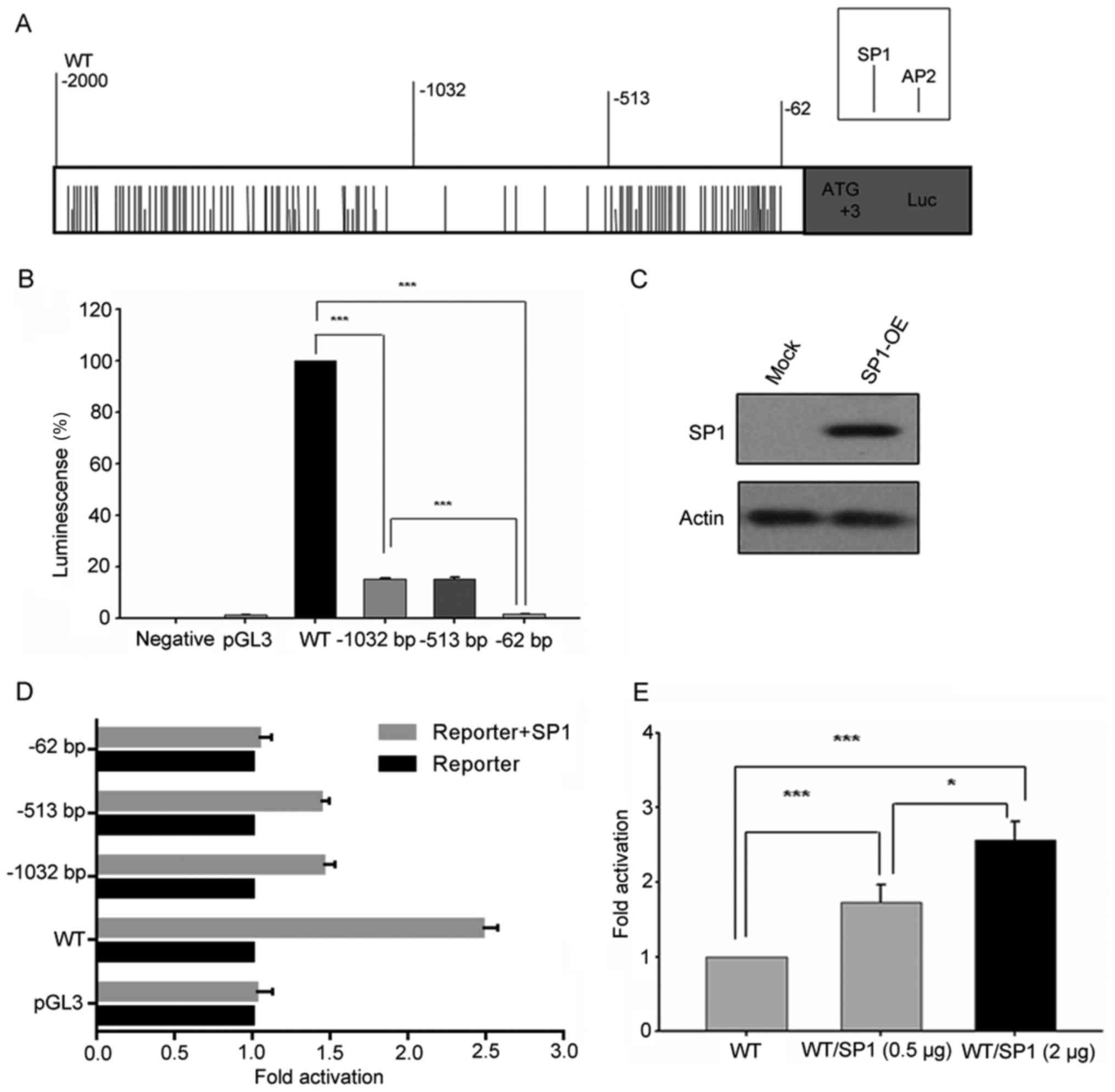
Analysis of putative transcription
factor binding sites in FoxO3a gene promoter
The transcription factor binding sites of the FoxO3a
gene promoter was analyzed using TFsearch program. The results
revealed that many potential SP1 and AP2 transcription factor
binding sites were found within the FoxO3a promoter (Fig. 5A). Furthermore, most of them were
centered on regions −2,000/-1,032 bp and −513/-62 bp (Fig. 5A). In addition, some other
transcription factor binding sites were also identified, such as
C/EBP β, NF-1, AP-1, SRF and NRF-1. To ascertain the regulation of
the FoxO3a gene transcription, full length of FoxO3a gene promoter
(−2,000 bp/0) and a series of 5′deletion fragment [-1,032 bp/0,
−513 bp/0, −62 bp/0] luciferase reporter plasmids were
constructed.
Activity analysis of FoxO3a promoter
luciferase reporter constructs
The activity of different lengths of FoxO3a promoter
luciferase reporter constructs was analyzed in HT29 cell lines. As
displayed in Fig. 5B, the
percentage of activity reduction was >80% in deletion construct
−1,032 bp/0 compared to wild-type (WT) (−2,000 bp/0). A similar
phenomenon was observed between deletion constructs −513 bp/0 and
−62 bp/0, accounting for the remaining 20% transcriptional
activity. Thus, these results indicated that the regions between
−2,000/-1,032 bp and −513/-62 bp of FoxO3a promoter play an
important role in regulating the transcription of FoxO3a
gene.
According to Fig.
5A, there were plenty of SP1 and AP2 transcription factor
binding sites between regions −2,000/-1,032 bp and −513/-62 bp.
These results indicated that SP1 and AP2 may play important roles
in FoxO3a gene transcription. In order to further identify
the major transcriptional factor, we transiently co-transfected the
FoxO3a promoter luciferase reporter constructs individually
combined with SP1 or AP2 expression plasmids. The expression of SP1
was examined by western blot analysis (Fig. 5C). It was revealed that the activity
of FoxO3a promoter luciferase constructs co-transfected with SP1
expression plasmid was obviously increased, except −62 bp/0
construct. Furthermore, luciferase activity of constructs
containing −513 bp/0 and −1032 bp/0 region of FoxO3a promoter
increased almost 1.5 times when co-transfected with SP1 expression
plasmid, compared with 2.5 times of increase in WT (Fig. 5D and E). However, co-transfection
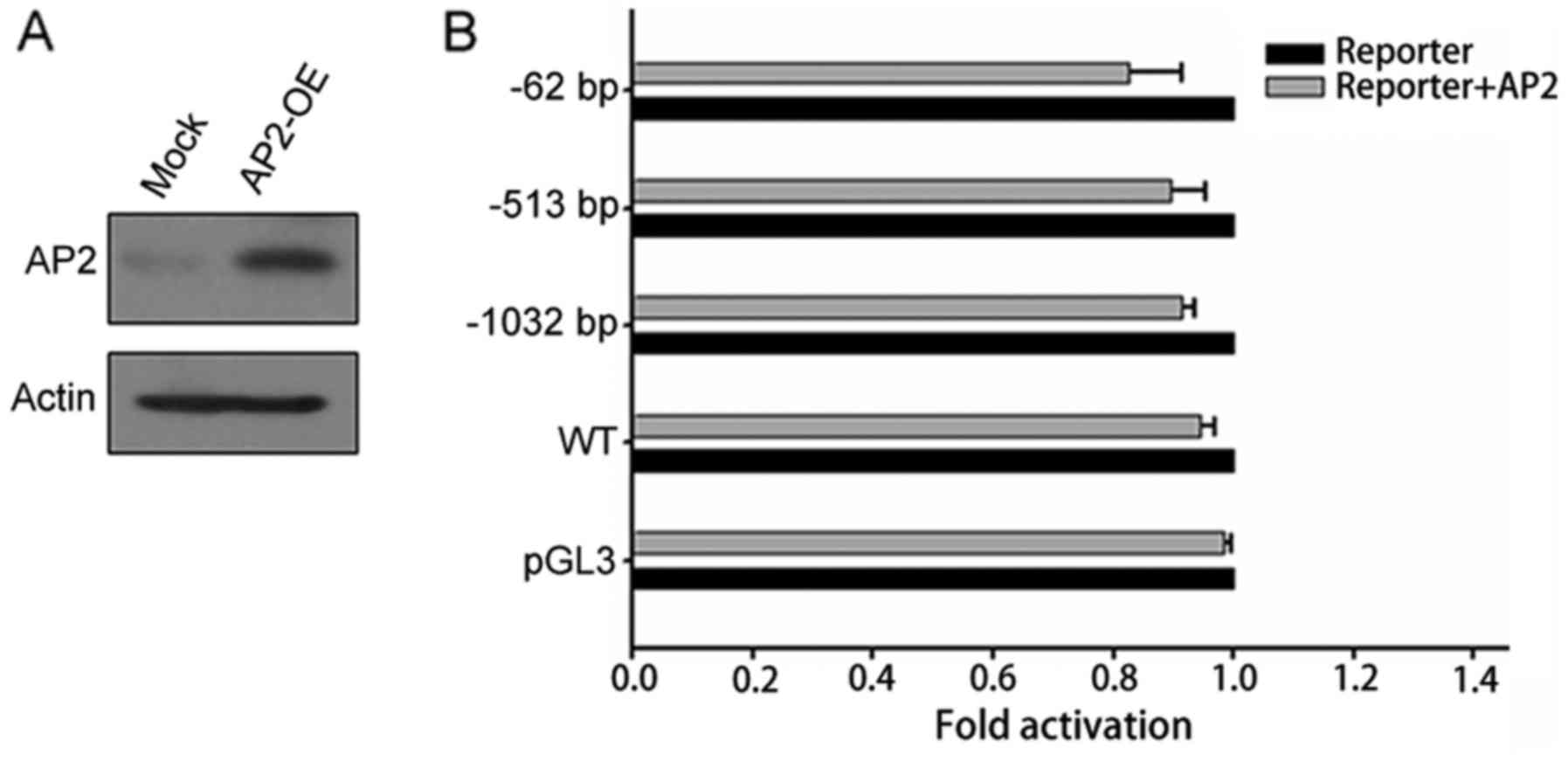
with AP2 did not promote FoxO3a promoter luciferase activity
(Fig. 6). These results confirmed
that −2,000/-1,032 bp and −513/-62 bp regions of FoxO3a promoter
were the critical regions for FoxO3a gene transcription and
that the transcription factor SP1 was an important regulator for
FoxO3a gene transcription.
Discussion
In the present study, we characterized the
transcriptional activity and regulation of the 5′-flank of human
FoxO3a gene located in the −2,000/-1,032 bp and −513/-62 bp
regions. This study demonstrated that the SP1 site plays an
important role in regulating the transcription of the FoxO3a
gene in CRC cells. Furthermore, we found that the FoxO3a
gene was upregulated in response to stress conditions, hypoxia and
oxidative stress and that the upregulated FoxO3a gene was important
for CRC progression in vivo and in vitro. These
findings may help to understand the complicated pathological
function of FoxO3a gene in tumor biology.
FoxO3a gene has an ambiguous function in CRC. A
study revealed that FoxO3a inactivation promoted tumor progression
(19). However, there are also
studies indicating that FoxO3a protected cells under stress
conditions, including oxidative stress, serum deprivation and
hypoxia (20). Consistent with
these studies, we found that the expression of FoxO3a in CRC cells
was increased significantly in response to oxidative stress and
hypoxia. However, the precise function of FoxO3a in tumor
progression and tumor microenvironment remains elusive. FoxO3a was
found to promote invasion of cancer cells (12). Therefore, caution should be taken
when FoxO3a is employed as a potential target for cancer
therapy.
The ubiquitous transcription factor SP1 is involved
in the regulation of many genes (21–23).
Although the expression of SP1 can be detected in most cells, its
expression varied during development (24,25).
SP1 is the founding member of SP/XKLF proteins, which contain three
highly homologous C-terminal zinc-finger motifs and are capable of
binding similar DNA sequences. The combination of SP/XKLF proteins
can either compete or interact with a given promoter element to
activate or suppress the transcription (21). Therefore, it is not easy to confirm
the function of SP1, due toå the potential functions of its related
family members. However, SP1 is essential for embryogenesis,
because SP1-/- mouse embryos display growth retardation and die
during gestation (26). In the
present study, we found that SP1 was the crucial regulator of
FoxO3a transcription in CRCs. More transcriptional factors need to
be identified in this complex regulation network.
In conclusion, our findings determined that the
crucial regions corresponding to the SP1 binding sites located
between −2,000 and −1,037 bp were essential for FoxO3a
transcriptional activity. Furthermore, FoxO3a transcription was
upregulated in response to hypoxic and oxidative stress in
colorectal tumor cells, indicating that the interaction between SP1
and FoxO3a may have important implications in CRC progression.
Acknowledgements
Not applicable.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gatenby RA and Gillies RJ: A
microenvironmental model of carcinogenesis. Nat Rev Cancer.
8:56–61. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Albini A and Sporn MB: The tumour
microenvironment as a target for chemoprevention. Nat Rev Cancer.
7:139–147. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosich L, Saborit-Villarroya I,
López-Guerra M, Xargay-Torrent S, Montraveta A, Aymerich M,
Villamor N, Campo E, Pérez-Galán P, Roué G, et al: The
phosphatidylinositol-3-kinase inhibitor NVP-BKM120 overcomes
resistance signals derived from microenvironment by regulating the
Akt/FoxO3a/Bim axis in chronic lymphocytic leukemia cells.
Haematologica. 98:1739–1747. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oellerich MF and Potente M: FOXOs and
sirtuins in vascular growth, maintenance, and aging. Circ Res.
110:1238–1251. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Becker T, Loch G, Beyer M, Zinke I,
Aschenbrenner AC, Carrera P, Inhester T, Schultze JL and Hoch M:
FOXO-dependent regulation of innate immune homeostasis. Nature.
463:369–373. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Monsalve M and Olmos Y: The complex
biology of FOXO. Curr Drug Targets. 12:1322–1350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hedrick SM, Michelini Hess R, Doedens AL,
Goldrath AW and Stone EL: FOXO transcription factors throughout T
cell biology. Nat Rev Immunol. 12:649–661. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seiler F, Hellberg J, Lepper PM,
Kamyschnikow A, Herr C, Bischoff M, Langer F, Schäfers HJ, Lammert
F, Menger MD, et al: FOXO transcription factors regulate innate
immune mechanisms in respiratory epithelial cells. J Immunol.
190:1603–1613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L, Yang X, Cao X, Liu F, Quan M and Cao
J: Casticin induces growth suppression and cell cycle arrest
through activation of FOXO3a in hepatocellular carcinoma. Oncol
Rep. 29:103–108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yung MM, Chan DW, Liu VW, Yao KM and Ngan
HY: Activation of AMPK inhibits cervical cancer cell growth through
AKT/FOXO3a/FOXM1 signaling cascade. BMC Cancer. 13:3272013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bullock MD, Bruce A, Sreekumar R, Curtis
N, Cheung T, Reading I, Primrose JN, Ottensmeier C, Packham GK,
Thomas G and Mirnezami AH: FOXO3 expression during colorectal
cancer progression: Biomarker potential reflects a tumour
suppressor role. Br J Cancer. 109:387–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tenbaum SP, Ordóñez-Morán P, Puig I,
Chicote I, Arqués O, Landolfi S, Fernández Y, Herance JR, Gispert
JD, Mendizabal L, et al: β-catenin confers resistance to PI3K and
AKT inhibitors and subverts FOXO3a to promote metastasis in colon
cancer. Nat Med. 18:892–901. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y,
Gong W, Chen Y, Cheng T, Zhi F, et al: Inhibition of the
transcription factor Sp1 suppresses colon cancer stem cell growth
and induces apoptosis in vitro and in nude mouse xenografts.
Oncol Rep. 30:1782–1792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim DW, Ko SM, Jeon YJ, Noh YW, Choi NJ,
Cho SD, Moon HS, Cho YS, Shin JC, Park SM, et al:
Anti-proliferative effect of honokiol in oral squamous cancer
through the regulation of specificity protein 1. Int J Oncol.
43:1103–1110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeon YJ, Ko SM, Cho JH, Chae JI and Shim
JH: The HDAC inhibitor, panobinostat, induces apoptosis by
suppressing the expresssion of specificity protein 1 in oral
squamous cell carcinoma. Int J Mol Med. 32:860–866. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karin M, Liu Z and Zandi E: AP-1 function
and regulation. Curr Opin Cell Biol. 9:240–246. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Greer EL and Brunet A: FOXO transcription
factors at the interface between longevity and tumor suppression.
Oncogene. 24:7410–7425. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z, Zhang H, Chen Y, Fan L and Fang J:
Forkhead transcription factor FOXO3a protein activates nuclear
factor κB through B-cell lymphoma/leukemia 10 (BCL10) protein and
promotes tumor cell survival in serum deprivation. J Biol Chem.
287:17737–17745. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Philipsen S and Suske G: A tale of three
fingers: The family of mammalian Sp/XKLF transcription factors.
Nucleic Acids Res. 27:2991–3000. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li L, He S, Sun JM and Davie JR: Gene
regulation by Sp1 and Sp3. Biochem Cell Biol. 82:460–471. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schreiber M, Baumann B, Cotton M, Angel P
and Wagner EF: Fos is an essential component of the mammalian UV
response. EMBO J. 14:5338–5349. 1995.PubMed/NCBI
|
|
24
|
Lania L, Majello B and De Luca P:
Transcriptional regulation by the Sp family proteins. Int J Biochem
Cell Biol. 29:1313–1323. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suske G: The Sp-family of transcription
factors. Gene. 238:291–300. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marin M, Karis A, Visser P, Grosveld F and
Philipsen S: Transcription factor Sp1 is essential for early
embryonic development but dispensable for cell growth and
differentiation. Cell. 89:619–628. 1997. View Article : Google Scholar : PubMed/NCBI
|