Introduction
Pressure ulcers (PUs) are frequent problems among
people who stay in hospital in chronic and acute care settings and
bring a heavy pressure on patients, their families and caregivers
(1). Today, PUs are one of the five
leading factors of damage to patients and preventable safety
condition in the world. Moreover, it is growingly considered as a
marker of the quality of care offered by health care institutions
(2). PUs in older inpatients can
cause substantial negative impacts on medical complications,
expense of care, duration of hospital stay, quality of life, pain
and death (3). The main risk factor
of PU is immobility; other patient features including poor
nutritional status and incontinence have also been discovered to
elevate the risk of PU (4). Older
patients with surgical repair of a hip fracture are at a high risk
because of the possibility for long-term immobility and other risk
factors of PU (5).
As a common complication of hip fracture, PU has a
frequency of 8.8–55%. It has substantial influence on the quality
of life, hospital care cost, and death (6). Some reports investigating biopsies and
wound fluids collected from PUs revealed that the healing process
might be impeded by the excessive levels of active forms of matrix
metalloproteinases (MMP)-2 and MMP9. These data indicate that the
proteinases could destruct extracellular proteins, receptors and
growth factors critical for PUs healing (7).
As endogenous ~22 nucleotide small non-coding RNAs,
microRNAs (miRNAs) regulate gene production by inducing either RNA
degradation or translation repression and impacting mRNAs (8). These non-coding RNAs bind to the
target mRNA at the 3′UT, and the seed region of the miRNA (from
nucleotide 2 to 8) mainly dictates the specificity of the binding.
Many target genes, usually sharing the same pathway, are modulated
by the same miRNA. A range of miRNAs can affect the same transcript
to make post-transcriptional modulation more complex (9). Sequence complementarity of just seven
nt located at the ‘seed region’ of miRNA (position 2 to position 8
of the miRNA) or even six nt (position 2 to position 7) with the
target mRNA is generally adequate to inhibit translation though
most human miRNAs consist of 22 nt on average (10). Moreover, SNPs and polymorphisms
located at the 3′UTR of genes that change miRNA binding may impact
the expression of protein related to the occurrence of a variety of
disorders (11).
It has been previously found that rs1056629 is
located within 3′UTR of MMP9, and potentially compromises the
interaction between MMP9 3′UTR and miR-491, as the presence of
minor allele of the SNP breaks the binding site of the miRNA
(12). rs1056629 has been reported
to be associated with an elevated risk of coronary heart disease
(12). Simultaneously, MMP9 has
been shown to be an important enzyme involved in the development of
pressure ulcer (13,14). In this study, we studied the
association between the rs1056629 and the risk of pressure ulcer in
the patients with hip fracture, as well as its effect on the serum
level of MMP9.
Materials and methods
Samples
Forty individuals comprising 22 subjects suffering
from PU and 18 healthy controls, were recruited for this study.
Additionally, the 22 subjects with PU were diagnosed just after
having hip fracture in our hospital. Tissue samples were collected
from all subjects, and stored in −80°C for further analysis.
Participants aged >90 years, malignant origin, psychiatric and
obstetric patients, allergy to wound products were excluded from
our study. All procedures and use of tissue samples were approved
by the Ethics Committee of our institute, and participants or their
first-degree relatives had already signed the informed consents
before start of the experiment after all potential risk factors
were carefully explained. The study was conducted according to the
Declaration of Helsinki.
Genotyping by TaqMan
DNA extraction kit (Qiagen, Dusseldorf, Germary) was
used to extract the DNA from peripheral blood samples in accordance
with the manufacturer's protocol. PCR was used to amplify the DNA
fragments including rs1056629. The PCR products were sequenced in
forward direction, and TaqMan genotyping kit (Qiagen) and BLAST
were used to determine the SNP genotypes. Each experiment was
performed three times.
In silico analysis
We search public online tool (http://www.bioguo.org) to find gene polymorphism in
MMP9 3′UTR that might break the interaction between MMP9 and
miRNA.
RNA isolation and real-time PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
utilized to extract total RNA from BJ fibroblast cells and tissue
samples according to the manufacturer's instructions. M-MLV
(Invitrogen) was utilized to synthesize the first stranded cDNA
from 2 µg total RNA in a 25 µl mixture following guideline
suggested by vendor. Agarose gels (2%) with 5 mg/ml nucleic acid
dye was utilized to electrophorese the PCR product, and AlphaEaseFC
software (Genetic Technologies, Miami, FL, USA) was utilized to
conduct expression analysis of MMP9. An iCycler iQ real-time PCR
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with SYBR
Green Master Mix (QPK-201; Toyobo, Co., Ltd., Osaka, Japan) was
utilized to perform real-time PCR, and the reaction was carried out
at 95°C for 5 min, 45 cycles of (25 sec at 95°C; 20 sec at 60°C; 30
sec at 72°C). U6 RNA and GAPDH served as internal controls.
2−ΔΔCt method was utilized to analyze the relative
expression of miR-194-3p, miR-491, miR-1915-3p, miR-941 and MMP9
mRNA. Three independent experiments were repeated.
Cell culture and transfection
BJ fibroblast cells were obtained from Lonza (Basel,
Switzerland), and RPMI-1640 medium (Gibco) containing 10% FBS
(fetal bovine serum) (Sijiqing, Hangzhou, China) and 1%
penicillin-streptomycin was utilized to culture these cells under a
humid atmosphere with 5% CO2/95% air at 37°C. Prior to
transfection, cell were seeded into 96-well plates at a final
density of 1×105 per well, when the cells grewn to 80%
confluence, they were co-transfected with miR-194-3p
mimics/inhibitors, miR-491 mimics/inhibitors, miR-1915-3p
mimics/inhibitors, miR-941 mimics/inhibitors using Lipofectamine
2000 (Invitrogen) according to the manufacturer's instructions.
Three independent experiments were repeated.
Vector construction and
mutagenesis
PCR was performed to amplify fragment of MMP9 cDNA,
and then PCR products were inserted into pGL3 vector (Promega
Corp., Madison, WI, USA), then direct sequencing was adopted to
confirm the accuracy. The QuikChange® Lightning
Site-Directed Mutagenesis kit (Agilent Technologies) was used to
perform the single-base mutation in gene polymorphism located in
binding site of miR-194-3p (A allele to C allele), miR-491 (A
allele to C allele), miR-1915-3p (G allele to A allele) or miR-941
(G allele to A allele) in MMP9 3′UTR according to the
manufacturer's instructions, and meanwhile QuikChange®
Lightning Site-Directed Mutagenesis kit (Agilent Technologies) with
carefully designed primers was used to perform the miR-194-3p,
miR-491, miR-1915-3p or miR-941 seed sequence mutation in MMP9
3′UTR. The above eight mutagenesis were performed for the same site
and introduced to the pGL3 vector (Promega Corp.) at the same time,
direct sequencing was adopted to confirm the mutation site
accuracy. Three independent experiments were performed.
Luciferase assay
Full length of MMP9 3′UTR containing predicted
binding site of miR-194-3p, miR-491, miR-1915-3p or miR-941 was
amplified using PCR, then the above PCR products were inserted into
pGL3-control vector (Promega Corp.), and located downstream of the
luciferase gene, and obtained pGL3-MMP9-3′UTR construct. Then the
‘seed sequence’ in MMP9 3′UTR was mutated, and PCR was also
performed to obtain point mutations, and cloned into pGL3-control
vector (Promega Corp.) to generate pGL3-MMP9 3′UTR Mut construct.
1×105 BJ fibroblast cells were seeded into 96-well
plates for 24 h. Lipofectamine 3000 (Invitrogen) was utilized to
transfect miR-194-3p, miR-491, miR-1915-3p or miR-941 mimic or NC
(negative control) RNA oligonucleotide and 0.4 mg of pGL3
constructs along with 0.07 mg of pRL-CMV plasmid-expressing renilla
luciferase into BJ fibroblast cells. Dual Luciferase Reporter assay
system (Promega Corp.) was utilized to detect luciferase activity
48 h post-transfection, and luciferase activity of firefly was
normalized to that of renilla luciferase. Three independent tests
were repeated.
Western blot analysis
The total protein was extracted from BJ fibroblast
cells and tissue samples using lysis buffer (Beyotime, Haimen,
China), and Enhanced BCA Protein assay kit (Byotime) was utilized
to measure protein concentration according to the manufacturer's
instructions. Polyacrylamide gel (10%) (w/v) with SDS-PAGE was
utilized to separate protein extracts, and then electro-transferred
onto PVDF (polyvinylidene difluoride) (PerkinElmer, Waltham, MA,
USA) membrane for 90 min. Non-fat milk (5%) was utilized to block
the membrane. Then monoclonal antibodies against anti-human MMP9
(1:5,000; Abcam, Cambridge, MA, USA) at 1:12,000 dilution β-actin
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) were utilized
to incubate the membranes for 12 h at 4°C, next, HRP (horse raddish
peroxidase)-linked mouse anti-goat secondary antibody at 1:15,000
dilution (1:3,000, Santa Cruz Biotechnology, Inc.) was utilized to
treat the membranes for another 1 h. The Fusion FX7 system
(VilberLourmat, Marne-la-Vallée, France) with SuperSignal™ West
Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was utilized to visualize the bands, and ImageJ
Analyst software [National Institutes of Health (NIH), Bethesda,
MD, USA) was utilized to quantify the level of MMP9. Three
independent experiments were carried out.
Immunohistochemistry
Anti-MMP9 antibody (1:500; Abcam) was utilized to
carry out immunohistochemistry for MMP9 on a 4-µm tissue sample.
Xylene was utilized to bake and deparaffinize the slides, and
graded alcohol was utilized to pass through the above slides, next
1 mM EDTA (Invitrogen) was utilized to retrieve antigen in a steam
pressure cooker (Decloaking Chamber; BioCare Medical, Walnut Creek,
CA, USA) for 30 sec at 125°C. The following experimentations were
performed in a hydrated chamber at room temperature. Peroxidase
Block (Dako, Carpentaria, CA, USA) was utilized to pretreat the
slides for 5 min to quench the activity of endogenous peroxidase,
and 50 mM Tris-Cl, pH 7.4 to wash the slides. 5 ml 50 mM Tris-Cl
containing 250 µl normal goat serum (Dako) was utilized to block
the slides, anti-MMP9 antibody (1:500; Abcam) to incubate the
slides for 60 min. Tris-Cl (50 mM), pH 7.4 was utilized to wash the
slides, rabbit HRP conjugated Signal stain boost IHC detection
reagent (Cell Signaling Technology, Inc., Danvers, MA, USA) was
utilized to treat the slides for 30 min, and Tris-Cl (50 mM), pH
7.4 to wash the slides, DAB (3,3′diaminobenzidine) chromogen (Dako)
was utilized to develop immunoperoxidase staining for 5 min,
followed by counterstaining with hematoxylin. Following staining,
two experienced pathologists scored immunostaining independently,
two pathologists were blinded to the clinical outcomes and
clinicopathological parameters of the subjects. The scores of the
two pathologists with any discrepancy were re-examining by both
pathologists to obtain a consensus score.
Statistical analysis
The data are presented as means ± SD (standard
deviation). GraphPad Prism 5.0 (GraphPad Software Inc. La Jolla,
CA, USA) was utilized to perform the statistical analyses. Unpaired
t-test was utilized for comparison of the result of real-time PCR.
Chi-square test was employed to analyze qualitative data, and
Pearson correlation analysis to determine the correlation.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-491 directly targets MMP9
We searched online miRNA database (http://www.bioguo.org) to identify the gene
polymorphisms in MMP9 3′UTR that might potentially interfere with
the interaction between MMP9 and miRNA, and found that gene
polymorphism MMP9 3′UTR might compromise the interaction between
MMP9 with miR-194-3p, miR-491, miR-1915-3p or miR-941, and
QuikChange® Lightning Site-Directed Mutagenesis kit
(Agilent Technologies, CA, USA) was used to introduce the mutations
(either the minor allele of the target polymorphism or the
complementary sequence of the seed sequence) located in binding
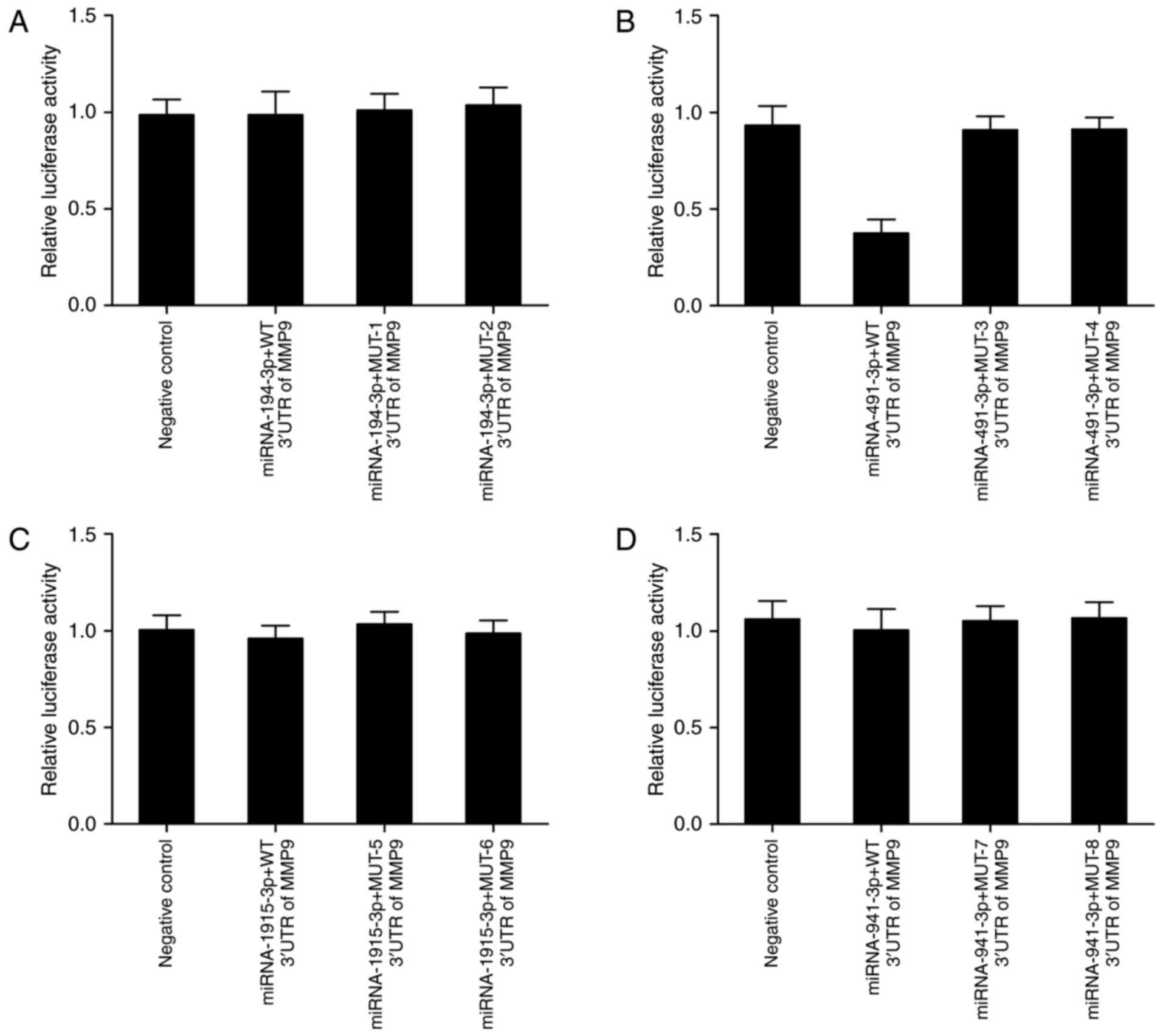
site of miR-194-3p (mutant-1 and mutant-2) (Fig. 1A), miR-491 (mutant-3 and mutant-4)
(Fig. 1B), miR-1915-3p (mutant-5
and mutant-6) (Fig. 1C) and miR-941
(mutant-7 and mutant-8) (Fig. 1D)
in MMP9 3′UTR according to the manufacturer's instructions.
Luciferase assay was performed to test whether
miR-194-3p, miR-491, miR-1915-3p or miR-941 directly regulated MMP9
expression or the effect of the candidate polymorphism on the
interaction between the miRNAs and MMP9. The cells were
co-transfected with constructs containing wild-type MMP9 3′UTR and
mutant-1–9 MMP9 3′UTR and miR-194-3p, miR-491, miR-1915-3p or
miR-941 mimic. As shown in Fig. 2,
only transfection with miR-491 (Fig.
2B) significantly reduced luciferase activity of wild-type MMP9
3′UTR, whereas luciferase activity of mutant-3 and mutant-4 MMP9
3′UTR showed no obvious difference in comparison with scramble
control. By contrast, luciferase activity of co-transfected
constructs containing wild-type MMP9 3′UTR and mutant-1/2/5/6/7/8
MMP9 3′UTR and miR-194-3p, miR-1915-3p or miR-941 mimic was
comparable with scramble control, respectively. The results
indicated that miR-491 directly targeted MMP9, and rs1056629
disrupted the interaction between MMP9 and miR-491.
Differential expression of miR-194-3p,
miR-491, miR-1915-3p or miR-941 and MMP9 in different groups
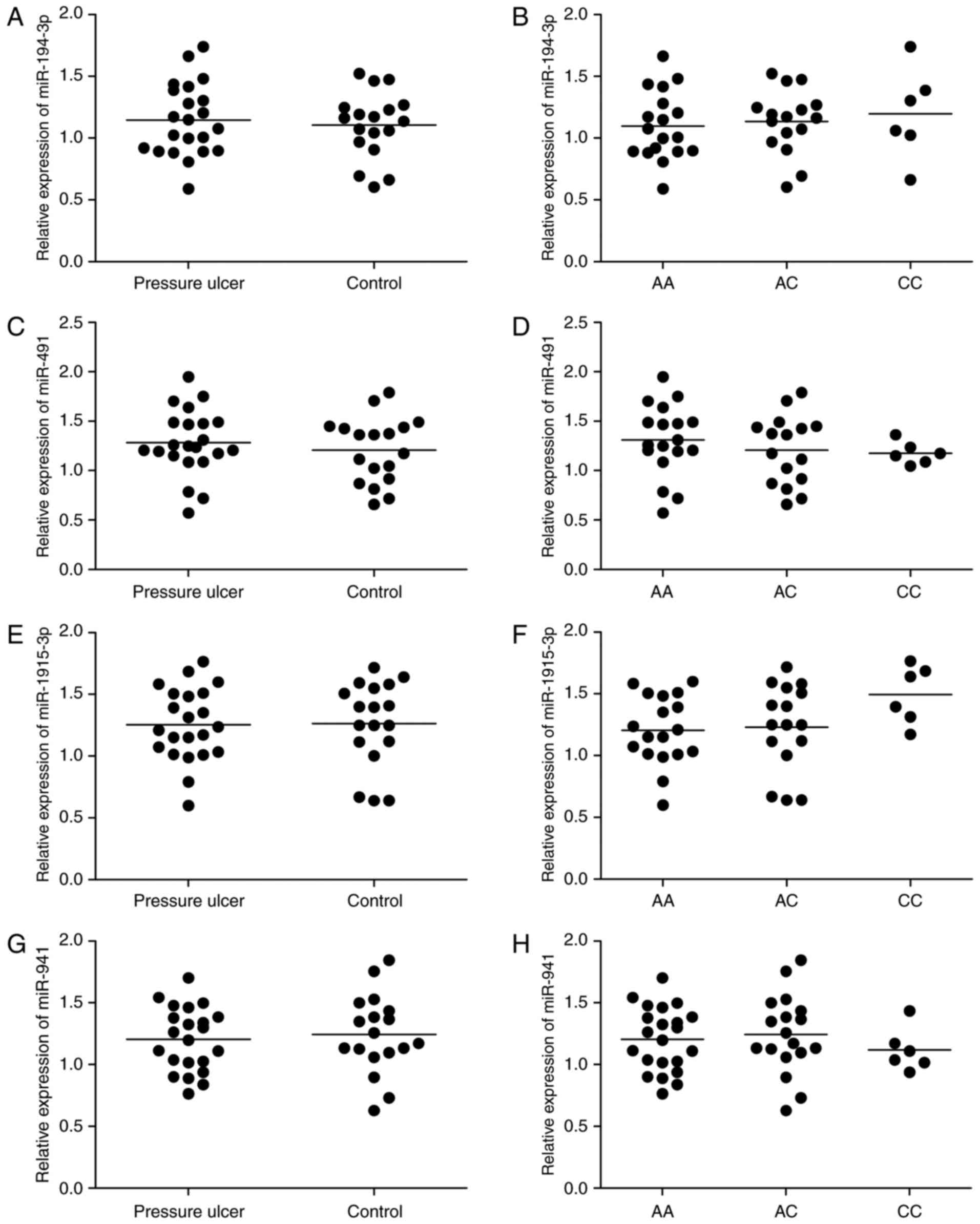
Forty individuals comprising with (N=22) or without
(N=18) PU were recruited for this study, and all those subjects
were rs1056629 genotype. Real-time PCR was performed to detect
expressions of miR-194-3p, miR-491, miR-1915-3p or miR-941 and MMP9
in these subjects. As shown in Fig.
3, miR-194-3p (Fig. 3A),
miR-491 (Fig. 3C), miR-1915-3p
(Fig. 3E) and miR-941 (Fig. 3G) levels in patients with PU was
similar with the controls, and miR-194-3p (Fig. 3B), miR-491 (Fig. 3D), miR-1915-3p (Fig. 3F) and miR-941 (Fig. 3H) levels are comparable with each
other among AA, AC and CC genotype groups. MMP9 mRNA level in
patients with PU (Fig. 4A) was much
higher than healthy controls, and also much higher in participants
carrying AA genotype (Fig. 4B) than
AC and CC genotypes, suggesting that interaction between miR-491
and MMP9 could be disrupted by the presence of the minor allele of
the rs1056629 polymorphism.
Differential protein levels of MMP9
and histology score in different groups
Immunohistochemistry was carried out to determine
MMP9 protein level among AA, AC and CC groups. As shown in Fig. 5A-C, MMP9 protein was highly
expressed in AA group compared to that in AC and CC group,
furthermore histology score in PU group was much higher in PU group
than control group (Fig. 5D), and
was also much higher in AA group than AC and CC groups (Fig. 5E), and histology score in AC and CC
was similar with each other.
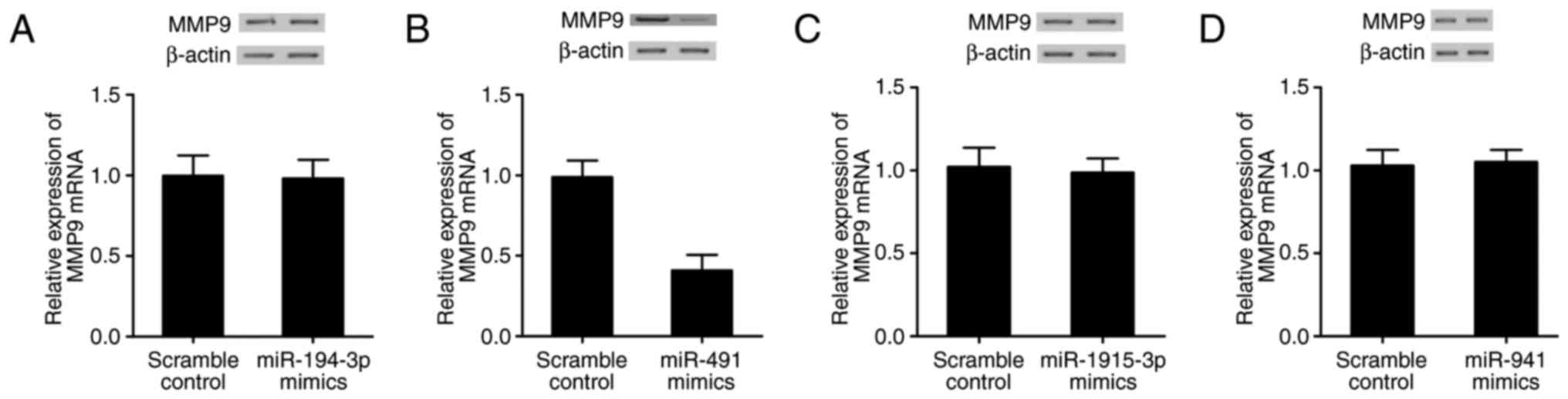
Effect of up- or downregulation of
four miRNAs on the expression of MMP9
Real-time PCR and western blot analysis were
performed to explore the effect of up- or downregulation of four
miRNAs on the expression of MMP9. MMP9 levels in BJ fibroblast
cells transfected with miR-194-3p, miR-491, miR-1915-3p or miR-941
mimic/inhibitor were measured. As shown in Fig. 6, only cells transfected with miR-491
mimic (Fig. 6B) showed evident
decrease in MMP9 mRNA and protein compared with scramble control,
while MMP9 mRNA and protein in BJ fibroblast cells transfected with
miR-194-3p (Fig. 6A), miR-1915-3p
(Fig. 6C) or miR-941 (Fig. 6D) mimic displayed no significant
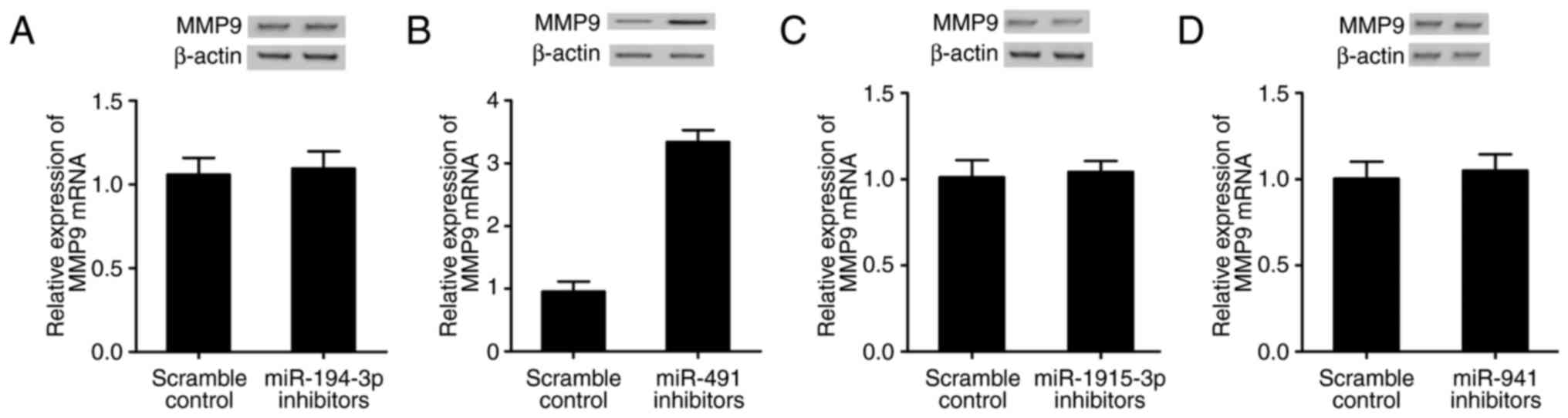
difference with scramble control. The MMP9 mRNA and protein levels
in cells transfected with miR-491 inhibitor (Fig. 7B) was substantially improved
compared with scramble control, and while MMP9 mRNA and protein
levels in cells transfected with miR-194-3p (Fig. 7A), miR-1915-3p (Fig. 7C) or miR-941 (Fig. 7D) inhibitor were comparable with
scramble control, verifying that miR-491 directly and negatively
regulated MMP9 expression.
Discussion
The loss of miR-491, has been identified in several
cancers, including breast cancer (colorectal cancer, glioblastoma
and hepatocellular carcinoma (HCC) (15–18).
miR-491-5p, a mature form of miR-491, has been demonstrated to
inhibit the growth and metastasis of cervical cancer, breast
cancer, pancreatic cancer and ovarian cancer by impacting hTERT
genes, JMJD2B, TP53 and Bcl-XL, respectively (19). However, a report indicated that the
level of miR-495-5p was elevated in colon cancer, particularly in
those aged ≥70 years, and high miR-491-5p expression related to
poor overall survival of patients with colon cancer indicating that
miR-491-5p acts as oncogene in colon cancer (20). miR-491 promotes cell apoptosis by
impacting Bcl-XL, and hence inhibits the proliferation of
colorectal cancer cells (17).
Moreover, in pancreatic cancer, miR-491 could act on TP53 and
Bcl-XL to trigger cell apoptosis via a pathway induced by
mitochondria (21). In addition,
miR-491 impacted a range of oncogenes, such as EGFR, Wnt signal and
GIT-1 to weaken the malignant features (16). Moreover, miR-491 functioned as an
anti-metastasis gene. G-protein-coupled receptor kinase-interacting
protein 1 and MMP9 could be targets of miR-491 (22). In this study, we identified that
MMP9 is a direct target of miR-491 by showing that only cells
transfected with miR-491 mimic showed evident decrease in MMP9 mRNA
and protein, and only cells transfected with miR-491 inhibitor
showed evident increase in MMP9 mRNA and protein compared with
scramble control.
MMP-induced proteolysis is critical in regulation of
cellular homeostasis: MMPs can start, enhance, or reduce signaling
cascades implicated in growth and inflammation by stimulating
cytokines and releasing free growth factors, and can degrade
structural compositions of the extracellular matrix (ECM) to modify
tissue architecture (23). MMP9
(known as gelatinase B is one of the 23 MMP family members and
exhibits promise as a therapeutic target, based on a great deal of
data showing its involvement in pathological processes accounting
for tumorigenesis, metastasis and chronic inflammation (24). Reduced MMP9 expression and activity
are related to a range of inflammatory problems, such as ulcerative
colitis (UC) (23,25). In areas of active disease, UC is a
recurrent/remitting autoimmune colonic inflammation characterized
by mediation of proteolytic activity and protein levels of MMP9
(26). MMP9 activity in UC is
involved in both perpetuation and generation of an inflammatory
state medicated by pro-inflammatory cytokines including IL1-α and
TNF-α and it can assist to maintain pro-inflammatory processes by
activating IL1-β, by potentiating IL-8, and by liberating TGF- and
TNF-α (27,28). MMP9 also accounts for the
inflammatory milieu via proteolysis of the basement membrane (BM)
compositions laminin and collagen IV (29). Epithelial cell apoptosis can be
caused by damage of epithelial BM, a defined character of UC, which
results in the loss of integrity of the colonic mucosal epithelial
barrier, thus worsening inflammation (26,30).
Likely, damage of the endothelial BM can promote lymphocyte and
neutrophil migration to the inflammatory site (31).
Intriguingly, a range of studies have reported the
activity and production of MMP9 in various pathological and
physiological processes (32). It
starts to impede wound closure, suppresses cell migration and
degrades the extracellular matrix in the bed skin in large
quantities, and for long periods in the incorrect site (33). However, Gumieiro et al showed
that pro-MMP9 was related to gait status recovery six months
following hip fracture but not related to PU and mortality in those
with hip fracture (13). Some
analysis of fluids and wound biopsies obtained from patients with
PU indicated increased expression of the activated MMP9. This
evidence indicated that this protease could destruct the
extracellular proteins, receptors and growth factors critical for
PU healing (7). It has been shown
that the activity of serum pro-MMP9 was elevated more in those with
PU than the healthy people (34).
In this study, we searched online miRNA database
(http://www.bioguo.org), and found rs1056629 in
MMP9 3′UTR that might break the interaction between MMP9 with
miR-194-3p, miR-491, miR-1915-3p or miR-941. We utilized
site-directed mutagenesis kit to obtain the 4 single-base mutations
in gene polymorphism and 4 seed sequence mutations located in
binding site of above 4 miRNAs in MMP9 3′UTR, and conducted
luciferase assay, then revealed that miR-491 directly targeted
MMP9, and rs1056629 broke the interaction between MMP9 and miR-491.
Furthermore, we enrolled 40 individuals comprising 22 subjects with
PU and 18 controls, and found that 4 miRNAs in patients with and
without PU, or in AA, AC and CC were comparable with each other.
And MMP9 mRNA in PU group was much higher than control, and was
also highly expressed in AA group. In addition, we investigated
MMP9 protein level among AA, AC and CC groups using
immunohistochemistry, and found that MMP9 protein was highly
expressed in AA group, and histology score in PU group was much
higher in PU group than control group, and was also much higher in
AA group than AC, CC groups. rs1056629 polymorphism is situated in
the 3′UTR of MMP9 at the miR-491 binding site, and it causes an
elevated risk for a cerebral infarction that is atherosclerotic
(12). It has been shown that
miR-491 is a new inhibitor of metastasis in OS via the modulation
of its downstream target, MMP9. While the minor allele of the
rs1056629 polymorphism could damage the interplay between MMP9 and
miR-491, it could be a new predictive biomarker of metastasis of OS
(35). All these piceces of
evidence are in line with our results in this study, and support
our speculation that the presence of rs1056629 polymorphism
interfered with the interaction between miR-491 and MMP9 and
altered the risk of PU in those patients.
Our study identified that miR-491 is involved in the
development of PU after hip fracture via targeting MMP9. Whereas
the interaction between miR-491 and MMP9 could be disrupted by the
presence of the minor allele of the rs1056629 polymorphism, the
rs1056629 polymorphism could be a novel biomarker for predicting
the occurrence of pressure ulcers after hip fracture.
References
|
1
|
Bours GJ, Halfens RJ, Abu-Saad HH and Grol
RT: Prevalence, prevention, and treatment of pressure ulcers:
Descriptive study in 89 institutions in the Netherlands. Res Nurs
Health. 25:99–110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Veronesi G, Harley K, Dugdale P and Short
SD: Governance, transparency and alignment in the Council of
Australian Governments (COAG) 2011 national health reform
agreement. Aust Health Rev. 38:288–294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Black J, Baharestani M, Cuddigan J, Dorner
B, Edsberg L, Langemo D, Posthauer ME, Ratliff C and Taler G:
National Pressure Ulcer Advisory Panel: National pressure ulcer
advisory panel's updated pressure ulcer staging system. Dermatol
Nurs. 19:343–349; quiz 350. 2007.PubMed/NCBI
|
|
4
|
Thompson D: A critical review of the
literature on pressure ulcer aetiology. J Wound Care. 14:87–90.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baumgarten M, Margolis DJ, Orwig DL,
Shardell MD, Hawkes WG, Langenberg P, Palmer MH, Jones PS, McArdle
PF, Sterling R, et al: Pressure ulcers in elderly patients with hip
fracture across the continuum of care. J Am Geriatr Soc.
57:863–870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lindholm C, Sterner E, Romanelli M, Pina
E, Torra y Bou J, Hietanen H, Iivanainen A, Gunningberg L, Hommel
A, Klang B and Dealey C: Hip fracture and pressure ulcers-the
Pan-European Pressure Ulcer Study-intrinsic and extrinsic risk
factors. Int Wound J. 5:315–328. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ladwig GP, Robson MC, Liu R, Kuhn MA, Muir
DF and Schultz GS: Ratios of activated matrix metalloproteinase-9
to tissue inhibitor of matrix metalloproteinase-1 in wound fluids
are inversely correlated with healing of pressure ulcers. Wound
Repair Regen. 10:26–37. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonaldo P and Sandri M: Cellular and
molecular mechanisms of muscle atrophy. Dis Model Mech. 6:25–39.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deveci M, Catalyürek UV and Toland AE:
mrSNP: Software to detect SNP effects on microRNA binding. BMC
Bioinformatics. 15:732014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan M, Zhan Q, Duan X, Song B, Zeng S,
Chen X, Yang Q and Xia J: A functional polymorphism at miR-491-5p
binding site in the 3′-UTR of MMP-9 gene confers increased risk for
atherosclerotic cerebral infarction in a Chinese population.
Atherosclerosis. 226:447–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gumieiro DN, Rafacho BP, Gonçalves AF,
Santos PP, Azevedo PS, Zornoff LA, Pereira GJ, Matsubara LS, Paiva
SA and Minicucci MF: Serum metalloproteinases 2 and 9 as predictors
of gait status, pressure ulcer and mortality after hip fracture.
PLoS One. 8:e574242013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao XL, Luo X, Wang ZX, Yang GL, Liu JZ,
Liu YQ, Li M, Chen M, Xia YM, Liu JJ, et al: Local blockage of
EMMPRIN impedes pressure ulcers healing in a rat model. Int J Clin
Exp Pathol. 8:6692–6699. 2015.PubMed/NCBI
|
|
15
|
Chen W and Qiu Y: Ginsenoside Rh2 targets
EGFR by up-regulation of miR-491 to enhance anti-tumor activity in
hepatitis B virus-related hepatocellular carcinoma. Cell Biochem
Biophys. 72:325–331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Liu Y, Granberg KJ, Wang Q, Moore
LM, Ji P, Gumin J, Sulman EP, Calin GA, Haapasalo H, et al: Two
mature products of MIR-491 coordinate to suppress key cancer
hallmarks in glioblastoma. Oncogene. 34:1619–1628. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakano H, Miyazawa T, Kinoshita K, Yamada
Y and Yoshida T: Functional screening identifies a microRNA,
miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal
cancer cells. Int J Cancer. 127:1072–1080. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Leivonen SK, Sahlberg KK, Mäkelä R, Due
EU, Kallioniemi O, Børresen-Dale AL and Perälä M: High-throughput
screens identify microRNAs essential for HER2 positive breast
cancer cell growth. Mol Oncol. 8:93–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Denoyelle C, Lambert B, Meryet-Figuiere M,
Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH,
Gauduchon P, et al: miR-491-5p-induced apoptosis in ovarian
carcinoma depends on the direct inhibition of both BCL-XL and EGFR
leading to BIM activation. Cell Death Dis. 5:e14452014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401.
2014.PubMed/NCBI
|
|
21
|
Guo R, Wang Y, Shi WY, Liu B, Hou SQ and
Liu L: MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces
cell apoptosis in SW1990 pancreatic cancer cells through
mitochondria mediated pathway. Molecules. 17:14733–14747. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou Y, Li Y, Ye J, Jiang R, Yan H, Yang
X, Liu Q and Zhang J: MicroRNA-491 is involved in metastasis of
hepatocellular carcinoma by inhibitions of matrix metalloproteinase
and epithelial to mesenchymal transition. Liver Int. 33:1271–1280.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu J, Van den Steen PE, Sang QX and
Opdenakker G: Matrix metalloproteinase inhibitors as therapy for
inflammatory and vascular diseases. Nat Rev Drug Discov. 6:480–498.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Q, Meijer MJ, Kubben FJ, Sier CF,
Kruidenier L, van Duijn W, van den Berg M, van Hogezand RA, Lamers
CB and Verspaget HW: Expression of matrix metalloproteinases-2 and
−9 in intestinal tissue of patients with inflammatory bowel
diseases. Dig Liver Dis. 37:584–592. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ordás I, Eckmann L, Talamini M, Baumgart
DC and Sandborn WJ: Ulcerative colitis. Lancet. 380:1606–1619.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ben David D, Reznick AZ, Srouji S and
Livne E: Exposure to pro-inflammatory cytokines upregulates MMP-9
synthesis by mesenchymal stem cells-derived osteoprogenitors.
Histochem Cell Biol. 129:589–597. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rodriguez D, Morrison CJ and Overall CM:
Matrix metalloproteinases: What do they not do? New substrates and
biological roles identified by murine models and proteomics.
Biochim Biophys Acta. 1803:39–54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ram M, Sherer Y and Shoenfeld Y: Matrix
metalloproteinase-9 and autoimmune diseases. J Clin Immunol.
26:299–307. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grossmann J: Molecular mechanisms of
‘detachment-induced apoptosis-Anoikis’. Apoptosis. 7:247–260. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Delclaux C, Delacourt C, D'Ortho MP, Boyer
V, Lafuma C and Harf A: Role of gelatinase B and elastase in human
polymorphonuclear neutrophil migration across basement membrane. Am
J Respir Cell Mol Biol. 14:288–295. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mast BA and Schultz GS: Interactions of
cytokines, growth factors, and proteases in acute and chronic
wounds. Wound Repair Regen. 4:411–420. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jain A and Bahuguna R: Role of matrix
metalloproteinases in dental caries, pulp and periapical
inflammation: An overview. J Oral Biol Craniofac Res. 5:212–218.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Latifa K, Sondess S, Hajer G, Manel BH,
Souhir K, Nadia B, Abir J, Salima F and Abdelhedi M: Evaluation of
physiological risk factors, oxidant-antioxidant imbalance,
proteolytic and genetic variations of matrix metalloproteinase-9 in
patients with pressure ulcer. Sci Rep. 6:293712016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian X and Zhang X: A single nucleotide
polymorphism (rs1056629) in 3′-UTR of MMP-9 is responsible for a
decreased risk of metastatic osteosarcoma by compromising its
interaction with microRNA-491-5p. Cell Physiol Biochem.
38:1415–1424. 2016. View Article : Google Scholar : PubMed/NCBI
|