Introduction
Gastric cancer (GC) is one of the most aggressive
cancer worldwide, with approximately 951,600 new cases and 723,100
new deaths in 2012 (1). Even though
the global GC incidence is decreasing, the overall disease burden
still ranks 5th in incidence and 3rd in mortality (1). In particular, 40% of all GC cases
occur in China and other East Asian countries, which may be
associated with various unhealthy living habits (2). Recently, endoscopy examination is the
main approach for GC screening, but the high medical cost of
endoscopy and the shortage of endoscopic professionals in primary
medical centers largely hampers the early diagnosis of GC (3,4).
Advances in the identification of biomarkers for detecting
precancerous and cancerous gastric lesions have offered alternative
strategies for GC screening (5).
For example, serum levels of pepsinogen I and II have been used in
the identification of high GC risk individuals before endoscopic
examination in numerous studies (5,6).
GC is considered as a disease with high
heterogeneity, yet the majority of GC patients are treated with
similar chemotherapeutic drugs and surgical techniques, resulting
in unfavorable combined sequelae and side effects (7,8).
Biomarkers also hold high expectation in the preoperative
classification of GC. Actually, protein expression levels,
non-coding RNAs, gene copy variations, and single nucleotide
polymorphisms (SNPs) are all potential biomarkers of GC (9–11). For
example, high expression of serpin A1 was found to be an indicator
of a poor prognosis (12), and
rs629367 was identified as correlated with poor survival in GC
patients (13). However, apart from
the well-known HER2 amplification, there are few biomarkers that
can be utilized in GC patient classification (14). It is still urgent to discover new
biomarkers to facilitate clinical decision-making and avoid
unnecessary over-treatment.
Heat shock protein family B (small) member 8 (HSPB8)
is a member of the small heat shock protein superfamily, which
contains a conservative α-crystallin domain at the C-terminal
(15–17). The most well-known function of HSPB8
is acting as a chaperone in association with Bag3 in the regulation
of macroautophagy (15,18–20).
HSPB8 was found to be associated with many diseases, such as
cardiomyopathy (21), amyotrophic
lateral sclerosis (22) and
Alzheimer's disease (23). HSPB8
was also found to be associated with estrogen-related cancers.
Piccolella et al found that HSPB8 modulates the
proliferation and migration of breast cancer cells (24). Suzuki et al found that HSPB8
regulates TGF-α-induced ovarian cancer cell migration (25). However, there are few reports
concerning the role of HSPB8 in gastrointestinal cancers. Based on
our previous study of the molecular signature of GC subtypes, HSPB8
was found to be involved in both diffuse and intestinal GCs
(26). In the present study, we
conducted a series of further assays in vitro and in
silico to reveal the biological role and potential prognostic
value of HSPB8 in GC.
Materials and methods
Cell line culture and siRNA
transfection
Human gastric cancer cell lines (AGS, BGC-803,
BGC-823, SCG-7901 and N87) and normal gastric cell line GES-1 were
provided by the Cancer Institute and Hospital, Chinese Academy of
Medical Sciences (Beijing, China) and maintained by our laboratory.
All cells were maintained in Dulbecco's modified Eagle's medium
(DMEM) with 10% fetal bovine serum (FBS) (purchased from Gibco
(Grand Island, NJ, USA), in an incubator containing 5%
CO2 at 37°C. A total of 4×105 cells were
seeded in each well of 6-well plates. After culturing for 24 h,
cells were transfected with the siRNAs using Lipofectamine 2000
(Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. All siRNAs were de
novo synthesized by Gene Pharma Co. (Shanghai, China) and all the
sequences are listed in Table
I.
 | Table I.Sequences of the siRNAs used in the
present study. |
Table I.
Sequences of the siRNAs used in the
present study.
| ID | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
| HSPB8 siRNA1 |
GCATTGTTTCTAAGAACTTCATT |
TGAAGTTCTTAGAAACAATGCTT |
| HSPB8 siRNA2 |
GGTCCCTCCTTACTCAACATT |
TGTTGAGTAAGGAGGGACCTT |
| NC siRNA |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
Western blot analysis and
antibodies
Rabbit antibodies against HSPB8 (1:500 diluted; cat.
no. ab96837), CREB (1:1,000 diluted; cat. no. ab31387) and pCREB
(Ser113, 1:1,000 diluted; cat. no. ab32096) were purchased from
Abcam (Cambridge, MA, USA). Rabbit antibodies against GAPDH
(1:10,000 diluted; cat. no. 14C10), Erk (1:2,000 diluted; cat. no.
137F5) and p-Erk1/2 (Thr202/Tyr204; 1:1,000 diluted; cat. no.
D13.14.4E) were purchased from Cell Signaling Technology, Inc.
(Berkley, MA, USA). Proteins were extracted using lysis buffer (50
mM Tris-HCl, pH 7.4; 10 mM EDTA; 0.5% NP-40; 1% Triton X-100) with
a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) added. SDS-polyacrylamide gel electrophoresis
(PAGE) was performed and then proteins were transferred from gels
to PVDF membranes. The membranes were incubated with primary
antibodies overnight at 4°C, washed with TBST for 5 times, 5 min
each time, and then incubated for 45 min with HRP-labeled goat
anti-rabbit IgG antibodies (1:2,000 diluted; cat. no. ZDR-5306;
ZSGB-Bio Co., Beijing, China) at room temperature, washed with TBST
for 3 times, 5 min each time. All proteins were detected with an
ECL Plus system (Beyotime Institute of Biotechnology, Jiangsu,
China).
Cell proliferation and apoptosis
assays
Cells were seeded in 96-well plates at a density of
2×103 cells per well. The viability of cells was
determined using Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan). The optical density at 490 nm
was measured by Smartspec Model 450 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) after 0, 24, 48 and 72 h, in triplicate. The
apoptosis rate of the cells was measured by flow cytometry. Cells
were obtained and washed with pre-cooling PBS, and re-suspended in
binding buffer. Annexin V-FITC (5 µl) and 7AAD (5 µl) reagents were
added to each sample and incubated at 25°C for 15 min away from
light. An additional 400 µl binding buffer was added and then the
cells were analyzed by a flow cytometer (BD Biosciences, Franklin
Lakes, NJ, USA). All assays were repeated three times
independently.
Reverse transcription and real-time
quantitative PCR
Total RNA was extracted using TRIzol reagents and
reverse transcripted using SuperScript II reverse transcriptase
(all from Life Technologies; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol.
Real-time quantitative PCR (qPCR) was conducted in triplicates
using Applied Biosystems 7500 using the SYBR-Green PCR Master Mix
(Roche). Relative mRNA levels of each gene were normalized to
GAPDH. All primer sequences used in this study are listed in
Table II.
 | Table II.Primers for real-time PCR used in the
present study. |
Table II.
Primers for real-time PCR used in the
present study.
| Gene name | Primer
sequences |
|---|
| HSPB8 | F:
CTCCTGCCACTACCCAAGC |
|
| R:
GGCCAAGAGGCTGTCAAGT |
| GAPDH | F:
GGACCTGACCTGCCGTCTAGAA |
|
| R:
GGTGTCGCTGTTGAAGTCAGAG |
| MMP9 | F:
AGACCTGGGCAGATTCCAAAC |
|
| R:
CGGCAAGTCTTCCGAGTAGT |
| EGR1 | F:
GGTCAGTGGCCTAGTGAGC |
|
| R:
GTGCCGCTGAGTAAATGGGA |
| CCNA1 | F:
GAGGTCCCGATGCTTGTCAG |
|
| R:
GTTAGCAGCCCTAGCACTGTC |
| CCND1 | F:
CGGCAAGTCTTCCGAGTAGT |
|
| R:
CCTCCTTCTGCACACATTTGAA |
| CCN1 | F:
GGATGGTAGTTTTGAGTCACCAC |
|
| R:
CACGAGGATAGCTCTCATACTGT |
| BCL2 | F:
CCTCCTTCTGCACACATTTGAA |
|
| R:
CGGTTCAGGTACTCAGTCATCC |
Online patient data acquisition and
statistical analysis
Phenotype data with genetic information of the
patients were all downloaded from TCGA (http://cancergenome.nih.gov/) (27,28).
For Gene Set Enrichment Analysis (GSEA) (29), a Pearson correlation based method
(1000 permutations of phenotype were run as recommended) was
applied to evaluate the correlation between HSPB8 expression and
gene sets of Kyoto Encyclopedia of Genes and Genomes (KEGG)
(30) and BIOCARTA (31). A FDR q-value <0.25 was considered
significant. For survival analysis, the median level of
mRNA/methylation of HSPB8 was chosen as the cut-off to separate two
subgroups and the Log-rank test was applied and corresponding
Kaplan-Meier plots were drawn to show the results intuitionally.
Correlation between mRNA and methylation of HSPB8 was calculated by
Pearson Chi-square analysis. For cell proliferation, apoptosis and
gene expression analysis, unpaired Student's t-tests were used to
compare two groups when the means follow the normal distribution,
and P<0.05 was considered statistically significant.
Results
Knockdown of HSPB8 inhibits gastric
cancer cell proliferation
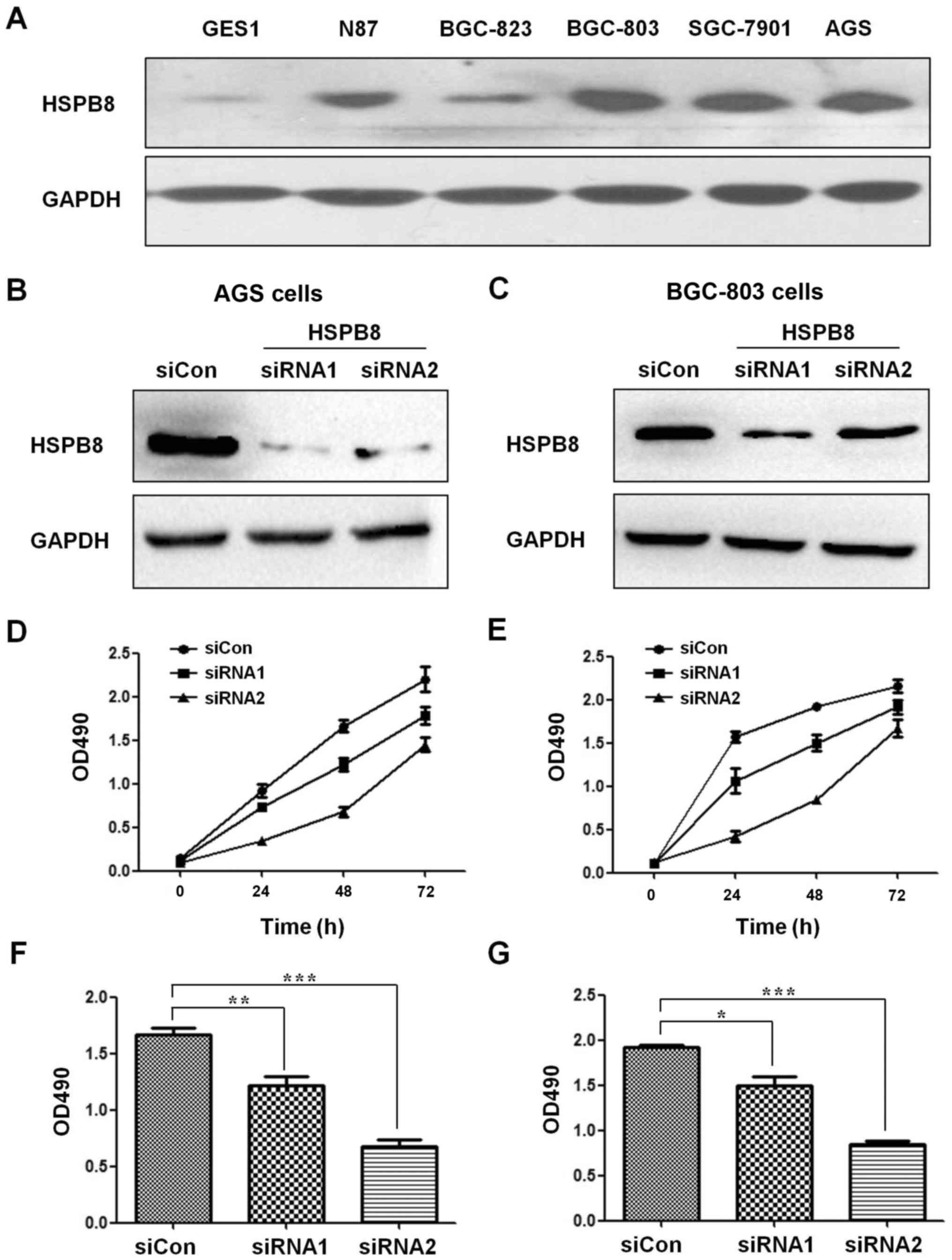
To evaluate the expression of HSPB8 in GC cells and
normal gastric epithelial cells, we detected HSPB8 in five GC cell
lines (AGS, BGC-803, BGC-823, SGC-7901 and N87) and one gastric
epithelial cell line (GES-1). The results demonstrated that HSPB8
was relatively higher in the GC cells than that noted in the normal
epithelial cells (Fig. 1A). To
examine the proliferation-promoting potential of HSPB8, two siRNAs
targeting HSPB8 were designed (sequences are shown in Table I) and transfected into AGS and
BGC-803 cells. The HSPB8 protein levels were decreased by both
siRNAs successfully (Fig. 1B and
C), and the proliferation rates of both AGS (Fig. 1D and F) and BGC-803 (Fig. 1E and G) cell lines were
significantly hampered by HSPB8 knockdown. Considering that the
efficiency of siRNA1 was higher than that of siRNA2, the subsequent
analyses were all performed using siRNA1.
Knockdown of HSPB8 promotes gastric
cancer cell apoptosis
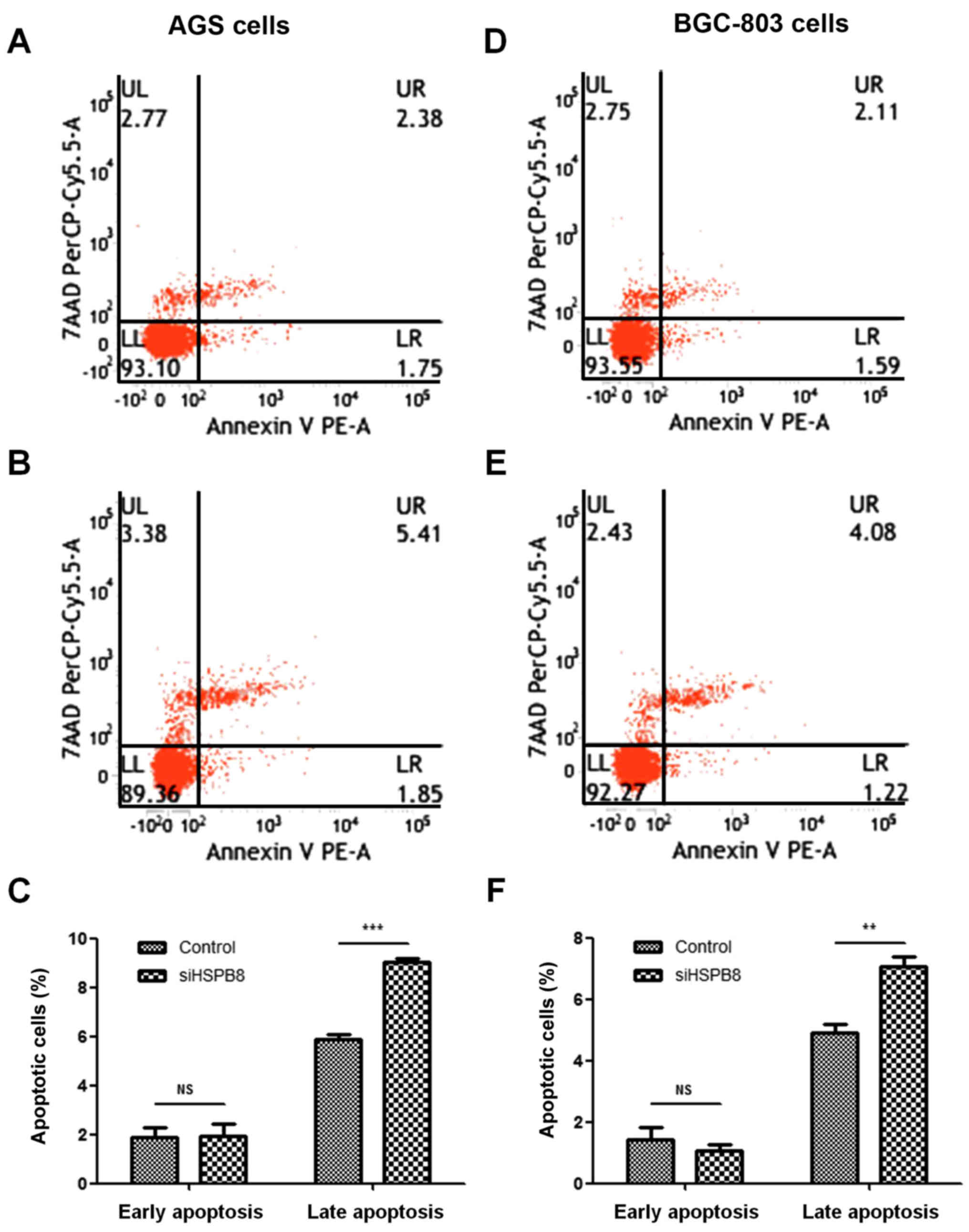
Suppression of cancer cell growth is usually
associated with activation of cellular apoptosis. To examine the
influence on apoptosis of HSPB8 knockdown, AGS and BGC803 cells
transfected with HSPB8-siRNA1 and NC were subjected to Annexin
V-FITC/7AAD staining and flow cytometry. The results indicated that
in both AGS (Fig. 2A-C) and BGC-803
(Fig. 2D-F) cell lines, HSPB8
knockdown had no significant influence on early apoptosis but
strongly increased the percentage of late apoptotic cells.
HSPB8 is positively correlated with
the ERK-CREB pathway
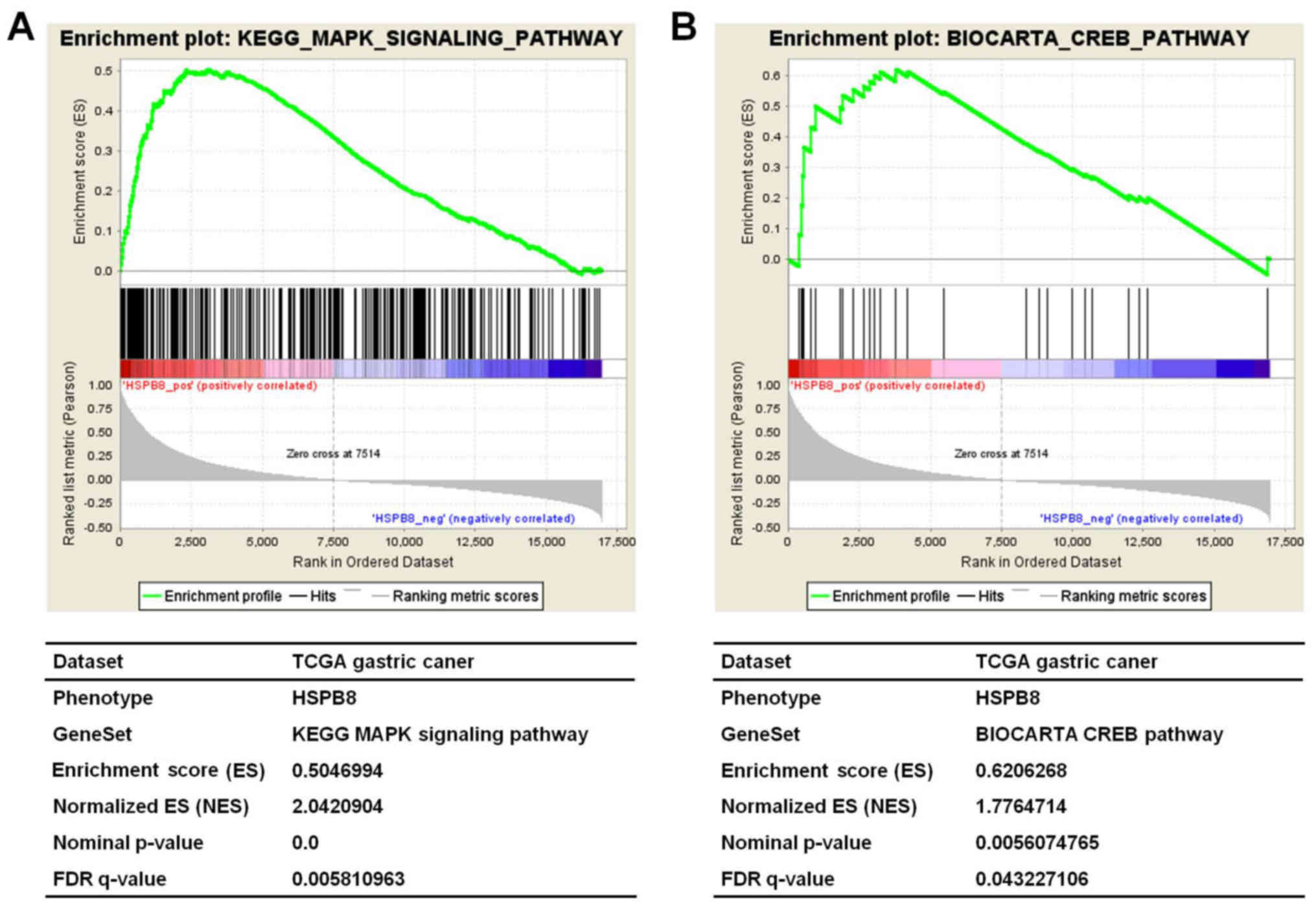
To further reveal the potential mechanism attributed
to the oncogenic role of HSPB8 in gastric cancer, the TCGA dataset
of gastric cancer expression data was downloaded and analyzed. The
GSEA analysis revealed that the HSPB8 expression level was strongly
positively correlated with the KEGG MAPK signaling pathway
(NES=2.042, P<0.001, FDR=0.006, Fig.
3A) and the BIOCARTA CREB pathway (NES=1.776, P=0.006,
FDR=0.043, Fig. 3B). Activation of
the ERK-CREB pathway would be a possible key mechanism of the
oncogenic activity of HSPB8.
Knockdown of HSPB8 decreases the
activity of the ERK-CREB pathway
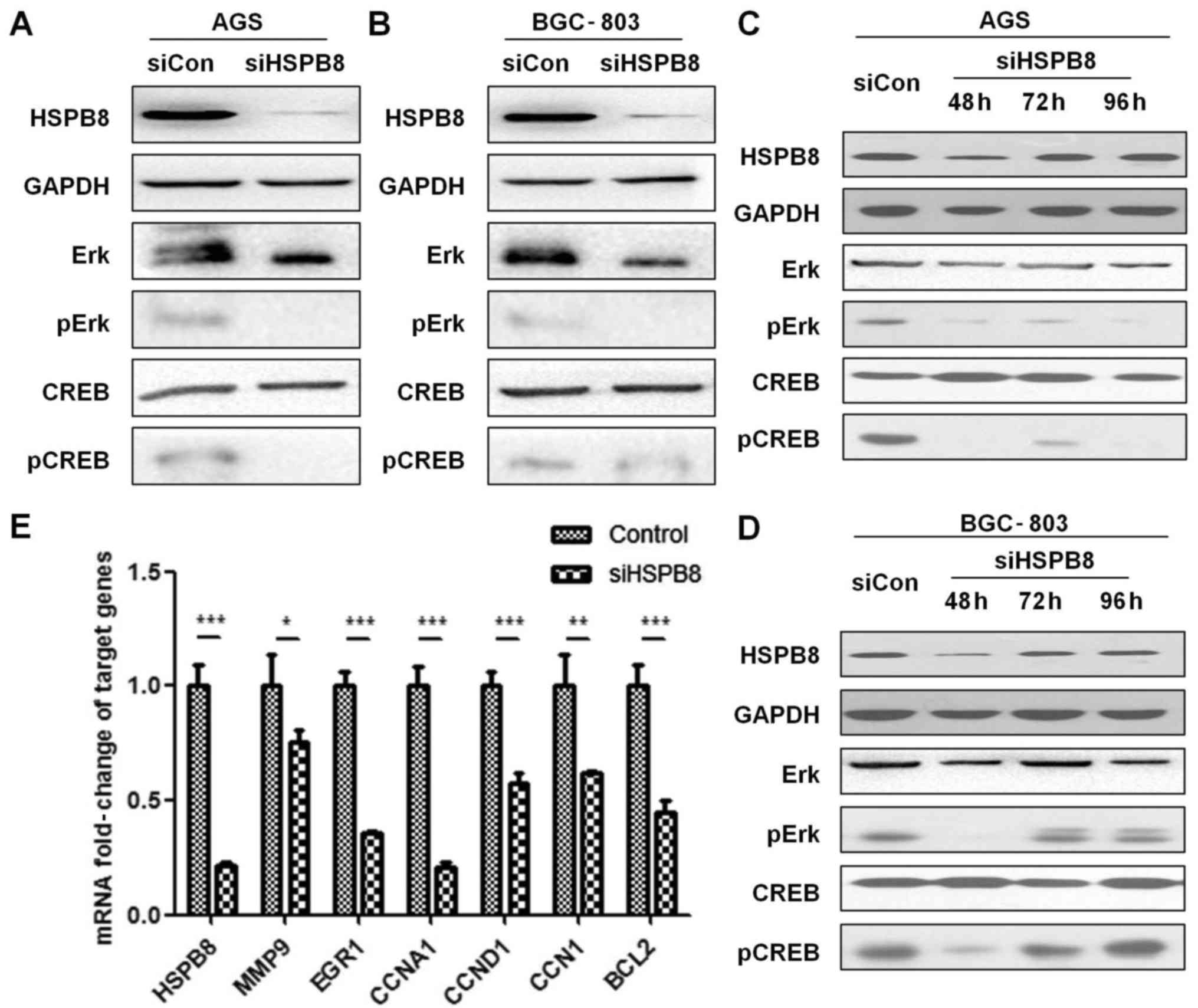
To verify our hypothesis of HSPB8's association with
the ERK-CREB pathway, we detected levels of phosphorylated and
total ERK and CREB proteins under siRNA interference of HSPB8 by
western blot analysis. Our results indicated that the total CREB
level was stable, while pErk and pCREB were significantly decreased
under HSPB8 knockdown (Fig. 4A and
B). To reveal whether the changes in pErk and pCREB were
time-dependent, we also detected the protein levels of these
proteins at 48, 72 and 96 h. The level of HSPB8 returned to a
normal level at 72 h after siRNA knockdown, suggesting that HSPB8
is crucial for cell survival and maintains a high synthesis rate in
these cells. pErk and pCREB remained suppressed at 96 h in AGS
cells, while in BGC-803 cells the levels of pErk and pCREB were
restored at 72 h (Fig. 4C and D).
Downstream genes of the ERK-CREB pathway were also detected by
RT-qPCR (primers are shown in Table
II), and we found that all the targeted genes we detected were
significantly decreased under HSPB8 knockdown (Fig. 4E).
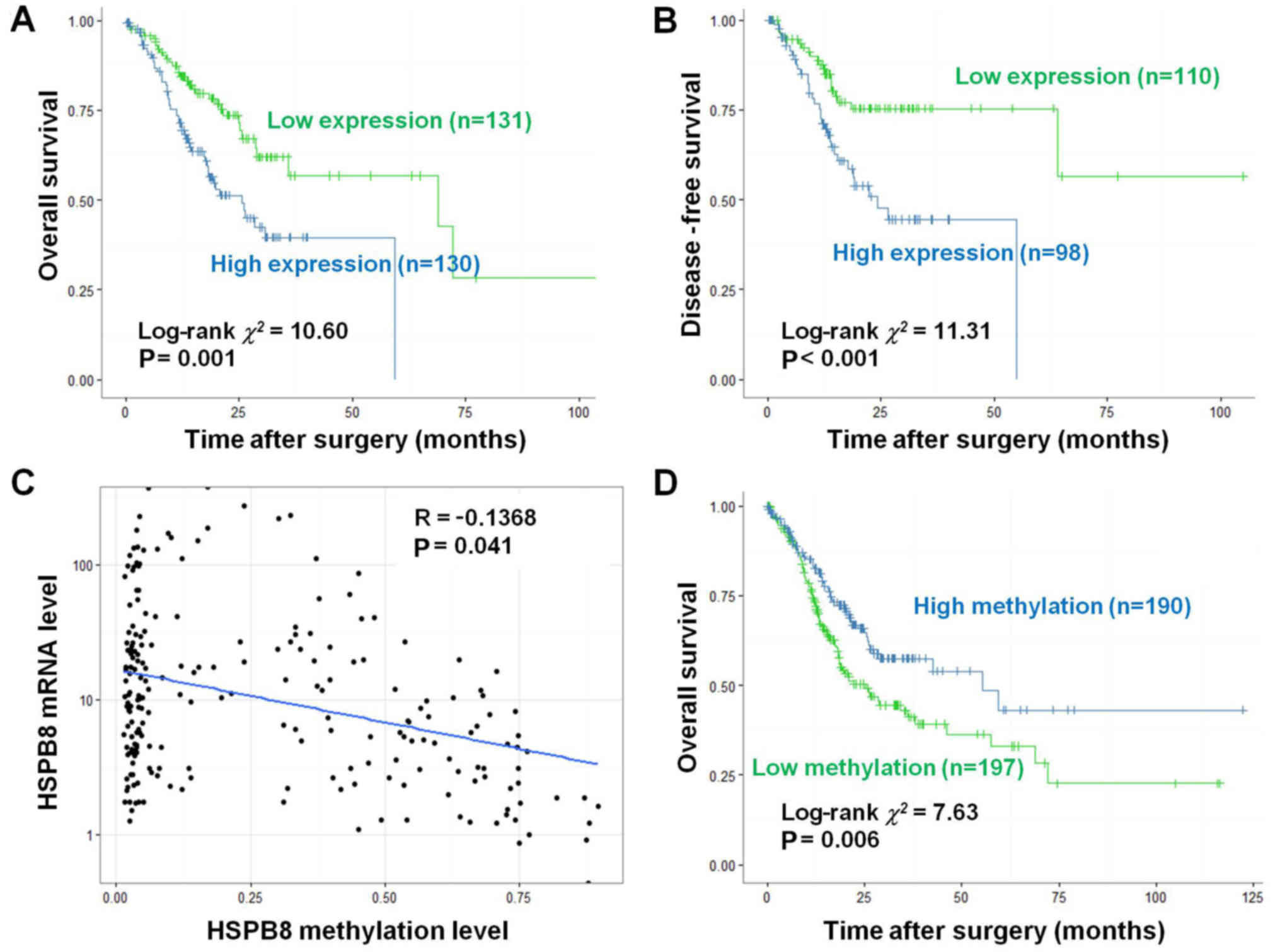
Expression and methylation levels of
HSPB8 are potential prognostic factors
To explore whether HSPB8 could be used as a
biomarker in gastric cancer, we further evaluated the survival of
TCGA gastric cancer patients. When divided by the median expression
level, the patients with a high HSPB8 level exhibited a
significantly worse prognosis than those with low HSPB8 in both
overall survival (OS) (log-rank χ2=10.60, P=0.001,
Fig. 5A) and disease-free survival
(DFS) (log-rank χ2=11.31, P<0.001, Fig. 5B).
DNA methylation data were also co-analyzed with
expression data, and indicated that the methylation level of HSPB8
DNA was significantly negatively associated with its expression (R=
−0.1368, P=0.041, Fig. 5C).
Patients with high HSPB8 methylation level exhibited a
significantly better prognosis than those with low methylation in
regards to OS (log-rank χ2=10.60, P=0.001, Fig. 5D).
Discussion
The small heat shock protein (sHsp) superfamily
consists of a series of 15–30 kDa proteins with a common
α-crystallin domain at the C-terminal. The most well-studied role
of sHsp is acting as molecular chaperones (32,33),
i.e. preventing the aggregation of enzymes under heat shock
conditions and stabilizing the proteins. There are also numerous
studies revealing that sHsp promote the functional refolding of
proteins after urea denaturation in an ATP-independent manner
(34). There are many members of
the sHsp superfamily involved in GC, such as HSP70 (35), HSP110 (36) and HSP27 (37). However, HSPB8 has drawn little
attention in this field.
In our previous study, we conducted a pilot
investigation of expression-based GC biomarker screening and found
that HSPB8 is a potential biomarker in both diffuse and intestinal
GC (26). In the present study, our
results indicated that HSPB8 is a proliferation-promoting protein
in GC cell lines, which could also aid in the evasion of apoptosis
by GC cells. The oncogenic role of HSPB8 is in accordance with most
other sHsp members, such as HSP70 (35), HSP110 (36) and HSP60 (38).
To reveal the possible mechanism of HSPB8 in GC, we
identified molecular pathways associated with HSPB8 expression by
analyzing the TCGA GC dataset. From all KEGG and BIOCARTA gene
sets, we found that the pathways of MAPK and CREB were positively
correlated with HSPB8, which was also validated by western blot
analysis in two GC cell lines. Furthermore, we found that the
targeted genes of the ERK-CREB pathway (MMP9, EGR1, CCNA1, CCND1,
CCN1 and BCL2) were significantly decreased under HSPB8 knockdown.
The positive correlation between CCN1 and HSPB8 (39) and the association between BCL2 and
HSPB8 (40), were both reported in
other cell lines, which may partially verify our finding of HSPB8's
role in the regulation of the ERK-CREB pathway.
It is worth noting that HSPB8 was restored to a
normal level at 72 h after siRNA knockdown, suggested that HSPB8 is
crucial for cell survival and maintains a high synthesis rate in
these cells. However, pErk and pCREB levels were restored along
with HSPB8 at 72 h in BGC-803 cells, but maintained a suppressed
level in AGS cells until 96 h. Thus, we suggest that HSPB8
knockdown may have a phenotypic effect on pErk and pCREB, but this
appears to be cell line-specific (i.e. this consistent change of
HSPB8 and pERK/pCREB was observed for the BGC-803 cell line but not
for the AGS cell line at different time-points). It is also
possible that HSPB8 knockdown resulted in a more far-reaching
effect on AGS cells (i.e. even when HSPB8 was restored quickly
after siRNA expired at 72 and 96 h, pErk and pCREB remained
suppressed). In all, any phenotypic effects observed after
transfection at 72 h should be interpreted with caution, and
further investigation of how HSPB8 promotes pErk and pCREB in a
cell line-specific manner is warranted.
Additionally, we also explored the potential of
HSPB8 as a prognostic biomarker in GC. Based on our results, both
the expression level and the methylation level of HSPB8 could be
used as unfavorable prognostic factors. Although the expression
level and the methylation level of HSPB8 were significantly
correlated, the expression factor still achieved a higher
discrimination than the methylation factor in GC patients
stratified based on survival data. However, further studies with a
larger population are needed before developing reagents for
clinical use. In our opinion, the effect of HSPB8 in GC is only
partially dependent on its methylation level. Transcriptional
factor level and activity, histone modification (methylation and
acetylation) and chromatin folding are all possible regulators of
HSPB8 expression. For TCGA data analysis, most GC patients had a
very low HSPB8 level (Fig. 5C),
indicating that the differential expression of HSPB8 was only
partially determined by its methylation level.
In conclusion, we demonstrated that HSPB8 could
promote the proliferation and inhibit the apoptosis of GC cells by
activating ERK-CREB signaling. We also demonstrated that high
expression of HSPB8 was indicative of a poor prognosis in GC
patients. These findings suggest new evidence to improve our
understanding of the mechanisms attributed to the carcinogenesis of
GC, and provide insight into the development of patient
stratification strategies for GC treatment.
Acknowledgements
Not applicable.
Funding
This study was supported by the Funding Program for
Excellent Talents of Beijing (2017000021469G212).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LM designed the study. JS and ML performed the
experiments. JS and LM analyzed the data. JS and LM draft the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study does not contain any research using human
participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in china,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi KS, Jun JK, Suh M, Park B, Noh DK,
Song SH, Jung KW, Lee HY, Choi IJ and Park EC: Effect of endoscopy
screening on stage at gastric cancer diagnosis: Results of the
national cancer screening programme in korea. Br J Cancer.
112:608–612. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei WQ, Yang CX, Lu SH, Yang J, Li BY,
Lian SY and Qiao YL: Cost-benefit analysis of screening for
esophageal and gastric cardiac cancer. Chin J Cancer. 30:213–218.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yeh JM, Hur C, Ward Z, Schrag D and Goldie
SJ: Gastric adenocarcinoma screening and prevention in the era of
new biomarker and endoscopic technologies: A cost-effectiveness
analysis. Gut. 65:563–574. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
den Hoed CM, van Eijck BC, Capelle LG, van
Dekken H, Biermann K, Siersema PD and Kuipers EJ: The prevalence of
premalignant gastric lesions in asymptomatic patients: Predicting
the future incidence of gastric cancer. Eur J Cancer. 47:1211–1218.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alsina M, Gullo I and Carneiro F:
Intratumoral heterogeneity in gastric cancer: A new challenge to
face. Ann Oncol. 28:912–913. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye P, Zhang M, Fan S, Zhang T, Fu H, Su X,
Gavine PR, Liu Q and Yin X: Intra-tumoral heterogeneity of HER2,
FGFR2, cMET and ATM in gastric cancer: Optimizing personalized
healthcare through innovative pathological and statistical
analysis. PLoS One. 10:e01432072015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Yang R, Lian J and Xu H: LncRNA
Sox2ot overexpression serves as a poor prognostic biomarker in
gastric cancer. Am J Transl Res. 8:5035–5043. 2016.PubMed/NCBI
|
|
10
|
Higaki E, Kuwata T, Nagatsuma AK, Nishida
Y, Kinoshita T, Aizawa M, Nitta H, Nagino M and Ochiai A: Gene copy
number gain of EGFR is a poor prognostic biomarker in gastric
cancer: Evaluation of 855 patients with bright-field dual in situ
hybridization (DISH) method. Gastric Cancer. 19:63–73. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yao YS, Yao RY, Zhuang LK, Qi WW, Lv J,
Zhou F, Qiu WS and Yue L: MOR1 expression in gastric cancer: A
biomarker associated with poor outcome. Clin Transl Sci. 8:137–142.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY,
Jo HJ, Kim DH, Kim GH and Park DY: Serpin peptidase inhibitor clade
A member 1 is a biomarker of poor prognosis in gastric cancer. Br J
Cancer. 111:1993–2002. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Q, Dong Q, He C, Liu W, Sun L, Liu J,
Xing C, Li X, Wang B and Yuan Y: A new polymorphism biomarker
rs629367 associated with increased risk and poor survival of
gastric cancer in Chinese by up-regulated miRNA-let-7a expression.
PLoS One. 9:e952492014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiu M, Zhou Y, Zhang X, Wang Z, Wang F,
Shao J, Lu J, Jin Y, Wei X, Zhang D, et al: Lauren classification
combined with HER2 status is a better prognostic factor in Chinese
gastric cancer patients. BMC Cancer. 14:8232014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carra S, Seguin SJ, Lambert H and Landry
J: HspB8 chaperone activity toward poly(Q)-containing proteins
depends on its association with Bag3, a stimulator of
macroautophagy. J Biol Chem. 283:1437–1444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamouda MA, Belhacene N, Puissant A,
Colosetti P, Robert G, Jacquel A, Mari B, Auberger P and Luciano F:
The small heat shock protein B8 (HSPB8) confers resistance to
bortezomib by promoting autophagic removal of misfolded proteins in
multiple myeloma cells. Oncotarget. 5:6252–6266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ke L, Meijering RA, Hoogstra-Berends F,
Mackovicova K, Vos MJ, Van Gelder IC, Henning RH, Kampinga HH and
Brundel BJ: HSPB1, HSPB6, HSPB7 and HSPB8 protect against RhoA
GTPase-induced remodeling in tachypaced atrial myocytes. PLoS One.
6:e203952011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carra S, Seguin SJ and Landry J: HspB8 and
Bag3: A new chaperone complex targeting misfolded proteins to
macroautophagy. Autophagy. 4:237–239. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hishiya A, Salman MN, Carra S, Kampinga HH
and Takayama S: BAG3 directly interacts with mutated
alphaB-crystallin to suppress its aggregation and toxicity. PLoS
One. 6:e168282011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marsh NM, Wareham A, White BG, Miskiewicz
EI, Landry J and MacPhee DJ: HSPB8 and the cochaperone BAG3 are
highly expressed during the synthetic phase of rat myometrium
programming during pregnancy. Biol Reprod. 92:1312015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sanbe A, Daicho T, Mizutani R, Endo T,
Miyauchi N, Yamauchi J, Tanonaka K, Glabe C and Tanoue A:
Protective effect of geranylgeranylacetone via enhancement of HSPB8
induction in desmin-related cardiomyopathy. PLoS One. 4:e53512009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Crippa V, Sau D, Rusmini P, Boncoraglio A,
Onesto E, Bolzoni E, Galbiati M, Fontana E, Marino M, Carra S, et
al: The small heat shock protein B8 (HspB8) promotes autophagic
removal of misfolded proteins involved in amyotrophic lateral
sclerosis (ALS). Hum Mol Genet. 19:3440–3456. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilhelmus MM, Boelens WC, Otte-Höller I,
Kamps B, Kusters B, Maat-Schieman ML, de Waal RM and Verbeek MM:
Small heat shock protein HspB8: Its distribution in alzheimer's
disease brains and its inhibition of amyloid-beta protein
aggregation and cerebrovascular amyloid-beta toxicity. Acta
Neuropathol. 111:139–149. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Piccolella M, Crippa V, Cristofani R,
Rusmini P, Galbiati M, Cicardi ME, Meroni M, Ferri N, Morelli FF,
Carra S, et al: The small heat shock protein B8 (HSPB8) modulates
proliferation and migration of breast cancer cells. Oncotarget.
8:10400–10415. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suzuki M, Matsushima-Nishiwaki R,
Kuroyanagi G, Suzuki N, Takamatsu R, Furui T, Yoshimi N, Kozawa O
and Morishige K: Regulation by heat shock protein 22 (HSPB8) of
transforming growth factor-α-induced ovary cancer cell migration.
Arch Biochem Biophys. 571:40–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Min L, Zhao Y, Zhu S, Qiu X, Cheng R, Xing
J, Shao L, Guo S and Zhang S: Integrated analysis identifies
molecular signatures and specific prognostic factors for different
gastric cancer subtypes. Transl Oncol. 10:99–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nature Genet. 45:1113–1120.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nishimura D: BioCarta. Biotech Software
& Internet Report. 2:117–120. 2004. View Article : Google Scholar
|
|
32
|
Shimizu M, Tanaka M and Atomi Y: Small
heat shock protein αB-crystallin controls shape and adhesion of
glioma and myoblast cells in the absence of stress. PLoS One.
11:e01681362016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu J, Luo Z, Zhang L, Wang L, Nie Q, Wang
ZF, Huang Z, Hu X, Gong L, Arrigo AP, et al: The small heat shock
protein αA-crystallin negatively regulates pancreatic
tumorigenesis. Oncotarget. 7:65808–65824. 2016.PubMed/NCBI
|
|
34
|
Nakamoto H, Fujita K, Ohtaki A, Watanabe
S, Narumi S, Maruyama T, Suenaga E, Misono TS, Kumar PK,
Goloubinoff P and Yoshikawa H: Physical interaction between
bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates
their cooperative action to refold denatured proteins. J Biol Chem.
289:6110–6119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bodoor K, Jalboush SA, Matalka I,
Abu-Sheikha A, Waq RA, Ebwaini H, Abu-Awad A, Fayyad L, Al-Arjat J
and Haddad Y: Heat shock protein association with
clinico-pathological characteristics of gastric cancer in jordan:
HSP70 is predictive of poor prognosis. Asian Pac J Cancer Prev.
17:3929–3937. 2016.PubMed/NCBI
|
|
36
|
Kimura A, Ogata K, Altan B, Yokobori T,
Ide M, Mochiki E, Toyomasu Y, Kogure N, Yanoma T, Suzuki M, et al:
Nuclear heat shock protein 110 expression is associated with poor
prognosis and chemotherapy resistance in gastric cancer.
Oncotarget. 7:18415–18423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Giaginis C, Daskalopoulou SS, Vgenopoulou
S, Sfiniadakis I, Kouraklis G and Theocharis SE: Heat shock
protein-27, −60 and −90 expression in gastric cancer: Association
with clinicopathological variables and patient survival. BMC
Gastroenterol. 9:142009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li XS, Xu Q, Fu XY and Luo WS: Heat shock
protein 60 overexpression is associated with the progression and
prognosis in gastric cancer. PLoS One. 9:e1075072014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J, Xu W, Sun T, Wang F, Puscheck E,
Brigstock D, Wang QT, Davis R and Rappolee DA: Hyperosmolar stress
induces global mRNA responses in placental trophoblast stem cells
that emulate early post-implantation differentiation. Placenta.
30:66–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kapila N, Sharma A, Kishore A, Sodhi M,
Tripathi PK, Mohanty AK and Mukesh M: Impact of heat stress on
cellular and transcriptional adaptation of mammary epithelial cells
in riverine buffalo (Bubalus bubalis). PLoS One.
11:e01572372016. View Article : Google Scholar : PubMed/NCBI
|