Introduction
AKT kinase, also known as protein kinase B, is an
important regulator for several growth factors and signaling
pathways, and activates a series of downstream substrates involving
cell proliferation, cell survival, stress response and metabolism
(1). Molecular elucidation of the
AKT signaling pathways and epidemiological investigations have
firmly established a critical role for AKT in human tumorigenesis
and chemoresistance (2–4). Therefore, AKT is a potential target
for developing new antitumor agents. Four types of AKT specific
inhibitors have been developed, which include the compounds that
target the ATP binding pocket, allosteric inhibitors, inhibitors
targeting the PH domain or pseudosubstrate inhibitors. However, due
to poor selectivity, toxicity, poor solubility, unacceptable
pharmacokinetics or inhibitory inactivition in phase II, none of
the AKT inhibitors has been approved for clinical application up to
date (2,5,6).
Xiakemycin A (XKA) is a new member of the
pyranonaphthoquinone (PNQ) family of antibiotics. As the first
Chinese karst cave-originated new antibiotic, XKA is isolated from
the fermentation broth of Streptomyces sp. CC8-201 (7). PNQ antibiotics have the basic skeleton
of naphtho[2,3-ϲ]pyran-5, 10-dione ring system and display a range
of interesting biological activities, including inhibitory
activities against tumor, fungi, bacteria, insect, virus and
mycoplasma (8,9). Concerning antitumor action, some PNQ
compounds were revealed to target several important molecules, such
as AKT, protein kinase A and DNA topoisomerase II (10–12).
However, the mechanisms underlying the signaling pathway and tumor
cell deaths induced by any of the PNQ antibiotics remain
unclear.
In the present study, tumor cell death induced by
XKA and its action on AKT and p53 were investigated. The results
firstly showed that XKA degraded cellular AKT protein, and that
cellular endogenous p53 protein levels were among the determining
factors for the action of XKA.
Materials and methods
Drugs and chemicals
XKA at >95% purity was isolated from the
fermentation broth of Streptomyces sp. CC8-201 and purified
by the previously described method (7). It was prepared into a 5 mM solution
with methanol and stored at −20°C before use. Dimethyl sulfoxide
(DMSO), 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium
bromide (MTT), triciribine hydrate (TCN) and propidium iodide (PI)
were all purchased from Sigma-Aldrich (Merck KGaA, Darmstadt,
Germany). In addition, 2,7-dichlorfluorescin diacetate
(H2DCFDA), RPMI-1640 and Dulbecco's modified Eagle's
medium (DMEM) were obtained from Invitrogen (Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Cell lines and cultures
Human hepatoblastoma HepG2, non-small lung cancer
A549, breast cancer MCF-7, prostate carcinoma PC-3, colon carcinoma
HCT 116, cervical cancer HeLa and human neuroblastoma SH-SY5Y cell
lines were all purchased from Shanghai Cell Bank, Institute of
Biochemistry and Cell Biology, Chinese Academy of Sciences. The STR
profiles of all the cell lines were confirmed by the provider.
Concerning the HepG2 cell line it is misidentified according to:
(https://www.ncbi.nlm.nih.gov/pubmed/19751877). This
cell line was thought to be derived from a hepatocellular carcinoma
(HCC) tumor but it has since been shown to be from hepatoblastoma.
It has not affected the outcomes of this study.
All the cells were cultured in RPMI-1640 or DMEM
supplemented with 10% fetal bovine serum (FBS; Tianjin Haoyang
Biotech Co., Tianjin, China). The cells were incubated at 37°C with
5% CO2 in a humidified atmosphere.
MTT assay
Cell viability was assessed by an MTT assay. Cells
were seeded in a 96-well plate for 24 h at a cell density of
3×103/well and then treated with XKA for 72 h. The final
concentration of methanol in the culture media was <0.1%. MTT
method was performed as previously described (13). The control group was set as 100%,
and IC50 values were assessed using the non-linear
regression analysis.
Western blot analysis
Western blot assay was performed as previously
described (14). The antibodies
against cleaved caspase-3 (1:1,000; cat. no. 9664), caspase-9
(1:1,000; cat. no. 9505), AKT (1:1,000; cat. no. 9272), p-AKT
(1:1,000; cat. no. 9271) and PARP-1 (1:1,000; cat. no. 9542) were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
The antibodies against p53 (1:2,000; cat. no. sc-126) and β-actin
(1:5,000; cat. no. sc-1616) were obtained from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA).
Detection of apoptotic cells by flow
cytometry
The apoptotic cells were stained with Annexin
V-FITC/PI apoptosis kit (BD Biosciences, San Jose, CA, USA),
following the manufacturer's protocol. The fluorescence intensities
were assessed using a BD FACSCalibur flow cytometer (BD
Biosciences).
RNA interference
Cells were transfected with Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions at a concentration of 100 pmol
siRNA/gene. The sequences of AKT and p53 siRNA were as follows:
AKT, 5′-ACGAGUUUGAGUACCUGAA-3′ and p53,
5′-GACTCCAGTGGTAATCTAC-3′.
Assessement of reactive oxygen species
(ROS)
Following treatment with 2 µM XKA for different
time-points, the cells were stained with 5 mM H2DFFDA
for 1 h at 37°C. The cells were harvested, and then flow cytometry
was performed with excitation and emission wavelengths of 488 and
530 nm, respectively.
PI staining
To determine the type of cell death, cells
(5×104 cells/well) were cultured in 24-well plates and
then treated with XKA. Cells were washed with PBS twice and stained
with 50 µg/ml PI for 15 min. The cells were observed and imaged
under a fluorescence microscope (Olympus Corporation, Tokyo,
Japan).
AKT1 activity assay in vitro
AKT1 assay kit was purchased from BioVision, Inc.
(Mountain View, CA, USA). The assay was performed according to the
manufacturer's protocol. In brief, the activity of the
immunoprecipitated AKT1 was assessed using exogenously recombinant
GSK3α as the substrate. The bands of phosphorylated GSK3α were
determined by western blot analysis.
Statistical analysis
Data are expressed as the means ± standard
deviations (SD). One-way analysis of variance (ANOVA) was used for
the inter-group comparison. Statistical analysis was performed
using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant result.
Results
The growth-inhibitory activity of XKA
on human tumor cells
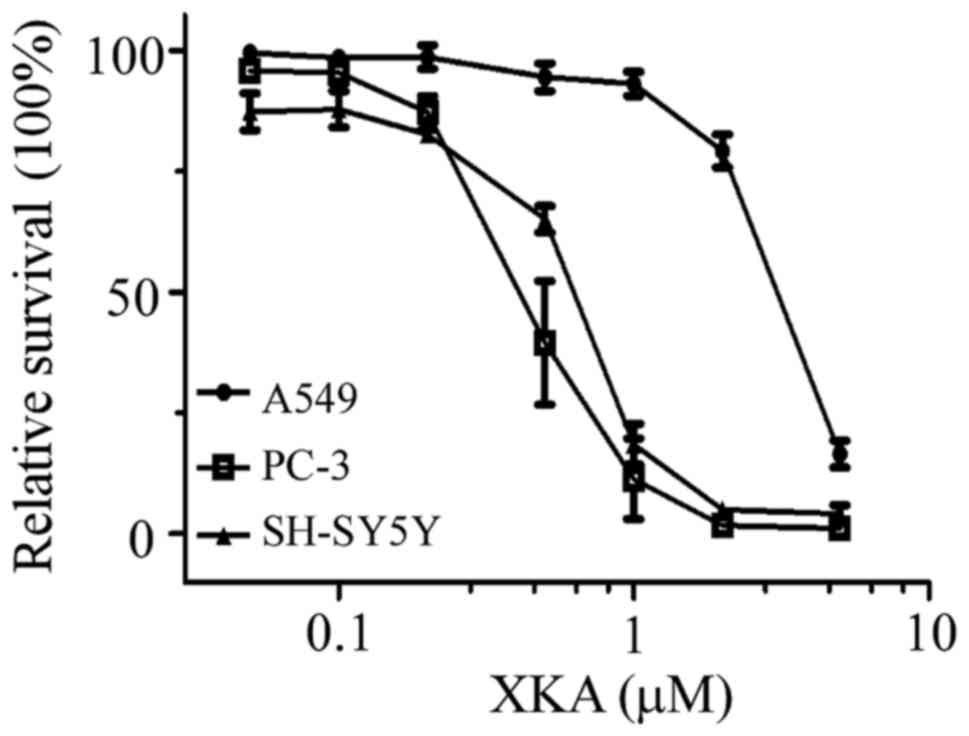
In order to assess the cytotoxic activity of XKA,
seven human tumor cell lines were used for the MTT assay. As
displayed in Fig. 1, the
growth-inhibitory activity of XKA on PC-3, SH-SY5Y and A549 cells
was concentration-dependent. The IC50 values of XKA on
the seven cell lines have been listed in a previous study (7), with a range of 0.43 to 2.77 µM. The
cell line most susceptible to XKA was PC-3.
Inhibition of AKT1 activity by XKA in
vitro
In a previous study, it was demonstrated that
selective inhibition of AKT kinase activity required the
3,6-dihydro-2H-pyran ring of PNQ (11). XKA possesses this basic structure
and possibly exerts an inhibitory activity of AKT. The AKT1
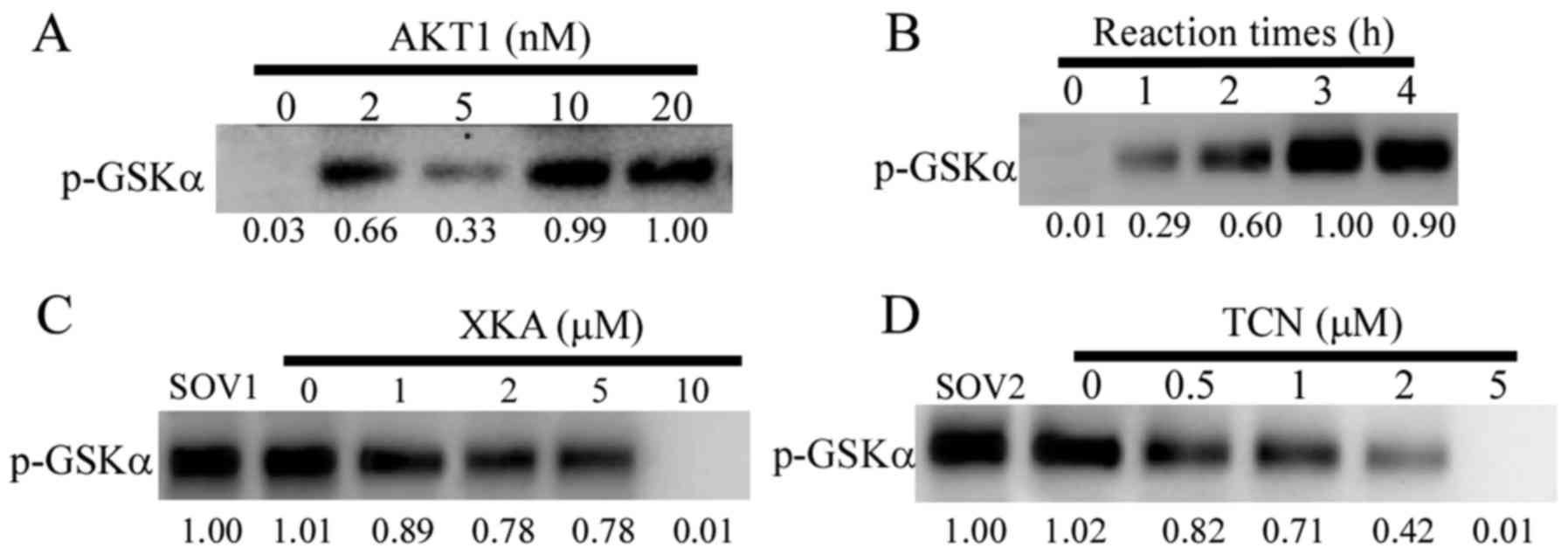
activity assay kit was used to verify this hypothesis. The reaction
conditions of the assay kit were optimized to 4 h for the reaction
time and 10 nM AKT1 for the following determinations (Fig. 2A and B). The suppression of AKT1
activity by TCN was concentration-dependent. In contrast to XKA,
there were small changes of the phosphorylated bands from the
concentrations of 1 to 5 µM (Fig.
2C). The IC50 values for inhibiting AKT1 activity by
TCN was 1.8 µM (Fig. 2D). However,
the IC50 values for inhibiting AKT1 activity by XKA was
over 5 µM.
Targeting cellular AKT by XKA
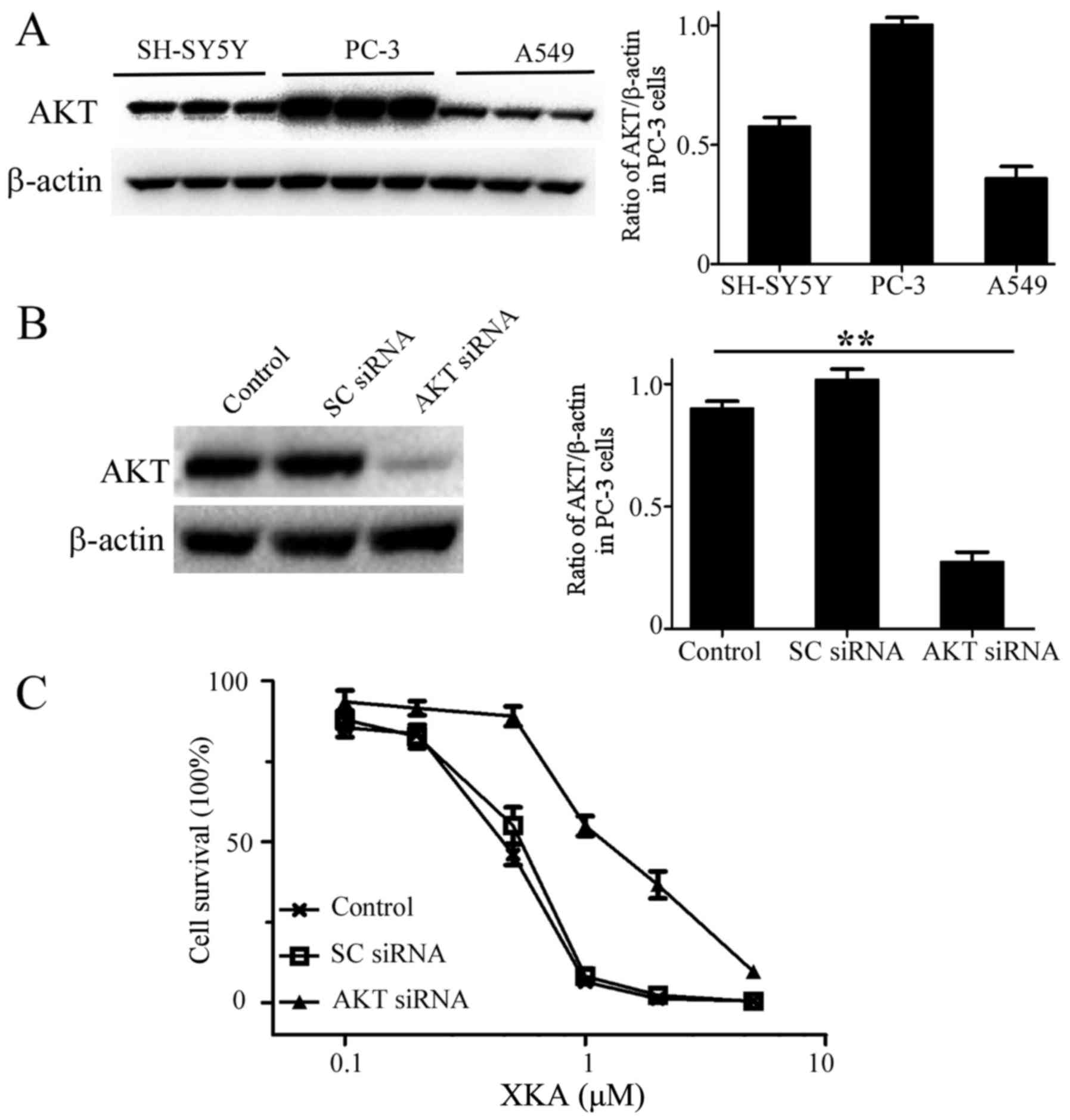
In order to clarify whether AKT is a target of XKA,
AKT protein levels from PC-3, SH-SY5Y and A549 cells were detected
by western blotting and quantified (Fig. 3A and B). The highest AKT content and
potent inhibition of XKA in PC-3 cells indicated that AKT involves
XKA action on tumor cells. RNA interference was used to confirm the
relationship. AKT protein levels were markedly knocked down in PC-3
cells (Fig. 3C). The XKA
susceptibility was significantly reduced and the IC50
value rised to 1.5 µM, indicating that AKT is a bona fide
target of XKA.
Degradation of AKT protein by XKA in
PC-3 cells
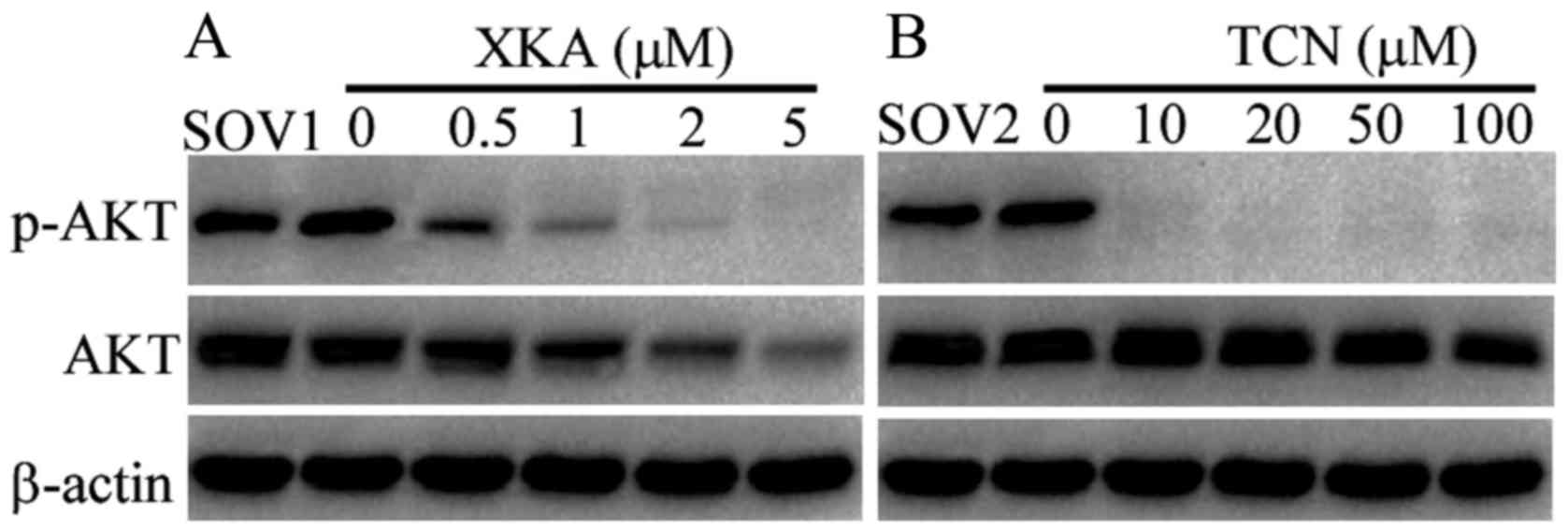
In order to decipher the difference between XKA and
TCN, AKT phosphorylation was detected by western blotting following
exposure to both drugs for 24 h. The phosphorylated AKT disappeared
after treatment with 10 µM TCN, while the total AKT protein levels
remained unchanged when the cells were treated with TCN from 10 to
100 µM (Fig. 4). Conversely, 0.5 µM
XKA treatment significantly reduced phosphorylated AKT levels and
the degradation of AKT was obvious following exposure to 1 µM XKA.
The results demonstrated that suppression of the AKT activity by
XKA is related to protein degradation, not kinase inhibition.
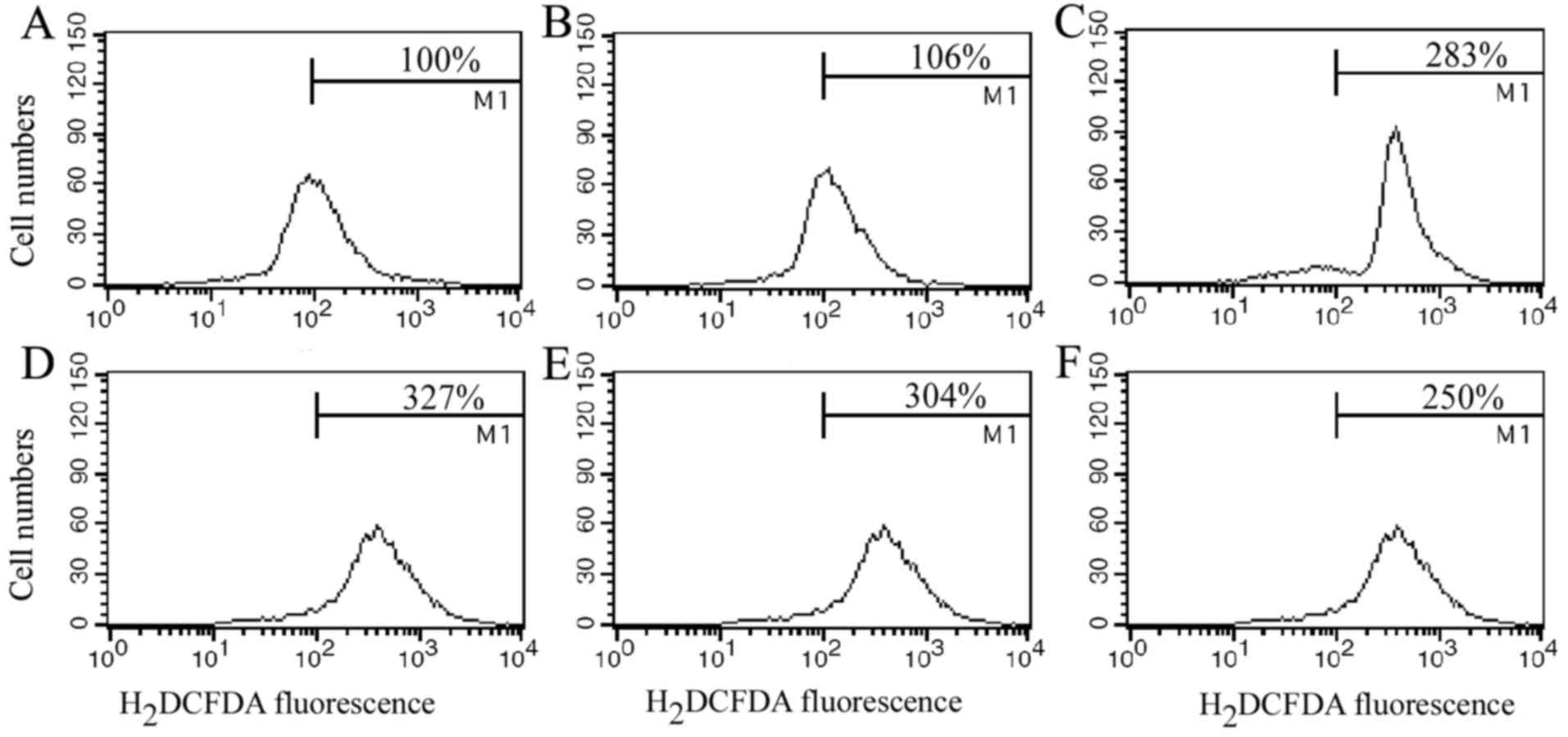
Generation of ROS triggered by
XKA
In order to explore the mechanism of the potent
inhibition of XKA towards tumor cells, ROS was determined with flow
cytometry. ROS generation markedly increased after the PC-3 cells
were treated with 2 µM XKA for 1 h and higher at 6 h treatment
(Fig. 5), indicating that the XKA
treatment triggered oxidant response.
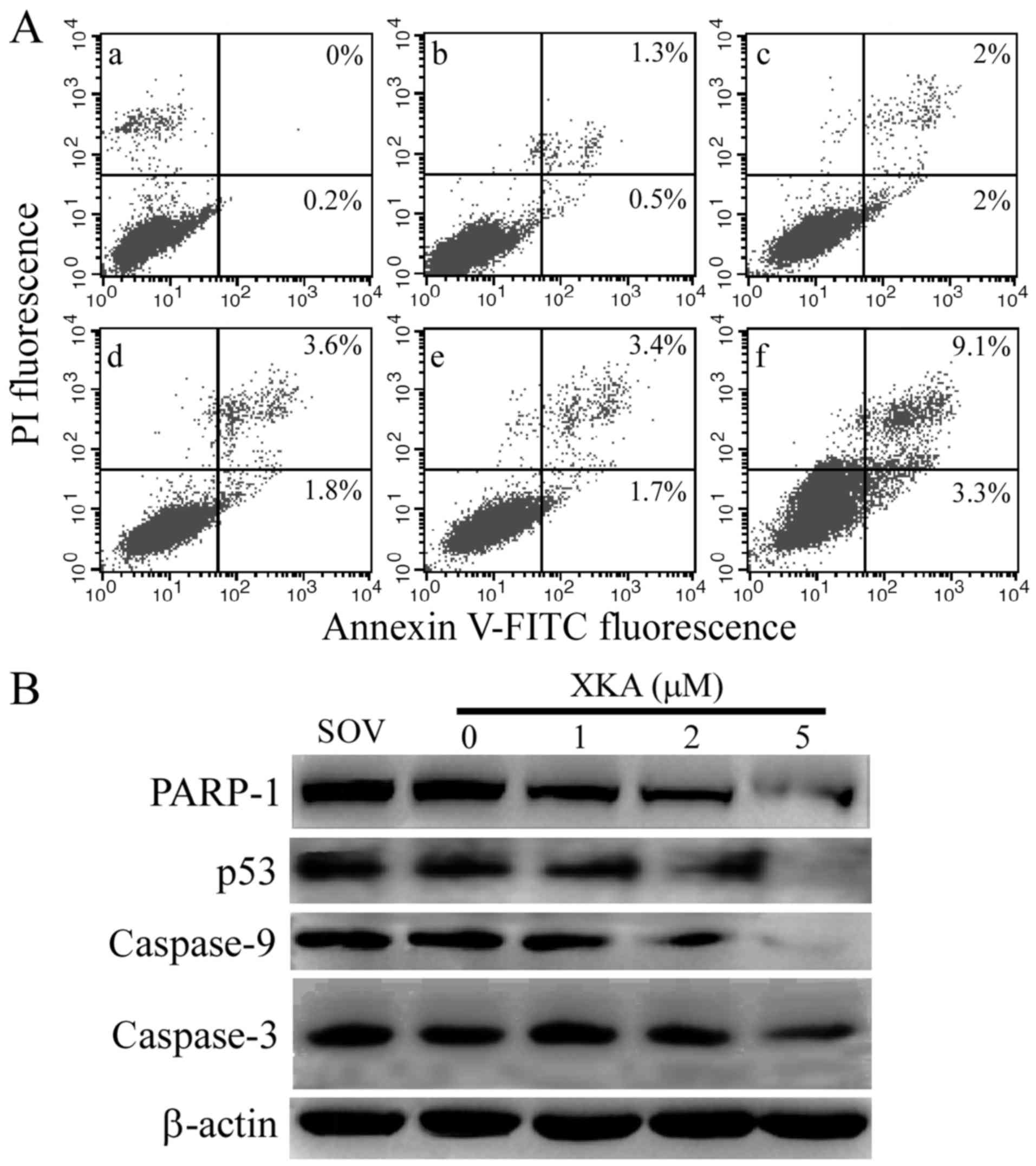
Small ammount of typical apoptotic
cells induced by XKA
ROS generation triggered by 2 µM XKA verified that
the cells were damaged. The ratio of apoptotic cells was detected
with Annexin V/PI staining in the XKA-treated PC-3 cells.
Approximately 12.4% apoptotic cells were detected after the PC-3
cells were treated with XKA for 24 h (Fig. 6A) and it did not increase after
exposure to XKA for 48 h, indicating that the majority of cells did
not undergo typical apoptosis.
To confirm the Annexin V/PI staining result, PARP-1
and p53 levels were detected by western blotting in PC-3 cells
following exposure to 2 µM XKA for 48 h (Fig. 6B). The cleaved PARP-1 fragment was
not observed and degradation of p53 occurred at 6-h treatment. The
results further ruled out the possibility that the XKA-induced cell
death was apoptosis.
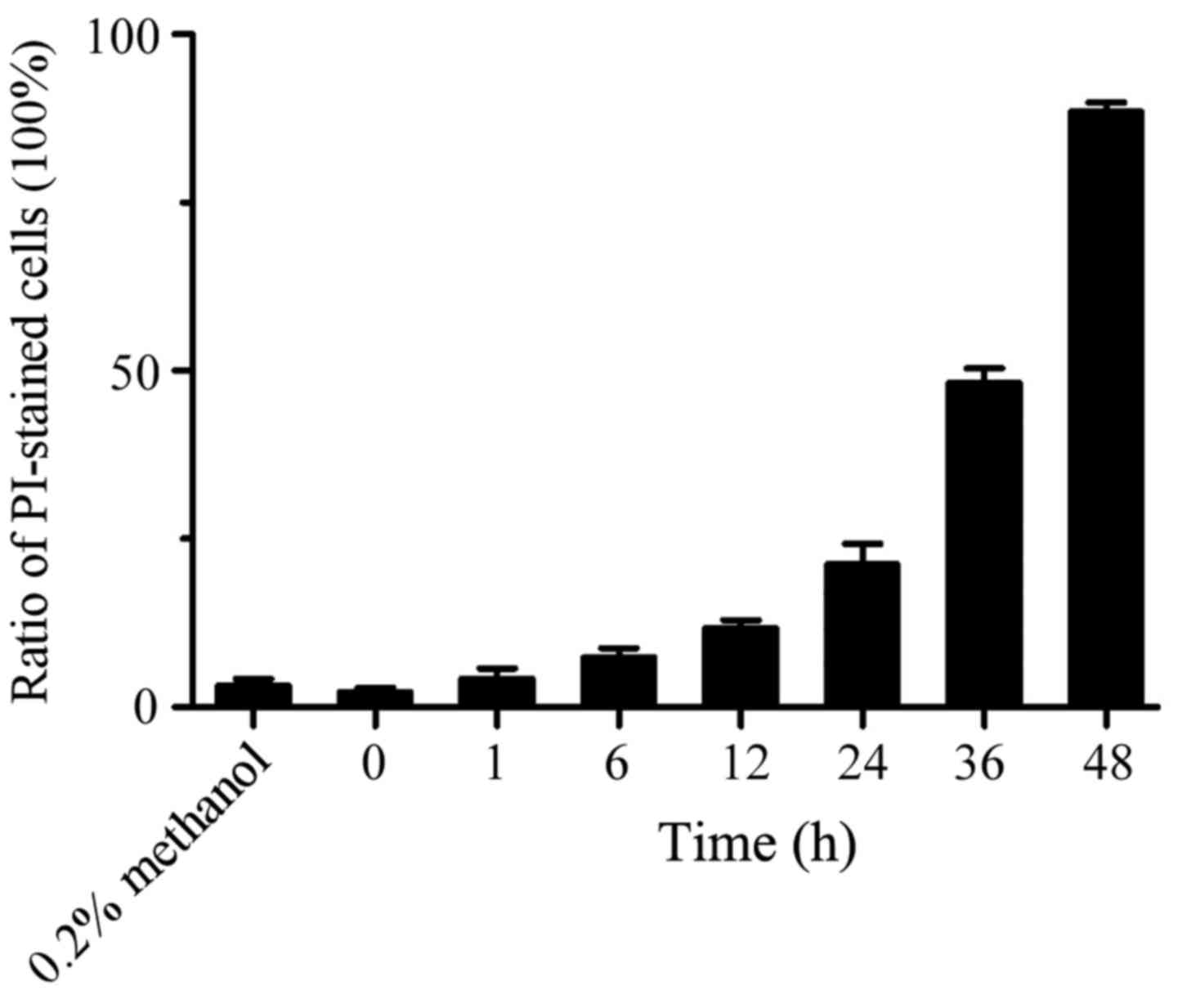
Significant increase of PI-staining
cells treated by XKA
In order to identify the type of cell death, PC-3
cells were incubated with 5 µM XKA from 1 to 48 h and PI-stained
cells were counted. With the extension of treatment time, the red
fluorescent cells increased significantly. At 48 h of treatment,
the cell ratio stained with PI reached 89% (Fig. 7).
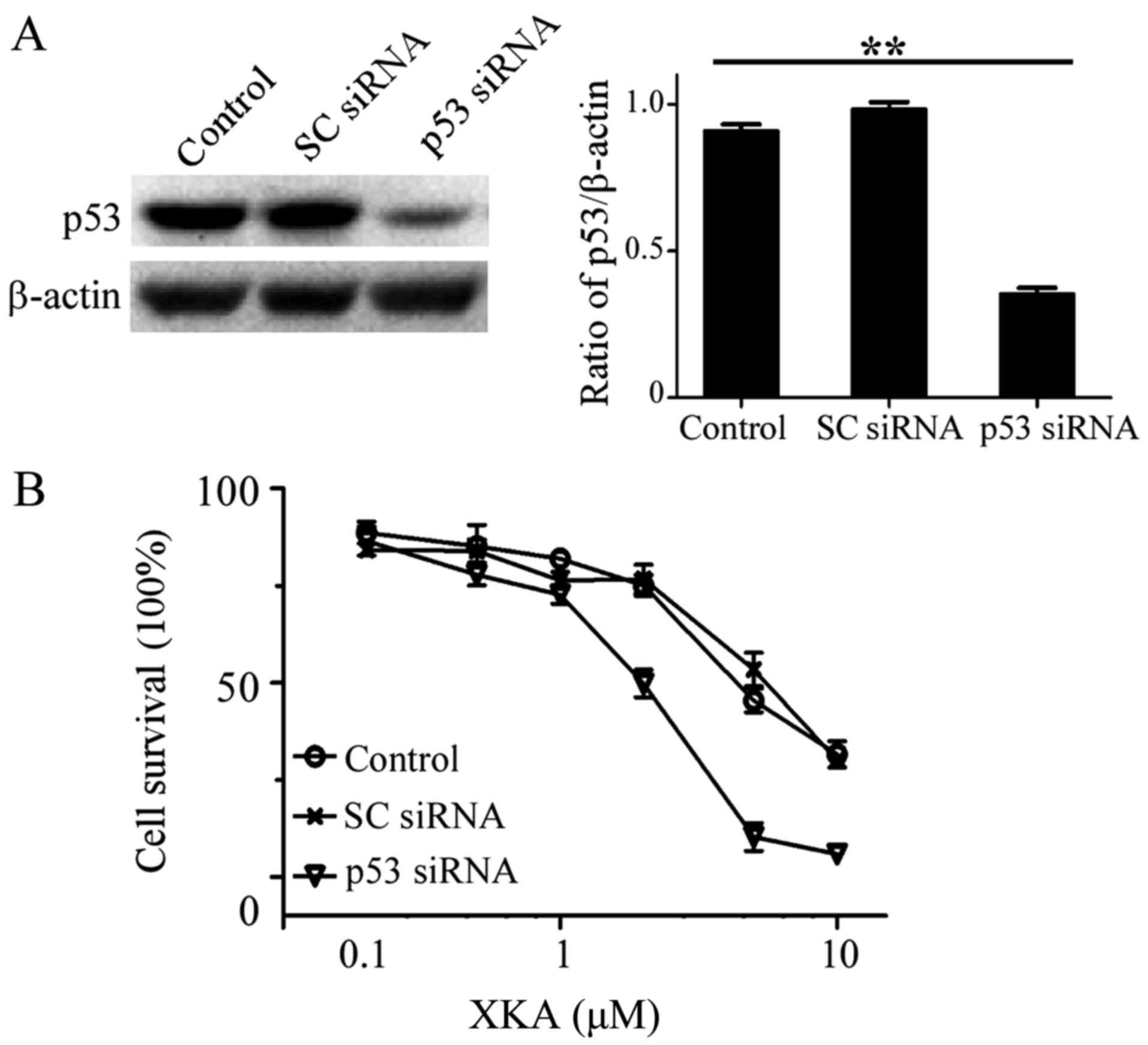
Effect of endogenous p53 levels on XKA
action
In order to search other molecules influencing XKA
action, p53 protein levels from seven cell lines were detected. The
highest level of p53 protein was observed in A549 and HCT-116 cell
lines (data not shown), indicating that p53 may be a protein
related with the action of XKA.
Knockdown of p53 in the A549 cells was performed and
MTT assay was used for determining the susceptibility to XKA. The
sensitivity to XKA increased after the reduction of p53 protein
levels. The IC50 value decreased to 1.2 µM in the p53
siRNA-treated cells (Fig. 8).
Discussion
In the present study, we presented evidence that the
susceptibility to XKA is positively correlated with the endogenous
level of AKT protein and inversely related to that of p53 in tumor
cells. The type of cell death induced by XKA may be necroptosis,
not apoptosis. To the best of our knowledge, this is the first
study to reveal that degradation of AKT was caused by PNQ
antibiotics.
PNQ antibiotics are a class of AKT selective kinase
inhibitors and the 3,6-dihydro-2H-pyran ring of the PNQ
lactone is an essential structure for potency and selectivity
(10,11). Reduction of hydroquinone can
subsequently form a quinone methide and this reactive intermediate
could then covalently alkylate Cys310 on the kinase activation,
resulting in PNQ lactone-AKT adduct and leading to inhibition of
AKT activity. In addition, the T-loop Cys310 residues
are critical for sensitivity to the PNQ inhibitors and the T-loop
Cys296 residues are indispensable. Through
deconstructing analogues to design different steric configuration
of basic skeleton and chain structure, researchers have proved that
the absolute configuration of PNQ affects its inhibitory activity
(15). XKA has the essential
structural feature for potent and selective inhibition of PNQ, but
weak action on inhibiting AKT kinase activity. It is valuable to
investigate their structure and AKT inhibition. In the present
study, we have only demonstrated AKT degradation in PC-3 cells.
Whether AKT is a real target of XKA should be further confirmed in
subsequent experiments. The overexpression of AKT protein in A549
or SH-SY5Y cells would clarify this issue.
In the present study, we did not demonstrate why
lower AKT protein levels in SH-SY5Y cells have similar
susceptibility to XKA as the PC-3 cells. Perhaps there are other
molecular factors for the determination of the action of XKA. We
aim to further study this difference in our laboratory.
The present study revealed that ROS generation
induced by XKA was very rapid due to unique chemical structure.
Lactoquinomycin A and XKA have the same basic skeleton (16). The former contains nitrogen in sugar
group at C8, while the latter contains unsaturated sugar at C6. XKA
has a higher degree of unsaturation than lactoquinomycin A and has
a high degree of ability as an electron acceptor, leading to
generation of superoxide radicals.
According to the results of the present study,
reduction of p53 protein levels has enhanced XKA cytotoxicity to
tumor cells, indicating one of the biomarkers for predicting the
action of XKA. Lower endogenous p53 protein levels had been
determined in SH-SY5Y cells (data not shown), which can partially
explain its susceptibility to XKA. However, our data are very
primary as we did not detect XKA action influenced by the wild-type
p53 or mutant p53. In further experiments, transfection of HPV E6
protein or addition of p53 degradation inhibitor may elucidate this
issue.
Tumor suppressor protein p53 and AKT kinase play
important roles in pro-apoptotic and anti-apoptotic signaling
pathways, and determine cellular survival. Accumulating evidence
indicates crosstalk between these pathways (17). Ubiquitin ligase Mdm2, the critical
p53-regulated enzyme, can be phosphorylated by AKT. Conversely, the
activated p53 can also act on AKT in several ways such as
caspase-mediated cleavage and self-degradation of AKT, induction of
PTEN expression and dephosphorylation of PI3K to suppress AKT
activity (18). It has been
reported that PTEN deficiency is a biomarker of good response to
AKT inhibitor MK-2206 (19). Based
on the interaction between p53, PTEN and AKT, rational combination
of biomarkers could be promising for predicting AKT inhibitors in
different types of tumor cells.
In the present study, the cell death induced by XKA
did not present typical apoptosis as the PI-stained cells increased
in a concentration-dependent manner. It had been partly suppressed
by specific necroptotic inhibitor (data not shown). Therefore, the
type of cell death induced by XKA may be necroptosis, which has
important roles in physiology as well as diseases (20). The exploration of the underlying
mechanism of XKA-induced cell death has been undertaken in our
laboratory.
Notably, XKA can degrade the cellular AKT and PARP-1
proteins, indicating a new mode of PNQ antibiotic action on tumor
cells. Suppression of the proteasome pathway is an important target
of drugs such as bortezomib (21).
It is valuable to study the underlying mechanism of protein
degradation by XKA.
In conclusion, the underlying mechanism of the
potent XKA inhibition of tumor cells is due to specific degradation
of the AKT protein and low cellular p53 levels. These finding will
aid to the development of AKT inhibitors as antitumor agents.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Natural Scientific Foundation of China (nos. 81373308 and 31471150)
and the Jiangxi Provincial Department of Science and Technology
(grant no. 2016BAB204165).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
QH and CS conceived and designed the study. CC, ZJ,
XO and HZ performed the experiments. QH, CC and CS wrote the paper.
QH and CS reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Review Board.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Manning BD and Toker A: AKT/PKB signaling:
Navigating the Network. Cell. 169:381–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mundi PS, Sachdev J, McCourt C and
Kalinsky K: AKT in cancer: New molecular insights and advances in
drug development. Br J Clin Pharmacol. 82:943–956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan CH, Jo U, Kohrman A, Rezaeian AH,
Chou PC, Logothetis C and Lin HK: Posttranslational regulation of
Akt in human cancer. Cell Biosci. 4:592014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim D, Dan HC, Park S, Yang L, Liu Q,
Kaneko S, Ning J, He L, Yang H, Sun M, et al: AKT/PKB signaling
mechanisms in cancer and chemoresistance. Front Biosci. 10:975–987.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nitulescu GM, Margina D, Juzenas P, Peng
Q, Olaru OT, Saloustros E, Fenga C, Spandidos DΑ, Libra M and
Tsatsakis AM: Akt inhibitors in cancer treatment: The long journey
from drug discovery to clinical use (Review). Int J Oncol.
48:869–885. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar CC and Madison V: AKT crystal
structure and AKT-specific inhibitors. Oncogene. 24:7493–7501.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang ZK, Guo L, Chen C, Liu SW, Zhang L,
Dai SJ, He QY, You XF, Hu XX, Tuo L, et al: Xiakemycin A, a novel
pyranonaphthoquinone antibiotic, produced by the
Streptomyces sp. CC8-201 from the soil of a karst cave. J
Antibiot (Tokyo). 68:771–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sperry J, Bachu P and Brimble MA:
Pyranonaphthoquinones - isolation, biological activity and
synthesis. Nat Prod Rep. 25:376–400. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brimble MA, Duncalf LJ and Nairn MR:
Pyranonaphthoquinone antibiotics - isolation, structure and
biological activity. Nat Prod Rep. 16:267–281. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiménez-Alonso S, Orellana HC,
Estévez-Braun A, Ravelo AG, Pérez-Sacau E and Machín F: Design and
synthesis of a novel series of pyranonaphthoquinones as
topoisomerase II catalytic inhibitors. J Med Chem. 51:6761–6772.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Toral-Barza L, Zhang WG, Huang X, McDonald
LA, Salaski EJ, Barbieri LR, Ding WD, Krishnamurthy G, Hu YB, Lucas
J, et al: Discovery of lactoquinomycin and related
pyranonaphthoquinones as potent and allosteric inhibitors of
AKT/PKB: Mechanistic involvement of AKT catalytic activation loop
cysteines. Mol Cancer Ther. 6:3028–3038. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Korwar S, Nguyen T and Ellis KC:
Preparation and evaluation of deconstruction analogues of
7-deoxykalafungin as AKT kinase inhibitors. Bioorg Med Chem Lett.
24:271–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Chen Y and He Q: Action of
bleomycin is affected by bleomycin hydrolase but not by caveolin-1.
Int J Oncol. 41:2245–2252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Zhang H and He Q: Involvement of
bleomycin hydrolase and poly(ADP-ribose) polymerase-1 in
Ubc9-mediated resistance to chemotherapy agents. Int J Oncol.
50:223–231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salaski EJ, Krishnamurthy G, Ding WD, Yu
K, Insaf SS, Eid C, Shim J, Levin JI, Tabei K, Toral-Barza L, et
al: Pyranonaphthoquinone lactones: A new class of AKT selective
kinase inhibitors alkylate a regulatory loop cysteine. J Med Chem.
52:2181–2184. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Léo PM, Morin C and Philouze C: Structure
revision of medermycin/lactoquinomycin a and of related C-8
glycosylated naphthoquinones. Org Lett. 4:2711–2714. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gottlieb TM, Leal JF, Seger R, Taya Y and
Oren M: Cross-talk between Akt, p53 and Mdm2: Possible implications
for the regulation of apoptosis. Oncogene. 21:1299–1303. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mayo LD, Dixon JE, Durden DL, Tonks NK and
Donner DB: PTEN protects p53 from Mdm2 and sensitizes cancer cells
to chemotherapy. J Biol Chem. 277:5484–5489. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sangai T, Akcakanat A, Chen H, Tarco E, Wu
Y, Do KA, Miller TW, Arteaga CL, Mills GB, Gonzalez-Angulo AM, et
al: Biomarkers of response to Akt inhibitor MK-2206 in breast
cancer. Clin Cancer Res. 18:5816–5828. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weinlich R, Oberst A, Beere HM and Green
DR: Necroptosis in development, inflammation and disease. Nat Rev
Mol Cell Biol. 18:127–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Csizmar CM, Kim DH and Sachs Z: The role
of the proteasome in AML. Blood Cancer J. 6:e5032016. View Article : Google Scholar : PubMed/NCBI
|